ABSTRACT.
Plasmodium lactate dehydrogenase (pLDH) is a common target in malaria rapid diagnostic tests (RDTs). These commercial antibody capture assays target either Plasmodium falciparum–specific pLDH (PfLDH), P. vivax–specific pLDH (PvLDH), or a conserved epitope in all human malaria pLDH (PanLDH). However, there are no assays specifically targeting P. ovale, P. malariae or zoonotic parasites such as P. knowlesi and P. cynomolgi. A malaria multiplex array, carrying the specific antibody spots for PfLDH, PvLDH, and PanLDH has been previously developed. This study aimed to assess potential cross-reactivity between pLDH from various Plasmodium species and this array. We tested recombinant pLDH proteins, clinical samples for P. vivax, P. falciparum, P. ovale curtisi, and P. malariae; and in vitro cultured P. knowlesi and P. cynomolgi. P. ovale-specific pLDH (PoLDH) and P. malariae-specific pLDH (PmLDH) cross-reacted with the PfLDH and PanLDH spots. Plasmodium Knowlesi-specific pLDH (PkLDH) and P. cynomolgi-specific pLDH (PcLDH) cross-reacted with the PvLDH spot, but only PkLDH was recognized by the PanLDH spot. Plasmodium ovale and P. malariae can be differentiated from P. falciparum by the concentration ratios of PanLDH/PfLDH, which had mean (range) values of 4.56 (4.07–5.16) and 4.56 (3.43–6.54), respectively, whereas P. falciparum had a lower ratio of 1.12 (0.56–2.61). Plasmodium knowlesi had a similar PanLDH/PvLDH ratio value, with P. vivax having a mean value of 2.24 (1.37–2.79). The cross-reactivity pattern of pLDH can be a useful predictor to differentiate certain Plasmodium species. Cross-reactivity of the pLDH bands in RDTs requires further investigation.
INTRODUCTION
Global malaria programs have been focused largely on Plasmodium falciparum in sub-Saharan Africa and to a lesser extent P. vivax in Asia and South America.1 However, the success of malaria elimination requires stopping transmission of all human-infecting Plasmodium species—such as P. falciparum, P. vivax, P. malariae, and P. ovale spp. (P. ovale curtisi of classic type and P. ovale wallikeri of variant type)—that cause natural human malaria via parasite speciation and treatment of infection.2–4 P. malariae and P. ovale spp. are prevalent in Africa and other malaria-endemic regions but receive little attention because of clinically mild symptoms.5,6
In addition, malaria can be caused in humans by simian parasites such as P. knowlesi and P. cynomolgi. P. knowlesi, commonly found in natural hosts including long-tailed macaques (Macaca fascicularis) and pig-tailed macaques (M. nemestrina) in Southeast Asian countries, has been documented in humans as a result of increased human–macaque contacts as a result of habitat disruption and destruction by extensive population growth and deforestation in the areas where natural hosts and the anopheline mosquito vectors carrying P. knowlesi exist.7,8 P. cynomolgi, often detected with other simian parasites such as P. inui, P. coatneyi, or P. fieldi in the macaque population in Southeast Asia, is a close relative of P. vivax.9,10 Natural human infection with P. cynomolgi has been reported in Southeast Asia.11–13
Severe malarial symptoms and even death have been described with human P. knowlesi infection.14–16 P. knowlesi has a relatively short asexual life cycle of approximately 24 hours, compared with approximately 48 hours for P. falciparum, P. vivax, and P. ovale, and 72 hours for P. malariae.17 Consequently, P. knowlesi infection can rapidly replicate to reach a high parasite density and progress to severe malaria similar to that from P. falciparum if there is no prompt medical treatment.18,19 Although cases of P. knowlesi (misdiagnosed as P. malariae) may have been prevalent at low levels for many decades, a relative surge has arisen as a result of a dramatic reduction in prevalence of P. vivax and P. falciparum over the past decade.20,21 Identification of P. knowlesi by microscopy is unreliable largely because of its similar morphological features with P. falciparum at the early trophozoite stage and to P. malariae at the other stages, notably trophozoite band forms.22,23
In addition to the limitations of microscopy to accurately identify P. knowlesi, there are no rapid diagnostic tests (RDTs) specifically designed for P. knowlesi identification. Currently available malaria RDTs use several key biomarkers to differentiate between species: P. falciparum-specific histidine-rich protein 2 (HRP2) or/and lactate dehydrogenase (PfLDH) for P. falciparum, and P. vivax-specific lactate dehydrogenase (PvLDH) for P. vivax. Some malaria RDTs detect pan-malarial biomarkers such as the pan-Plasmodium lactate dehydrogenase (pLDH) or Plasmodium aldolase (pAldo) that can, in principle, detect but not differentiate all Plasmodium species.24 Several studies demonstrated a risk of cross-reactivity in commercial malaria RDTs (OptiMAL-IT® [Bio-Rad, Hercules, CA], Entebe Malaria Cassette, BinaxNOWTM Malaria [Abbott, Abbott Park, IL], and Paramax-3TM [Tulip Diagnostics Ltd, Goa, India]) between parasites of two major Plasmodium species and P. knowlesi parasites, and poor sensitivity in cases of low parasitemia with P. knowlesi infection, revealing the need for caution in data interpretation if confirmatory testing is not conducted.25,26 For more accurate identification of P. knowlesi, several groups reported the stronger performance of molecular techniques, such as nested polymerase chain reaction (PCR) assay, loop-mediated isothermal amplification (LAMP), and real-time PCR assays.27–30 However, these molecular tests are not simple to use or cost-effective. Similarly, there are no P. ovale- and P. malariae-specific RDTs, also leading to the underreporting of these more widely spread forms of human malaria.31,32 Interestingly, an immunocapture assay has been used to evaluate specificity of monoclonal antibodies against the pLDH isoforms from Plasmodium species.33,34 A similar reactivity pattern with the pLDH proteins of P. cynomolgi (PcLDH) and P. vivax (PvLDH) indicated the close similarity of pLDH isoforms, whereas the absence of reactivity of the P. falciparum-specific antibodies with other simian parasites except P. knowlesi suggested the possible use of these antibodies to distinguish P. knowlesi from the other simian parasites.
The pLDH is the primary target analyte for P. vivax diagnosis with RDTs.35 With the emergence of hrp2/hrp3 gene deletions in P. falciparum infections leading to false negative RDT results, pLDH is increasingly a key biomarker for RDT-based diagnosis of malaria.36,37 It is important to understand the reactivity of different Plasmodium species to the different pLDH assays used in RDTs—namely P. falciparum pLDH, P. vivax pLDH, and PanLDH. In previous studies, we described the development and validation of a laboratory-based multiplex platform, the Human Malaria Array (5-Plex; Quansys Biosciences, Logan, UT), that can quantify HRP2 and pLDH from P. falciparum and P. vivax.38,39 This multiplex immunoassay, if applied to characterize clinical as well as in vitro cultured samples from these closely related but less-common Plasmodium species, may increase our understanding of the performance of current RDTs in these species.
Here, analysis of cross-reactivity of this reference assay was investigated with clinical samples of P. malariae and P. ovale and by using culture-adapted strains of both P. cynomolgi and P. knowlesi.40,41 We also explored the indicators that can differentiate these less-common parasites from P. falciparum and P. vivax parasites, and the time-course expression of P. knowlesi-specific pLDH (PkLDH) protein in culture by a multiplex reference assay.
MATERIALS AND METHODS
Clinical samples.
A total of 116 clinical blood samples (29 P. falciparum, 62 P. vivax, 17 P. malariae, and 8 P. ovale) from febrile patients with malaria mono-infection were obtained from the Foundation for Innovative New Diagnostics (FIND) Specimen Bank. Nested PCR with microscopy was used to identify the species of human malaria parasites in the samples. Parasite density in clinical samples was confirmed by either microscopy or quantitative PCR (qPCR) methods.
In vitro Plasmodium parasite culture.
The P. knowlesi strain A1-H.1 was cultured, as previously described.41 Parasites were grown in RPMI-1640 (Sigma-Aldrich, St. Louis, MO), supplemented with 2 g/L dextrose anhydrous, 0.05 g/L hypoxanthine, 0.5% AlbuMAX II (Invitrogen, Waltham, MA), and 10% (v/v) horse serum (Invitrogen). Human red blood cells (RBCs) were obtained from Duffy antigen-positive individuals from PlasmaLab (Everett, WA), and washed RBCs were added to 2% hematocrit in the culture media. The culture was maintained at 37°C with a hypoxic gas mixture of 90% N2, 5% CO2, and 5% O2. For time-course studies, high parasite density cultures were synchronized in three replicates twice using centrifugation with Nycodenz AG® (Axis-shield Diagnostics Ltd, Dundee, Scotland) gradient solution 4 hours apart, retaining the upper brown portion in the first centrifugation and the lower RBC portion for the second centrifugation, to achieve the highest synchronization rate. After synchronization, synchrony was confirmed, and parasite density in the culture was determined by staining thin-film smear using a Hemacolor® staining kit (HARLECO, Sigma Aldrich, St. Louis, MO) and counting parasites in a minimum of 2,000 RBCs using a microscope. Following the second synchronization, the culture was incubated and harvested at different time points. For kinetics of PkLDH, infected RBC (iRBC) cultures that were incubated in the culture tubes in three replicates were harvested at 0, 1, 2, 6, 19, 21, 25, 27, 28, 29, and 30 hours. After centrifugation, the cell culture supernatant was collected, and the remaining iRBC pellets were washed with phosphate-buffered saline (PBS) and resuspended in PBS at the original culture volume. The total number of iRBCs was counted using an Accuri™ C6‐UV flow cytometer (BD Biosciences, San Jose, CA)
Plasmodium cynomolgi parasites were cultured in complete RPMI media supplemented with GlutaMAXTM (Gibco, Waltham, MA) and 200 µM hypoxanthine (Calbiochem, San Diego, CA) as described previously.40 Rhesus monkey blood (BioIVT, Westbury, NY) was added to 5% hematocrit, and parasites in a plate were cultured at 37°C in a hypoxic gas mixture of 90% N2, 5% CO2, and 5% O2. Three lots of in vitro culture as both iRBC pellet and culture supernatant were prepared on the second day after synchronization and frozen prior to shipment to PATH (Seattle, WA) for the antigen quantification.
Production of Plasmodium species-specific pLDH proteins.
PATH collaborated with the University of Queensland Protein Expression Facility (University of Queensland, St. Lucia, Australia) to develop Plasmodium species-specific pLDH proteins and human LDH protein for use as reference proteins. Based on the DNA sequence information available from GenBank, the full-length ldh genes specific for P. falciparum, P. vivax, P. ovale curtisi, and P. malariae and the human full-length ldh gene were synthesized and cloned into the prokaryotic expression vector pOPIN-NHis3C to express in RosettaTM (DE3)pLysS competent cells (Novagen, Madison, WI). Bacterial culture was performed in 1 L TB media (Novagen) containing antibiotics at 30°C for around 24 hours. Culture was harvested by centrifugation at 5,000 × g for 30 minutes at 4°C and resuspended in lysis buffer. After lysing using sonication, the lysate was centrifuged at 20,000 × g for 30 minutes at 4°C, and the supernatant representing the soluble fraction was isolated. The soluble fraction was subjected to an immobilized metal affinity chromatography using a HisTrapTM Fast Flow column (GE Healthcare, Chicago, IL) followed by size-exclusion chromatography (SEC)4 using a HiLoad® 26/600 Superdex® 200 column (GE Healthcare, Chicago, IL). The SEC elution fractions were analyzed by resolving on 12% SDS-PAGE. The SEC elution fractions of interest were pooled and concentrated using an Amicon® centrifuge filter (Sigma-Aldrich, St. Louis, MO). The sample was filtered through a 0.22 µm filter and stored at −80°C. The amount and purity of His-tag-containing recombinant pLDH and human LDH proteins were estimated by absorbance at 280 nm using NanoDropTM (Thermo Fisher Scientific, Waltham, MA) spectrophotometer and SDS-PAGE, respectively. All recombinant pLDH and human LDH proteins showed more than 95% purity (Supplemental Figure 1) and were kept at −80°C for long-term storage until future use.
Quantification of malaria antigens.
The presence and quantification of PfLDH, PvLDH, and PanLDH in iRBC pellets, culture supernatant, clinical samples, and samples spiked with recombinant pLDH proteins were tested using the Q-PlexTM Human Malaria Array (Quansys Biosciences, Logan, UT; hereafter the 5-Plex) to target five different analytes—HRP2, PfLDH, PvLDH, PanLDH, and C-reactive protein (CRP)—according to the manufacturer’s instructions.39 These analytes are identified using pairs of specific antibodies in which capture antibodies are printed as defined spots in a geometric planar array on the surface of each well and a mix of detection antibodies are used to identify the analytes captured on the defined spots. For the pLDH assays, capture antibodies shown to be specific for P. vivax and P. falciparum parasites were individually spotted for the capture of PvLDH and PfLDH, respectively, with a third spot representing a capture antibody to Plasmodium LDH as a PanLDH determinant. A broadly reactive PanLDH antibody was used as the detector antibody for all three assays (PfLDH, PvLDH, and PanLDH). For comparison of reactivity pattern, iRBC pellets of synchronized P. cynomolgi and P. knowlesi cultures and P. vivax clinical samples were diluted at 200 and 40 p/µL of parasite density and tested on the 5-Plex. For the samples at high concentration, which falls outside the standard range, the samples were subsequently diluted 1:20 and 1:400 with assay diluent, and the dilution factors were applied for the final concentration of antigens.
Statistical analysis.
All data were analyzed using GraphPad Prism (version 5.0; La Jolla, CA). Pearson’s correlation coefficient was calculated for the PanLDH level based on the 5-Plex and for parasite density based on microscopy or qPCR. The results relative to ratios of PanLDH/PfLDH and PanLDH/PvLDH are presented as mean ± SD. Student’s t-test was used to determine significant differences between ratio values of PanLDH and species-specific pLDH of recombinant and native proteins, estimated by reference assay. In addition, a two-way analysis of variance (ANOVA) with Sidak multiple comparison test was used to establish statistical significance between groups (P < 0.05).
RESULTS
Performance of the 5-Plex for reactivity to recombinant and native pLDH proteins for P. falciparum, P. vivax, P. malariae, and P. ovale.
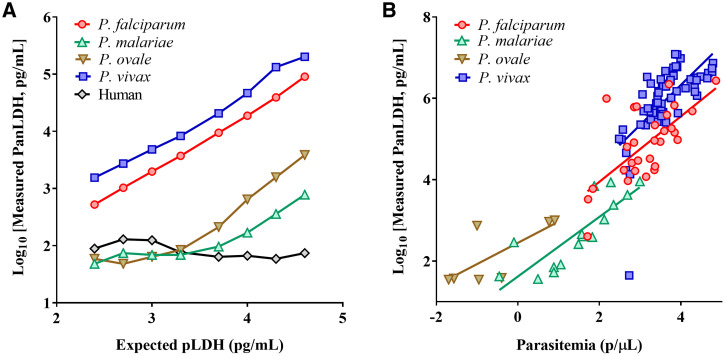
Dilution series of whole blood samples spiked with recombinant pLDH proteins specific for P. falciparum, P. vivax, P. malariae, and P. ovale curtisi, and clinical whole blood samples infected with each of four Plasmodium species were tested on the 5-Plex to examine the presence and level of reactivity to the assays for PanLDH, PfLDH, and PvLDH. Analysis showed that good specificity on PfLDH and PvLDH spots was found with recombinant PfLDH and PvLDH proteins, respectively (Supplemental Figure 2A). Interestingly, recombinant P. ovale-specific pLDH (PoLDH) and P. malariae-specific pLDH (PmLDH) proteins showed reactivity with both PanLDH and PfLDH spots of the 5-Plex. However, a notable difference was observed with the relative signal from the PanLDH spot, in which both PoLDH and PmLDH recombinant proteins showed relatively less signal, resulting in lower measured concentration as compared with recombinant PfLDH or PvLDH proteins for a given concentration (Figure 1A).
Figure 1.
Binding reactivity of recombinant or native pLDH proteins to PanLDH spot. (A) Expected concentration vs. measured concentration of recombinant pLDH and human LDH proteins. Depicted are line charts including eight data points generated with 2-fold serial dilutions of pLDH and human LDH proteins. (B) Scatter plots and best fit lines between parasite density and PanLDH level in clinical blood samples estimated by microscopy or qPCR, and the 5-Plex, respectively. Any samples with P. malariae, P. ovale, or P. vivax infections, which had no available data for parasite density, were excluded from analysis. Pearson correlation coefficient was calculated using GraphPad Prism. This figure appears in color at www.ajtmh.org.
For the reactivity to native pLDH from four Plasmodium species, the reactivity with PanLDH, PfLDH, and PvLDH spots of the 5-Plex was examined. Native PoLDH and PmLDH in clinical samples behaved similarly to their recombinant counterparts with both the PanLDH and PfLDH spots on the 5-Plex (Supplemental Figure 2B). Furthermore, the relation between level of PanLDH and parasite density was assessed with clinical samples infected by each Plasmodium species (Figure 1B). The correlation analysis demonstrated that PanLDH level showed significant positive correlations with parasite density determined by either microscopy or qPCR for P. falciparum (N = 29, r = 0.670, P < 0.0001), P. vivax (N = 60, r = 0.674, P < 0.0001) and P. malariae (N = 16, r = 0.834, P < 0.0001) but relatively poor correlation with parasite density for P. ovale (N = 7, r = 0.737, P = 0.059).
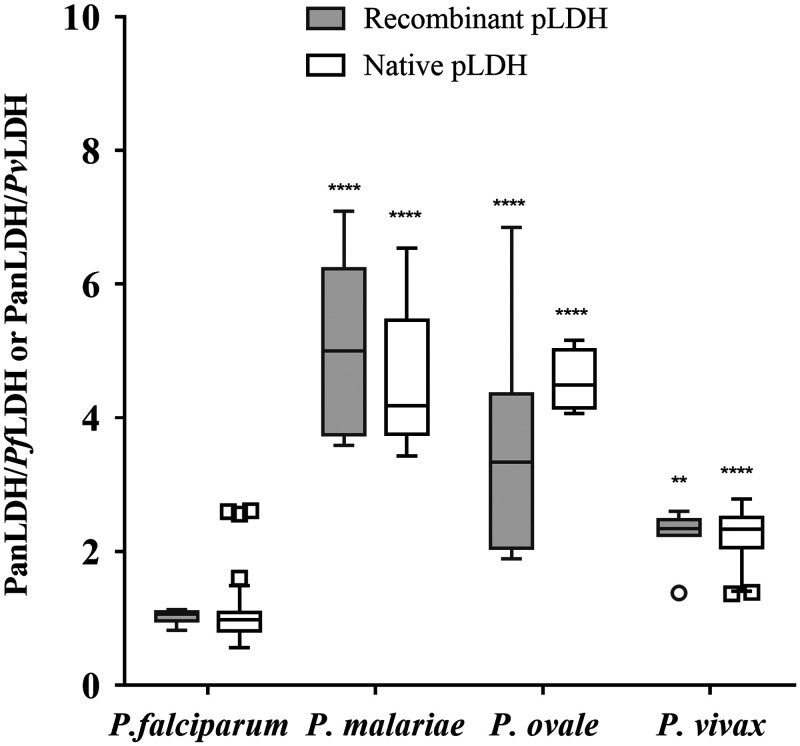
The ratio of PanLDH to species-specific pLDH was similar for both native pLDH from clinical samples and the same recombinant proteins across the four Plasmodium species (Table 1, Figure 2). Specifically, the ratio was 1.03 ± 0.1 and 1.12 ± 0.55 for recombinant and native PfLDH proteins, 4.25 ± 0.78 and 4.56 ± 1.0 for recombinant and native PmLDH proteins, 3.10 ± 1.04 and 4.56 ± 0.46 for recombinant and native PoLDH proteins, and 2.26 ± 0.38 and 2.24 ± 0.37 for recombinant and native PvLDH proteins (all P values statistically not significant). The PanLDH concentration measurement in the 5-Plex is calibrated to recombinant PfLDH and so the ratio of close to 1 for PanLDH/PfLDH is to be expected.
Table 1.
Comparison of four Plasmodium-specific recombinant pLDH proteins against their corresponding native counterparts related to their binding reactivity
| Recombinant protein | Clinical sample | |||||||
|---|---|---|---|---|---|---|---|---|
| Reactivity | PfLDH (n = 8) | PmLDH (n = 8) | PoLDH (n = 8) | PvLDH (n = 8) | P. falciparum (n = 29) | P. malariae (n = 15) | P. ovale (n = 4) | P. vivax (n = 58) |
| Pan/Pf ratio |
Pan/Pv ratio |
Pan/Pf ratio |
Pan/Pv ratio |
|||||
| Range | 0.82–1.13 | 3.59–5.49 | 1.89–4.38 | 1.38–2.6 | 0.56 − 2.61 | 3.43–6.54 | 4.07–5.16 | 1.37–2.79 |
| Mean | 1.03 | 4.25 | 3.10 | 2.26 | 1.12 | 4.56 | 4.56 | 2.24 |
| SD | 0.1 | 0.78 | 1.04 | 0.38 | 0.55 | 1.0 | 0.46 | 0.37 |
| t-test | – | – | – | – | P = 0.63 | P = 0.33 | P = 0.27 | P = 0.89 |
Pan/Pf = PanLDH/PfLDH; Pan/Pv = PanLDH/PvLDH. Descriptive statistics for the ratio of PanLDH concentration to PfLDH or PvLDH concentration as indicated, determined via the 5-Plex. Any clinical samples that showed unquantifiable or infection-negative test results by the 5-Plex were excluded from analysis. P values correspond to comparison of different antigen sources.
Figure 2.
Reactivity of recombinant and native pLDH proteins. Any samples showing antigen-negative results by the 5-Plex were excluded from analysis. Depicted are box and whisker plots for ratios of PanLDH/PfLDH for the P. falciparum, P. malariae, and P. ovale columns or ratio of PanLDH/PvLDH for the P. vivax column for each source of pLDH (recombinant and native). The ends of the boxes correspond to the 25th and 75th percentiles, respectively, and the middle line corresponds to the median. Two-way ANOVA plus Sidak multiple comparison test was used to determine the statistical significance against ratio values of P. falciparum. Significant differences (**** P < 0.0001, ** P < 0.001).
Performance of reference assay evaluated with in vitro cultures of P. cynomolgi and P. knowlesi.
To examine the binding patterns of pLDH from P. knowlesi and P. cynomolgi on each assay for PanLDH, PfLDH, and PvLDH, a series of dilutions (200 and 40 p/µL) of whole blood sample spiked with synchronized P. cynomolgi and P. knowlesi iRBC pellets were tested with the 5-Plex. The results were comparable to that of a clinical P. vivax blood sample. The results showed the similar reactivity between PkLDH and PvLDH proteins for binding to the PanLDH and PvLDH spots, and with comparable PanLDH/PvLDH ratio values between these two Plasmodium species at 2.7 for PkLDH and 2.8 for PvLDH at 200 p/µL of parasite density (Table 2). However, we found that PcLDH of P. cynomolgi only reacted with the PvLDH spot and not to the PanLDH spot.
Table 2.
Concentration estimates for PanLDH and PvLDH determined from cell cultures of P. cynomolgi and P. knowlesi and a clinical sample of P. vivax
| P. cynomolgi | P. knowlesi | P. vivax | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Parasitemia (p/mL) | PanLDH (pg/mL) | PvLDH (pg/mL) | Pan/Pv ratio | PanLDH (pg/mL) | PvLDH (pg/mL) | Pan/Pv ratio | PanLDH (pg/mL) | PvLDH (pg/mL) | Pan/Pv ratio |
| 200 | 28.8* | 4,603.2 | 0.01 | 890.3 | 332.1 | 2.7 | 30,758.1 | 11,183.0 | 2.8 |
| 40 | 29.6* | 1,102.5 | 0.03 | 207.2 | 78.6 | 2.6 | 8,064.1 | 2,805.8 | 2.9 |
Pan/Pv = PanLDH/PvLDH.
For the Pan/Pv calculation, the PanLDH value from background binding was used.
Time-course study on PkLDH expression over parasite stages of P. knowlesi.
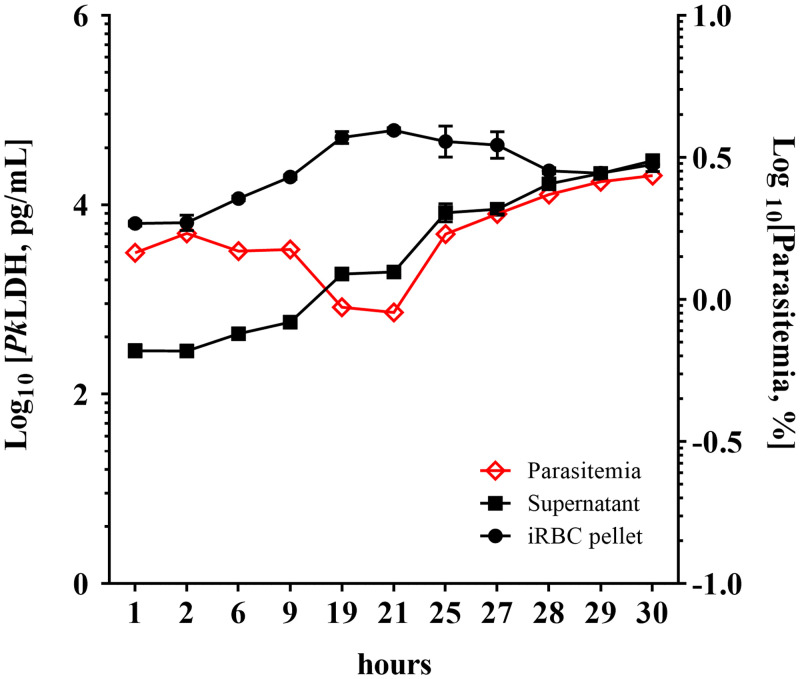
For the time-course study, PkLDH protein in the iRBC pellet and supernatant obtained from in vitro culture over 30 hours after synchronization were tested by the 5-Plex. Our results (Figure 3) demonstrated a life cycle for P. knowlesi of approximately 28 hours based on the time gap between the first ring stage and the second ring stage (Supplemental Table 1), similar to the previously published finding of 27 hours.39 Although most of the PkLDH was found in iRBC pellets rather than in the supernatant, both pellets and supernatant showed progressively increased concentration of PkLDH (∼8 times more after 19 hours compared with 1 hour for both pellets and supernatant) during ring and trophozoite stages (Figure 3, Supplemental Table 1). The maximal amount of PkLDH is reached in the pellet during the schizont stage (21 to 27 hours), but a continuous accumulation of PkLDH released from the schizont rupture in the supernatant was observed over this period of time. Interestingly, during the first life cycle, parasite density seems to be decreased during the schizont stage, and the number of parasites in the second life cycle is increased after the released cells infect the fresh cells.
Figure 3.
PkLDH accumulation in P. knowlesi cultures. Percentage parasitemia in synchronized cultures was monitored using microscopy, and the PkLDH level in the iRBC pellet and supernatant from 1 mL of synchronized culture was quantified using PvLDH assay and in three replicates over 30 hours. For estimating parasitemia, one of three replicates was examined. Time zero represents time of synchronization. Error bars indicate standard deviation of the mean. This figure appears in color at www.ajtmh.org.
DISCUSSION
The human malaria array (5-Plex) was developed to provide quantitative measurement of the malaria antigens HRP2, PfLDH, and PvLDH and a qualitative measure for all malaria species (PanLDH) as well as measure the human biomarker CRP.39 The pLDH spots used in this reference assay were screened for specificity against PfLDH and PvLDH38,39 as well as characterized for their utility to screen for hrp2/hrp3 deletions both in whole blood pellets39 and dried blood spots.42 We demonstrated that the malaria array is a valuable tool in the detection and diagnosis of P. falciparum and P. vivax, using the described biomarkers.39,42 However, its capability to diagnose less-common Plasmodium species such as P. malariae and P. ovale, and two simian Plasmodium species, P. knowlesi and P. cynomolgi, which have been recognized to cause human infection, has not been fully explored. As the 5-Plex includes a PanLDH spot detecting all Plasmodium species, in this study, we first evaluated the potential cross-reactivity of pLDH proteins from these parasites against pan-malaria and species-specific spots using both recombinant and native proteins.
The recombinant pLDH proteins of P. falciparum, P. malariae, P. ovale curtisi, and P. vivax offer great diagnostic value for the appropriate evaluation of the assay performance when there are limited clinical blood samples. Our study demonstrated that these recombinant pLDH proteins behave similarly to native pLDH proteins from corresponding Plasmodium species as demonstrated by PanLDH/species-specific pLDH ratio while being distinct from the equivalent host LDH protein (Table 1). However, there seemed to be different binding characteristics resulting in different signal responses from the PanLDH spot on the 5-Plex: the highest signal was observed with recombinant PvLDH followed by recombinant PfLDH, recombinant PoLDH, and then recombinant PmLDH (Supplemental Figure 2A).
The significant variation of PanLDH values in the estimated concentration of recombinant pLDH proteins suggest two possibilities: 1) some minor variance in the amino acid sequence of the conserved region or that of the surrounding area that enhances or reduces the antigen–antibody reaction, and 2) inefficient protein folding during protein expression/purification might have occurred when using a prokaryotic expression system. The second possibility may be addressed by evaluating the native pLDH proteins purified from different Plasmodium species, but obtaining native proteins from live parasites is a major challenge, especially with species of P. vivax, P. ovale, and P. malariae, for which continuous culture systems have not been established unlike P. falciparum strains.43–45 Alternatively, nonhuman primate models could offer the strategies of producing non-P. falciparum parasites and their respective native proteins.46
The pLDH amount measured as PanLDH correlates to parasite density equally across species, as shown by the normalized binding reactivity of native pLDH proteins with respect to the given parasite number (Supplemental Figure 3). The results suggest that this could be an artifact derived from the protein purification processes.
Additionally, both PmLDH and PoLDH were found to react not only to the PanLDH spot but also to the PfLDH spot, suggesting an overlap in antigenic epitopes of pLDH possessed by P. falciparum, P. malariae, and P. ovale. In our preliminary study, cross-reactivity between P. malariae and the PfLDH band on a few commercial RDTs was also observed (data not shown). It will therefore be of interest to investigate the extent of the level of cross-reactivity, which may have a significant influence on interpreting diagnostic test results. The PanLDH/species-specific pLDH ratio values for P. malariae, P. ovale, and P. vivax were significantly different from that for P. falciparum (Figure 1), suggesting the potential to use this ratio difference in differentiating Plasmodium species on the 5-Plex.
The reactivity patterns of pLDH from P. vivax and P. cynomolgi were compared with P. knowlesi, which is more distantly related to P. vivax. P. cynomolgi causes periodic relapses by dormant hypnozoites, early infectious gametocyte formation, and invasion of Duffy blood group-positive reticulocytes, and thus offers a robust model for P. vivax infection in the rhesus monkey model.40,47,48 This study revealed that P. cynomolgi failed to exhibit cross-reactivity to the PanLDH spot of the 5-Plex but did show reactivity to the PvLDH spot (Table 2). Unfortunately, there is no information available on the specific epitopes for PanLDH-specific capture and detector antibodies used in the 5-Plex. We aligned the amino acid sequences of the pLDH proteins from four different Plasmodium species (obtained from the GeneBank database) and found a conserved region (the region forming the loop structure) as the potential site recognized by pan-malaria antibodies and several species-specific regions as described previously (Supplemental Figure 4).49–51 We did not find any obvious sequence variance in the potential pan-epitope region of PcLDH protein as compared with three other parasite species including P. falciparum, P. vivax, and P. knowlesi that would explain our observation. It would be interesting to explore binding characteristics of commercially available pan-malaria antibodies to the laboratory-adopted P. cynomolgi strain or recombinant PcLDH reagents and investigate the extent of sequence conservation within the conserved region of PcLDH protein from the cultured P. cynomolgi in a future study.
PkLDH exhibited significant cross-reactivity to both the PanLDH and PvLDH spots, but no or little cross-reactivity with the PfLDH spot on the 5-Plex. A previous study looking at cross-reactivity between pLDH proteins from different Plasmodium species and different antibodies to pLDH highlighted that PkLDH can react to antibodies specific to both PfLDH and PvLDH.33 In the 5-Plex, only cross-reactivity between PkLDH and the PvLDH antibody pair was observed. Additionally, the PanLDH/PvLDH ratio for P. knowlesi was similar to that seen when measuring PvLDH from the P. vivax clinical samples. Although the PanLDH and PvLDH spots on the 5-Plex can detect the presence of the PkLDH antigen, they were unable to distinguish P. knowlesi from P. vivax infection. To differentiate P. knowlesi from other Plasmodium species, development of an expanded array that hosts a P. knowlesi-specific antigen assay is required.
This is the first study to demonstrate the dynamic kinetic expression of PkLDH in a culture system. The results exhibited accumulation of PkLDH over time in culture, except with a slight decrease in PkLDH concentration seen in samples from the late schizont stage at around 25–28 hours. The pellet fraction contains the largest total amount of PkLDH until 29 hours after synchronization. At an inflection point where more PkLDH is contained in the supernatant, the erythrocytic cycle is completed through the rupturing of the schizonts to create new ring-stage parasites. The overall decrease in PkLDH seen starting at hour 25 may be explained by the lability of PkLDH and consequently an overall drop in PkLDH concentration until the parasites mature again. It should also be noted that the reactivity pattern of PkLDH proteins as a function of PanLDH/PvLDH ratio was maintained at 2.11 ± 0.24 and 2.35 ± 0.24 in both cell pellet and supernatant compartments, respectively, throughout all development stages (Supplemental Table 2).
The differential binding of pLDH from the different Plasmodium species investigated and observed in this study across the 5-Plex PfLDH, PvLDH, and PanLDH assays echoes results from previous studies investigating both antibody reactivity to pLDH and RDT reactivity to pLDH.26,33,52,53 The results presented here are most likely specific to the particular anti-pLDH antibodies used in this 5-Plex assay. In a similar manner, the pLDH assay components of widely used RDTs should be evaluated for cross-reactivity to the different human malaria species, especially to P. ovale, P. malariae, and P. knowlesi, and also to the less-common species known to infect humans, including P. cynomolgi and P. simium. This will be particularly relevant to countries approaching elimination and/or that have a significant relative burden of P. knowlesi.54,55 Between the emergence of hrp2/hrp3 deletions in P. falciparum infections and pLDH as the most commonly used target analyte for non-P. falciparum malaria RDTs, understanding the dynamics and cross-reactivity of pLDH between the different human Plasmodium species is essential. Additionally, given the known cross-reactivities between different Plasmodium species, new diagnostic markers for better differentiating Plasmodium species should be explored.
Supplemental tables
ACKNOWLEDGMENTS
We thank the University of Queensland Protein Expression Facility for production of recombinant proteins and FIND for providing the clinical samples for this study. We thank Drs. David Boyle and Xavier C. Ding for critical review of the manuscript draft.
Note: Supplemental tables and figures appear at www.ajtmh.org.
REFERENCES
- 1.World Health Organization, 2018. World Malaria Report. Geneva, Switzerland: WHO. Available at: http://apps.who.int/iris/bitstream/handle/10665/275867/9789241565653-eng.pdf. Accessed February 12, 2021.
- 2.Sutherland CJ, 2016. Persistent parasitism: the adaptive biology of malariae and ovale malaria. Trends Parasitol 32: 808–819. [DOI] [PubMed] [Google Scholar]
- 3.Rabinovich RN. et al. , 2017. malERA: an updated research agenda for malaria elimination and eradication. PLoS Med 14: e1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diallo MA, Diongue K, Diagne G, Seck MC, Ndiaye M, Dieye B, Ndiaye YD, Badiane AS, Ndiaye D, 2017. Plasmodium ovale wallikeri and Plasmodium ovale curtisi Malaria in Senegal in 2016. Bull Soc Pathol Exot 110: 286–290. [DOI] [PubMed] [Google Scholar]
- 5.Nino CH, Cubides JR, Camargo-Ayala PA, Rodriguez-Celis CA, Quinones T, Cortes-Castillo MT, Sanchez-Suarez L, Sanchez R, Patarroyo ME, Patarroyo MA, 2016. Plasmodium malariae in the Colombian Amazon region: you don’t diagnose what you don’t suspect. Malar J 15: 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haiyambo DH, Uusiku P, Mumbengegwi D, Pernica JM, Bock R, Malleret B, Renia L, Greco B, Quaye IK, 2019. Molecular detection of P. vivax and P. ovale foci of infection in asymptomatic and symptomatic children in northern Namibia. PLoS Negl Trop Dis 13: e0007290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, Cox-Singh J, Singh B, 2011. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog 7: e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh B, Daneshvar C, 2013. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev 26: 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tachibana S. et al. , 2012. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet 44: 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Kadir KA, Quintanilla-Zarinan LF, Villano J, Houghton P, Du H, Singh B, Smith DG, 2016. Distribution and prevalence of malaria parasites among long-tailed macaques (Macaca fascicularis) in regional populations across Southeast Asia. Malar J 15: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM, 2014. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J 13: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grignard L, Shah S, Chua TH, William T, Drakeley CJ, Fornace KM, 2019. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J Infect Dis 220: 1946–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raja TN, Hu TH, Kadir KA, Mohamad DSA, Rosli N, Wong LL, Hii KC, Simon Divis PC, Singh B, 2020. Naturally acquired human Plasmodium cynomolgi and P. knowlesi infections, Malaysian Borneo. Emerg Infect Dis 26: 1801–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajahram GS, Cooper DJ, William T, Grigg MJ, Anstey NM, Barber BE, 2019. Deaths from Plasmodium knowlesi malaria: case series and systematic review. Clin Infect Dis 69: 1703–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B, 2008. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis 46: 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barber BE, William T, Grigg MJ, Menon J, Auburn S, Marfurt J, Anstey NM, Yeo TW, 2013. A prospective comparative study of knowlesi, falciparum, and vivax malaria in Sabah, Malaysia: high proportion with severe disease from Plasmodium knowlesi and Plasmodium vivax but no mortality with early referral and artesunate therapy. Clin Infect Dis 56: 383–397. [DOI] [PubMed] [Google Scholar]
- 17.Garnham PCC, 1988. Malaria parasites of man: life cycles and morphology. In: Wernsdorfer WH, McGregor I, eds. Malaria: Principles and Practice of Malariology, Edinburgh, Scotland: Churchill Livingstone, 61–69. [Google Scholar]
- 18.Knowles R, Gupta BMD, 1932. A study of monkey-malaria, and its experimental transmission to man. Ind Med Gaz 67: 301–320. [PMC free article] [PubMed] [Google Scholar]
- 19.Willmann M, Ahmed A, Siner A, Wong IT, Woon LC, Singh B, Krishna S, Cox-Singh J, 2012. Laboratory markers of disease severity in Plasmodium knowlesi infection: a case control study. Malar J 11: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.William T, Rahman HA, Jelip J, Ibrahim MY, Menon J, Grigg MJ, Yeo TW, Anstey NM, Barber BE, 2013. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax Malaria in Sabah, Malaysia. PLoS Negl Trop Dis 7: e2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.William T, Jelip J, Menon J, Anderios F, Mohammad R, Awang Mohammad TA, Grigg MJ, Yeo TW, Anstey NM, Barber BE, 2014. Changing epidemiology of malaria in Sabah, Malaysia: increasing incidence of Plasmodium knowlesi. Malar J 13: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KS, Cox-Singh J, Singh B, 2009. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J 8: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barber BE, William T, Grigg MJ, Yeo TW, Anstey NM, 2013. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson ML, 2012. Malaria rapid diagnostic tests. Clin Infect Dis 54: 1637–1641. [DOI] [PubMed] [Google Scholar]
- 25.Kawai S, Hirai M, Haruki K, Tanabe K, Chigusa Y, 2009. Cross-reactivity in rapid diagnostic tests between human malaria and zoonotic simian malaria parasite Plasmodium knowlesi infections. Parasitol Int 58: 300–302. [DOI] [PubMed] [Google Scholar]
- 26.Foster D, Cox-Singh J, Mohamad DS, Krishna S, Chin PP, Singh B, 2014. Evaluation of three rapid diagnostic tests for the detection of human infections with Plasmodium knowlesi. Malar J 13: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imwong M, Tanomsing N, Pukrittayakamee S, Day NP, White NJ, Snounou G, 2009. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J Clin Microbiol 47: 4173–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iseki H, Kawai S, Takahashi N, Hirai M, Tanabe K, Yokoyama N, Igarashi I, 2010. Evaluation of a loop-mediated isothermal amplification method as a tool for diagnosis of infection by the zoonotic simian malaria parasite Plasmodium knowlesi. J Clin Microbiol 48: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Divis PC, Shokoples SE, Singh B, Yanow SK, 2010. A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malar J 9: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babady NE, Sloan LM, Rosenblatt JE, Pritt BS, 2009. Detection of Plasmodium knowlesi by real-time polymerase chain reaction. Am J Trop Med Hyg 81: 516–518. [PubMed] [Google Scholar]
- 31.Makanjuola RO, Taylor-Robinson AW, 2020. Improving accuracy of ,alaria diagnosis in underserved rural and remote endemic areas of Sub-Saharan Africa: a call to develop multiplexing rapid diagnostic tests. Scientifica (Cairo) 2020: 3901409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotepui M, Masangkay FR, Kotepui KU, De Jesus Milanez G, 2020. Misidentification of Plasmodium ovale as Plasmodium vivax malaria by a microscopic method: a meta-analysis of confirmed P. ovale cases. Sci Rep 10: 21807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCutchan TF, Piper RC, Makler MT, 2008. Use of malaria rapid diagnostic test to identify Plasmodium knowlesi infection. Emerg Infect Dis 14: 1750–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpalipan P, Pattaradilokrat S, Harnyuttanakorn P, 2018. Global sequence diversity of the lactate dehydrogenase gene in Plasmodium falciparum. Malar J 17: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal R, Choi L, Johnson S, Takwoingi Y, 2020. Rapid diagnostic tests for Plasmodium vivax malaria in endemic countries. Cochrane Database Syst Rev 11: CD013218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatton ML, Dunn J, Chaudhry A, Ciketic S, Cunningham J, Cheng Q, 2017. Implications of parasites lacking Plasmodium falciparum histidine-rich protein 2 on malaria morbidity and control when rapid diagnostic tests are used for diagnosis. J Infect Dis 215: 1156–1166. [DOI] [PubMed] [Google Scholar]
- 37.Cunningham J. et al. , 2019. A review of the WHO malaria rapid diagnostic test product testing programme (2008–2018): performance, procurement and policy. Malar J 18: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang IK. et al. , 2019. Simultaneous quantification of Plasmodium antigens and host factor C-reactive protein in asymptomatic individuals with confirmed malaria by use of a novel multiplex immunoassay. J Clin Microbiol 57: e00948–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang IK. et al. , 2020. Multiplex human malaria array: quantifying antigens for malaria rapid diagnostics. Am J Trop Med Hyg 102: 1366–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chua ACY. et al. , 2019. Robust continuous in vitro culture of the Plasmodium cynomolgi erythrocytic stages. Nat Commun 10: 3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon RW, Hall J, Rangkuti F, Ho YS, Almond N, Mitchell GH, Pain A, Holder AA, Blackman MJ, 2013. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci USA 110: 531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang IK. et al. , 2021. Assessment of Plasmodium antigens and CRP in dried blood spots with multiplex malaria array. J Parasit Dis 45: 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava K, Singh S, Singh P, Puri SK, 2007. In vitro cultivation of Plasmodium falciparum: studies with modified medium supplemented with ALBUMAX II and various animal sera. Exp Parasitol 116: 171–174. [DOI] [PubMed] [Google Scholar]
- 44.Trager W, Jensen JB, 1976. Human malaria parasites in continuous culture. Science 193: 673–675. [DOI] [PubMed] [Google Scholar]
- 45.Haynes JD, Diggs CL, Hines FA, Desjardins RE, 1976. Culture of human malaria parasites Plasmodium falciparum. Nature 263: 767–769. [DOI] [PubMed] [Google Scholar]
- 46.Galinski MR, Barnwell JW, 2012. Non-human primate models for human malaria research. In: Abee CR, Christian KM, Tardif S, Morris T, eds. Nonhuman Primates in Biomedical Research. London, England: Academic Press, 299–323. [Google Scholar]
- 47.Barnwell JW, Galinski MR, 2014. Malarial liver parasites awaken in culture. Nat Med 20: 237–239. [DOI] [PubMed] [Google Scholar]
- 48.Dembele L. et al. , 2014. Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat Med 20: 307–312. [DOI] [PubMed] [Google Scholar]
- 49.Verma P, Biswas S, Mohan T, Ali S, Rao DN, 2013. Detection of histidine rich protein and lactate dehydrogenase of Plasmodium falciparum in malaria patients by sandwich ELISA using in-house reagents. Indian J Med Res 138: 977–987. [PMC free article] [PubMed] [Google Scholar]
- 50.Hurdayal R, Achilonu I, Choveaux D, Coetzer TH, Dean Goldring JP, 2010. Anti-peptide antibodies differentiate between plasmodial lactate dehydrogenases. Peptides 31: 525–532. [DOI] [PubMed] [Google Scholar]
- 51.Tomar D, Biswas S, Tripathi V, Rao DN, 2006. Development of diagnostic reagents: raising antibodies against synthetic peptides of PfHRP-2 and LDH using microsphere delivery. Immunobiology 211: 797–805. [DOI] [PubMed] [Google Scholar]
- 52.Barber BE, William T, Grigg MJ, Piera K, Yeo TW, Anstey NM, 2013. Evaluation of the sensitivity of a pLDH-based and an aldolase-based rapid diagnostic test for diagnosis of uncomplicated and severe malaria caused by PCR-confirmed Plasmodium knowlesi, Plasmodium falciparum, and Plasmodium vivax. J Clin Microbiol 51: 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigg MJ, William T, Barber BE, Parameswaran U, Bird E, Piera K, Aziz A, Dhanaraj P, Yeo TW, Anstey NM, 2014. Combining parasite lactate dehydrogenase-based and histidine-rich protein 2-based rapid tests to improve specificity for diagnosis of malaria due to Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. J Clin Microbiol 52: 2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaw MT, Lin Z, 2019. Human Plasmodium knowlesi infections in South-East Asian countries. J Microbiol Immunol Infect 52: 679–684. [DOI] [PubMed] [Google Scholar]
- 55.Rahim M, Munajat MB, Idris ZM, 2020. Malaria distribution and performance of malaria diagnostic methods in Malaysia (1980–2019): a systematic review. Malar J 19: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.