Abstract
CtBP (carboxyl-terminal binding protein) participates in regulating cellular development and differentiation by associating with a diverse array of transcriptional repressors. Most of these interactions occur through a consensus CtBP-binding motif, PXDLS, in the repressor proteins. We previously showed that the CtBP-binding motif in E1A is flanked by a Lys residue and suggested that acetylation of this residue by the p300/CBP-associated factor P/CAF disrupts the CtBP interaction. In this study, we show that the interaction between CtBP and the nuclear hormone receptor corepressor RIP140 is regulated similarly, in this case by p300/CBP itself. CtBP was shown to interact with RIP140 in vitro and in vivo through a sequence, PIDLSCK, in the amino-terminal third of the RIP140 protein. Acetylation of the Lys residue in this motif, demonstrated in vivo by using an acetylated RIP140-specific antibody, dramatically reduced CtBP binding. Mutation of the Lys residue to Gln resulted in a decrease in CtBP binding in vivo and a loss of transcriptional repression. We suggest that p300/CBP-mediated acetylation disrupts the RIP140-CtBP complex and derepresses nuclear hormone receptor-regulated genes. Disruption of repressor-CtBP interactions by acetylation may be a general mode of gene activation.
CtBP (carboxyl-terminal binding protein) was initially identified through its ability to interact with a five-residue motif, PLDLS, in the carboxyl terminus of the adenoviral transforming protein E1A (21, 22). Mutation of this motif increases the level of E1A-directed cellular transformation (2, 21), suggesting that the CtBP interaction normally serves to dampen E1A function. In keeping with this model, mutation of the CtBP-binding motif decreases the ability of E1A to repress transcription (22). Like other transcriptional corepressors, CtBP does not interact with DNA directly but blocks gene expression when fused to a heterologous DNA binding domain (26). Precisely how CtBP blocks transcription has not been resolved, however. Interactions have been demonstrated between CtBP and the class I and class II histone deacetylases (HDACs), as well as with other proteins believed to be involved in transcriptional silencing (23, 29, 37).
Like the better-characterized E1A-binding proteins, CBP/p300 and retinoblastoma protein, CtBP has been shown to have critical functions in the regulation of cellular genes involved in growth and differentiation. Perhaps the most definitive demonstrations of these functions have been elucidated with Drosophila melanogaster. These studies have shown that CtBP is essential for short-range repression, a process involved in the establishment of localized stripes, bands, and tissue-specific expression in the syncytial embryo (12). Examples of CtBP-binding transcriptional repressors in Drosophila include snail, knirps, zhf-1, and kruppel (14, 15, 17). In general, these factors each interact with CtBP through a motif, PXDLS, that is highly related to that in E1A. In mammalian cells, transcription factors that function through CtBP include BKLF (basic kruppel-like factor), ZEB (zinc finger, E box binding factor, a homologue of zhf-1), Ikaros, and Net (5, 8, 18, 31). The effects of CtBP on gene expression are frequently cell and context dependent, however, suggesting that CtBP interactions with particular repressors may be subject to additional levels of regulation (16). We recently provided evidence that one such mode of regulation involves acetylation (39). The CtBP-binding motif in E1A is flanked by a Lys residue that we have shown to be acetylated in vitro and in vivo by the p300/CBP-associated factor, P/CAF. Of note, acetylation of this residue was found to decrease the CtBP interaction. Thus, in the context of E1A, acetylation disrupts CtBP binding and leads to a loss of transcriptional repression. Whether this model pertains to other CtBP complexes is unknown.
In this paper, we examine the interaction of CtBP with the nuclear hormone receptor interacting protein, RIP140 (40). We first identified RIP140 by searching a protein database for sequences containing an extended consensus CtBP-binding sequence, PXDLSXK/R. This sequence includes the core PXDLS motif in addition to the flanking basic residues (K/R) found in E1A and certain other known CtBP-binding repressors (19, 22, 31). RIP140 was also identified as a CtBP-binding partner in a Saccharomyces cerevisiae two-hybrid screen.
RIP140 is perhaps the most enigmatic of the nuclear hormone receptor-interacting proteins. In the unliganded or antagonist-bound state, nuclear hormone receptors are bound by corepressor complexes containing NCoR/SMRT, Sin3, and one or more of the HDACs (reviewed in reference 6). Upon ligand binding, nuclear receptors undergo a conformational change, resulting in displacement of the Sin3/NCoR complex, followed by recruitment of ligand-dependent coactivators in the p160 family, p300/CBP, and P/CAF (1, 3, 20, 24, 34). The intrinsic acetyltransferase activities of these coactivators are believed to modify the amino-terminal tails of nucleosomal histone proteins in a manner that facilitates transcription. In the sequential-step model of transcriptional activation initially proposed by Roeder and coworkers, other coactivators, such as TRAP/ARC/DRIP, are then recruited to mediate RNA polymerase II-dependent transcription (for a review, see reference 32). RIP140 does not appear to conform to the standard model because it associates with receptors in a ligand-dependent manner but is a corepressor rather than a coactivator (11, 13, 28, 30, 36).
Because of its ligand-dependent association with the nuclear hormone receptors, RIP140 was initially classified as a transcriptional coactivator (4). Subsequent studies refuted this role, however, and RIP140 is now generally acknowledged to function as a corepressor (10, 30). Deletional analyses have identified three domains within RIP140 that are capable of inhibiting gene expression (11), but the mechanisms of repression remain unclear. Initially, it was proposed that RIP140 competed with the p160 family of coactivators for binding to the ligand-activated conformation of the receptor (28, 30). More recent studies have suggested that HDACs might also contribute to repression. In support of this idea, a portion of the gene repression activity of RIP140 has been shown to be sensitive to trichostatin A, an HDAC inhibitor, and direct binding of RIP140 to the class I HDACs has been demonstrated (35, 36).
The studies in this paper indicate that the amino-terminal repression function of RIP140 is mediated by the corepressor CtBP. Of interest, the CtBP binding site in RIP140 is flanked by a Lys residue which, like the motif in E1A, is a potential site of acetylation. We demonstrate that RIP140 is acetylated at this site by CBP/p300 and that this modification abolishes the CtBP interaction, resulting in a loss of repression. These studies support the hypothesis that acetylation of transcriptional repressors may be a general mode of disrupting CtBP corepressor complexes.
MATERIALS AND METHODS
Plasmids.
pEFRIP140 (4) was a gift from F. Schaufele (University of California, San Francisco). The fragment of RIP140 spanning amino acids (aa) 1 to 495 [RIP140 (1-495)], RIP140 (623-951), and RIP140 (977-1158) were cloned by PCR into pET23b (Novagen) or pGEXKG (Promega). The PIDLSCK motif in RIP140 is located between aa 440 and 446. The K446Q, PIDL→AIAL, and PIDL→AAAA mutations were constructed by the Quikchange method (Stratagene). His-tagged human CtBP1 (hCtBP1) was cloned by PCR into pET23b (Novagen). Rc/CMV-hCtBP1 was a gift from G. Chinnadurai (St. Louis University Health Sciences Center). Gal4-RIP140 (386-470) was cloned by PCR into a pcDNA3 vector lacking the simian virus 40 (SV40) origin. The VP16-hCtBP1 construct was subcloned into pcDNA3-VP16 (provided by R. Maurer, Oregon Health Sciences University). FLAG-hCtBP1-pcDNA3 was cloned by PCR into pcDNA3. LexA-hCtBP1 was cloned by PCR into pBTM116 (7). The (Gal4)5-E1b-luciferase vector was from M. Green (University of Massachusetts Medical Center). The (Gal4)5-TK-luciferase vector and SV40-lacZ were gifts from L.-N. Wei (University of Minnesota Medical School). The Gal4-SV40-CAT vector and Gal4-ZEB (700-776) construct were gifts from D. Dean (Washington University School of Medicine). The (ERE)2-pS2-CAT reporter gene was a gift from W. L. Kraus (Cornell University).
Protein purification.
GST-RIP140 (1-495), (623-951), (977-1158) wild type and mutant constructs were expressed in bacteria and purified by glutathione Sepharose affinity chromatography (Sigma). His-tagged hCtBP1 was expressed in bacteria and purified by nickel-nitrilotriacetic acid affinity chromatography (Qiagen). Full-length FLAG-tagged p300 was expressed in Sf9 cells and purified using the M2 FLAG affinity matrix (Sigma).
Antibodies.
The acetylated K446 RIP140 antibody (αAcK446) was generated against the peptide PIDLSCacKHGTE. The antibody was affinity purified against the acetylated peptide and cross-absorbed against the nonacetylated peptide (Research Genetics, Huntsville, Ala.). A commercially available RIP140 antibody (αRIP140; Santa Cruz Biotechnology) was used for immunoprecipitation of endogenous RIP140. A monoclonal tetra-His antibody (Qiagen) was used to detect His-tagged hCtBP1. FLAG-tagged hCtBP1 was detected by using an antibody directed against the FLAG epitope (Upstate Biotechnology).
Yeast two-hybrid assay.
L40 yeast cells expressing LexA-hCtBP1 were transformed with an E9.5 mouse VP16 fusion cDNA library (7) as previously described (38). Yeast cells positive for histidine auxotrophy and β-galactosidase production were identified and confirmed by a second round of transformations. LexA-lamin was used as a negative control.
In vitro acetylation assays.
Recombinant His-tagged RIP140 fragments or GST-RIP140 fragments were incubated with full-length, baculovirus-purified p300 for 30 min at 30°C in a 30-μl reaction mixture containing acetylation buffer (10 mM Tris-HCl [pH 8.0], 10% glycerol, 1 mM EDTA, 10 mM sodium butyrate, 1 mM dithiothreitol [DTT]) and [3H]acetyl coenzyme A (AcCoA; Amersham Pharmacia Biotech). Reaction mixtures were electrophoresed on a 10% polyacrylamide gel and analyzed by fluorography. For glutathione S-transferase (GST) pull-down assays, acetylation reactions were performed as described previously using unlabeled AcCoA (10 μM). Mock acetylation reactions were performed using 1 pmol of p300 in the absence of 10 μM AcCoA or with AcCoA in the absence of p300.
GST pull-down assays. (i) Binding assays.
GST or GST-RIP140 fragments were acetylated or mock acetylated prior to immobilization onto glutathione beads. Equimolar amounts of GST or GST fusion proteins were linked to glutathione beads (Amersham Pharmacia Biotech) in the presence of binding buffer (20 mM HEPES [pH 7.6], 300 mM KCl, 10% glycerol, 0.5% NP-40, 1 mM DTT, 10 μM Na3VO4, 10 μM NaF, and complete protease inhibitors [Roche Molecular Biochemicals]) for 1 h at room temperature. Bovine serum albumin was then added to a final concentration of 0.5% for 30 min at 4°C. The beads were washed once with binding buffer and incubated for 1 h at room temperature with 35S-labeled in vitro-translated hCtBP1 (Promega) or recombinant His-tagged hCtBP1. The samples were washed three times with binding buffer, electrophoresed on a 10% polyacrylamide gel, and processed for autoradiography or Western analysis.
(ii) Peptide competition assays.
Recombinant His-tagged hCtBP1 (11.6 nmol) was preincubated with a 20 or 40 μM concentration of nonacetylated (SNCVPIDLSCKHGT) or acetylated (SNCVPIDLSCacKHGT) peptides in binding buffer for 30 min on ice and subsequently incubated with the immobilized GST or GST fusion proteins as described previously. After being washed in binding buffer, the proteins were eluted with 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer and electrophoresed on 10% polyacrylamide gels. The reaction mixtures were analyzed by Western blotting using a tetra-His antibody (Qiagen).
Cell culture and transfection assays. (i) Mammalian two-hybrid assays.
HepG2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; BRL) supplemented with 10% fetal bovine serum (FBS; HyClone). Transfections were performed with the Fugene reagent (Roche Molecular Biochemicals) as directed by the manufacturer. The cells were grown in 24-well plates and transfected with 100 ng of reporter, 50 ng of Gal4-RIP140 (386-470), and 50 ng of VP16-hCtBP1. The total amount of DNA per well (500 ng) was kept constant by the addition of an empty pcDNA3 vector. Luciferase assays were performed as described previously (39). Each condition was assayed in triplicate a minimum of three times.
(ii) Repression assays.
COS7 and HepG2 cells were cultured in DMEM supplemented with 10% FBS. Cells were cultured in six-well plates and transfected using the Fugene reagent with 300 ng of Gal4-SV40-CAT reporter, 20 ng of SV40-lacZ, and 700 ng of Gal4 fusion construct. The total amount of DNA was maintained at 1.5 μg using empty pcDNA3 vector. Protein levels of the Gal4-RIP140 constructs were determined by immunoprecipitation followed by Western analysis of cell extracts.
For studies of estrogen receptor-dependent transcription, HepG2 cells were cultured in phenol red free DMEM (BRL) supplemented with charcoal and dextran stripped FBS (HyClone) for a minimum of 2 weeks before culturing in 24-well plates. Transfections of 100 ng of (ERE)2-pS2-CAT reporter, 20 ng of SV40-lacZ, 100 ng of full-length pEFRIP140 wild type and mutants, and 100 ng of pCMV-hCtBP1 were performed using the Fugene reagent. Cells were treated with 100 μM CoCl2 for 16 h and either 10 nM 17β-estradiol or ethanol for 4 to 6 h. The total DNA concentration was kept constant by addition of empty pcDNA3 vector. Chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assays (Roche Molecular Biochemicals) were performed 48 h after transfection as directed by the manufacturer. β-Galactosidase assays were performed using the Emerald II reagent (TROPIX; PE Biosystems). Each condition was assayed in triplicate a minimum of three times.
In vivo acetylation assays.
COS7 and HepG2 cells were cultured in 10% FBS-supplemented DMEM and grown on 6-cm-diameter plates for transfection with 2.5 μg of full-length CBP. After 48 h, the cells were washed with cold phosphate-buffered saline (PBS) and lysed by incubation on ice for 10 min in 500 μl of lysis buffer (20 mM HEPES [pH 7.6], 300 mM KCl, 10% glycerol, 0.1 mM EDTA, 0.2 mM ZnAc2, 1% NP-40, 1 mM DTT, 10 μM Na3VO4, 10 μM NaF, and complete protease inhibitors). Cell debris was cleared by centrifugation at 20,000 × g for 10 min at 4°C. Approximately 0.8 μg of polyclonal RIP140 antibody (Santa Cruz Biotechnology) or normal rabbit immunoglobulin G (Sigma) was bound to protein G Sepharose beads (Amersham Pharmacia Biotech) and incubated with the cell lysates for 1 h at 4°C. The beads were washed with lysis buffer and eluted with 5× SDS-PAGE loading buffer. The immunoprecipitates were subjected to Western analysis using the affinity-purified acetylated K446 RIP140 antibody (αAcK446) or a commercially available RIP140 antibody.
Coimmunoprecipitation assays.
COS7 cells were grown to 50% confluency on 100-mm-diameter plates and transfected with 4 μg of FLAG-hCtBP1-pcDNA3. Cells were washed with cold PBS 48 h posttransfection, collected in lysis buffer (PBS, 0.1% NP-40, 1 mM DTT, 50 mM β-glycerophosphate, 10 mM NaF, 10 μM Na3VO4, and complete protease inhibitors), and sonicated. The cell lysates were then centrifuged, and the supernatants were added to an M2 FLAG affinity matrix (Sigma). Coimmunoprecipitations were performed for 1 h at 4°C. Subsequently, the beads were washed four times in lysis buffer and eluted by boiling for 5 min in 5× SDS-PAGE loading buffer. After gel electrophoresis and transfer to polyvinylidene difluoride membrane (Millipore), the immunoprecipitates were probed with an anti-RIP140 antibody (Santa Cruz Biotechnology).
RESULTS
Identification of potential CtBP-interacting proteins.
To determine whether acetylation might be a general mechanism for the regulation of CtBP interactions, we identified a family of CtBP-binding partners using a yeast two-hybrid screen. Approximately 68% of the 41 positive clones identified from a mouse embryonic day 9.5 cDNA library contained a sequence that was closely related to the PXDLS motif (Table 1). Of these, 75% contained a flanking Lys or Arg residue. Of the DNAs containing a PXDLS motif whose identities were known, 72% corresponded to known or suspected transcriptional repressors. Among this group were CtIP (CtBP-interacting protein), BKLF, and ZEB, which have previously been shown to repress transcription by binding to CtBP. The interaction of RIP140 with CtBP had not been described before.
TABLE 1.
Yeast two-hybrid CtBP interaction proteinsa
| Group no. | Motif | Name of protein |
|---|---|---|
| I | P X D L S X K | |
| P I D L S C K | RIP140 | |
| P L D L S S K | BCoR | |
| P T D L S M K | Unknown | |
| P L D L S A K | Unknown | |
| P E N L S T K | MyT-1 | |
| P Q D L P S K | MyT-1 | |
| V L D L S T K | Unknown | |
| V L D L S V K | RP58 | |
| II | P X D L S X R | |
| P L D L S D R | CtIPb | |
| P L D L S M R | Unknown | |
| P L D L S L R | Zinc finger protein 219 | |
| P L D L T V R | Unknown | |
| P L N L S S R | SRp55-3 | |
| III | P X D L S X X K | |
| P L D L S L P K | ZEBb | |
| P V D L T V N K | BKLFb | |
| P I D L T K S K | Teashirt2 | |
| P L D L S P V K | Unknown | |
| P M D L S T V K | RING 3, and unknown | |
| P L D I L Q S K | Retinoic acid-induced protein | |
| V V D L T L P K | Ku70 | |
| V L D L S V H K | RIZ | |
| IV | P X D L S X X X | |
| P L D L S S G V | RIZ | |
| V V D L S K A S | Unknown | |
| P L D L S C G S | Unknown | |
| P L N L S L G P | Zinc finger protein 217 | |
| P T D L S V N P | Unknown | |
| V I D L T I E S | Miz1 | |
| P L N L R I P S | NAB1 |
Two potential CtBP-binding motifs in MyT1 and RIZ are listed; 13 clones did not contain an identifiable PXDLS motif.
Protein previously shown to interact with CtBP.
Characterization of the RIP140-CtBP interaction.
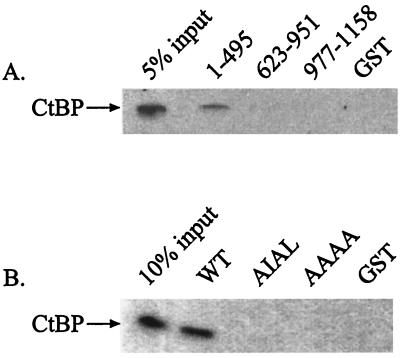
To identify the CtBP-binding site in RIP140, we generated GST-RIP140 constructs spanning the three previously characterized repression domains (11) (Fig. 1). GST pull-down assays verified that only the fragment containing the PXDLS motif, GST-RIP140 (1-495), was capable of interacting with CtBP (Fig. 2A). This fragment overlapped with the portion of RIP140 detected in the yeast two-hybrid assay. Mutation of the PXDLS motif completely abolished the interaction (Fig. 2B). Neither the portion of RIP140 containing the central cluster of LXXLL motifs, GST-RIP140 (623-951), nor the carboxyl-terminal portion, GST-RIP140 (977-1158), was capable of interacting with CtBP (Fig. 2A), suggesting that these domains may repress transcription through an alternate mechanism. Indeed, we showed that GST-RIP140 (977-1158) interacted with HDAC1 from HeLa nuclear extracts (data not shown).
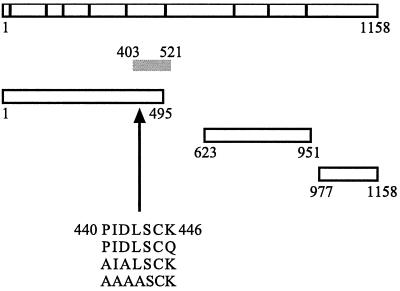
FIG. 1.
RIP140 contains a consensus CtBP-binding site. The nine LXXLL motifs present in RIP140 are represented by the vertical bars. The fragment of RIP140 (aa 403 to 521) isolated from an E9.5 mouse embryo cDNA library is indicated by the gray bar. Fragments of RIP140 used in GST pull-down assays are also depicted. Mutations in the consensus CtBP-binding motif (aa 440 to 446) are indicated.
FIG. 2.
RIP140 binds to hCtBP1 in vitro. (A) Equimolar amounts of GST-RIP140 (1-495), GST-RIP140 (623-951), GST-RIP140 (977-1158), and GST alone were incubated with in vitro-translated hCtBP1. The samples were electrophoresed on a 10% polyacrylamide gel and analyzed by autoradiography. (B) Mutation of the consensus hCtBP1 binding motif abolishes the RIP140-hCtBP1 interaction. Equimolar concentrations of GST-RIP140 (1-495) wild type (WT), PIDL→AIAL, PIDL→AAAA, and GST alone were incubated with recombinant, His-tagged hCtBP1. The samples were electrophoresed on a 10% polyacrylamide gel and analyzed by Western blotting using a monoclonal His tag antibody.
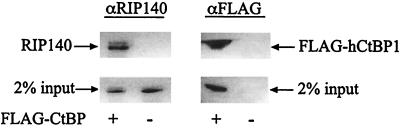
To determine whether the full-length RIP140 and CtBP proteins interact in vivo, we performed coimmunoprecipitation assays in COS7 cells, which contain low but detectable levels of RIP140. Cells were transiently transfected with FLAG-tagged hCtBP1, and lysates were incubated with an M2 FLAG affinity matrix. Immunoprecipitates containing FLAG-hCtBP1 were analyzed by Western blotting using an antibody directed against the amino-terminal portion of RIP140. These assays confirmed that endogenous RIP140 associates with CtBP in vivo (Fig. 3). Treatment of cells with estradiol did not affect the amount of CtBP that coimmunoprecipitated with RIP140 (data not shown).
FIG. 3.
RIP140 interacts with hCtBP1 in vivo. COS7 cells were transiently transfected (+) or mock transfected (−) with FLAG-hCtBP1. Immunoprecipitations were performed using an M2 FLAG affinity matrix. Western analyses were performed using a polyclonal RIP140 (left panel) or FLAG (right panel) antibody, as indicated at the top of the figure.
RIP140 is acetylated in vitro and in vivo.
Previous studies from our lab have demonstrated that acetylation of a Lys residue (Lys239) near the carboxyl terminus of E1A regulates the interaction with CtBP (39). The modified Lys residue in E1A is adjacent to the CtBP-binding site, an arrangement that is conserved in RIP140. We thus tested whether RIP140 could similarly become acetylated. Our previous studies showed that P/CAF and GCN5 were equally capable of acetylating E1A. To our surprise, neither enzyme was able to acetylate RIP140 in vitro (data not shown). In contrast, in vitro acetylation reactions using baculovirus-expressed, full-length mouse p300 and [3H]AcCoA demonstrated that GST-RIP140 (1-495) is readily acetylated (Fig. 4A). Mutation of the Lys residue adjacent to the CtBP-binding motif markedly attenuated acetylation (Fig. 4A). Although acetylation was not completely abolished, our results suggest that Lys446 is a major RIP140 acetylation site. Neither GST-RIP140 (623-951) nor GST-RIP140 (977-1158) could be acetylated by p300 (data not shown).
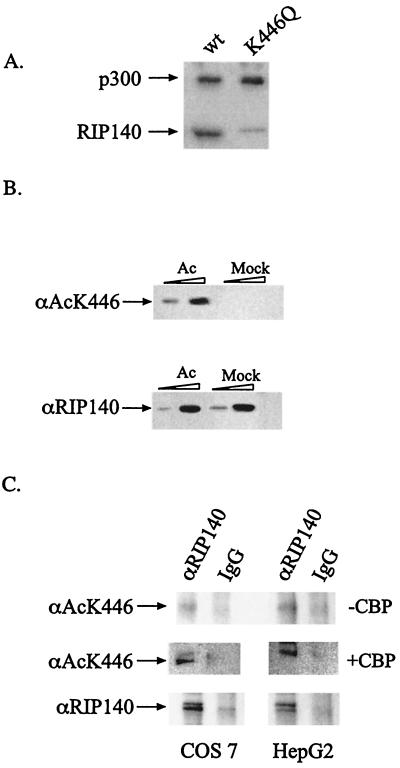
FIG. 4.
RIP140 is acetylated at lysine 446. (A) Eleven nanomoles of recombinant GST-RIP140 (1-495) wild type or K446Q mutant was incubated with full-length, baculovirus-purified p300 (1 pmol) in the presence of [3H]AcCoA. Reactions were analyzed by SDS-PAGE and visualized by fluorography. (B) Recombinant GST-RIP140 (1-495) (25 and 300 ng) was acetylated or mock acetylated with p300. Samples were electrophoresed on a 10% polyacrylamide gel and subjected to Western analysis using an antibody directed against acetylated (top) or unacetylated (bottom) RIP140. (C) In the top panels, endogenous RIP140 was immunoprecipitated from COS7 and HepG2 cells with a RIP140 antibody and subjected to Western analysis using the acetylated K446 RIP140 antibody. Middle panels show cells transfected with full-length CBP before immunoprecipitation. Total RIP140 proteins immunoprecipitated from these cells are shown in the bottom panels.
Commercially available antibodies do not recognize acetylated Lys residues in all contexts, and this appears to be the case for RIP140 (data not shown). Consequently, to determine whether acetylation of RIP140 at Lys446 occurs in vivo, we generated an antibody against a RIP140 peptide, PIDLSCacKHGTE, containing the acetylated Lys (αAcK446). After affinity purification, the αAcK446 antibody recognized the acetylated form of RIP140 but did not detect RIP140 which had been subjected to a mock-acetylation reaction (Fig. 4B, top). Specificity of the αAcK446 antibody was also demonstrated by showing that it did not recognize other acetylated proteins, such as p53 (data not shown). Under the conditions used, the αRIP140 and αAcK446 antibodies appear equally capable of recognizing their epitopes in the recombinant GST-RIP140 fusion proteins (Fig. 4B).
To determine whether RIP140 was acetylated in vivo, we transiently transfected COS7 and HepG2 cells, both of which contain endogenous RIP140, with a full-length mouse CBP expression vector. The cell lysates were immunoprecipitated using an antibody that recognizes RIP140 (αRIP140) or immunoglobulin G fractions from nonimmunized rabbits. Western analysis of the immunoprecipitates demonstrates that in the absence of exogenous CBP, only low levels of endogenous acetylated RIP140 were detected (Fig. 4C, top panels). However, in the presence of exogenous CBP, RIP140 is readily acetylated in vivo in both cell types (Fig. 4C, compare middle and bottom panels).
Acetylation of RIP140 abolishes its interaction with CtBP.
To determine whether the Lys residue adjacent to the CtBP-binding site participates in the RIP140-CtBP interaction, we mutated this residue to Gln, which resembles acetylated Lys in that it is uncharged at neutral pH. Mutation of Lys446 to Gln abolished the interaction (Fig. 5A), confirming the importance of the Lys residue for CtBP interaction and suggesting that acetylation of this residue might also regulate CtBP binding. To test this hypothesis, GST-RIP140 (1-495) was acetylated or mock acetylated (treated in the absence of AcCoA) with p300 and incubated with in vitro-translated hCtBP1. Acetylation abolished interaction of RIP140 with CtBP, while mock acetylation did not (Fig. 5A). These studies indicate that the ability of RIP140 to interact with CtBP was regulated by acetylation.
FIG. 5.
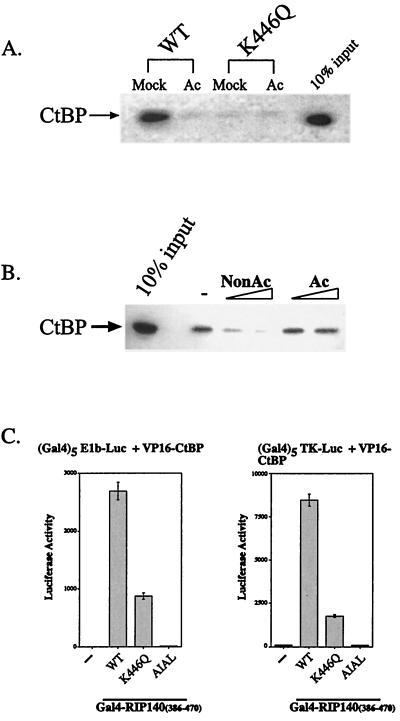
Acetylation of RIP140 disrupts its interaction with hCtBP1. (A) Mutation or acetylation of K446 disrupts the interaction of RIP140 with hCtBP1. GST pull-down assays were performed using acetylated or mock-acetylated GST-RIP140 (1-495) wild type (WT) or K446Q. Samples were incubated with in vitro-translated hCtBP1, electrophoresed on a polyacrylamide gel, and analyzed by autoradiography. (B) Peptide competition assays. GST-RIP140 (1-495) was incubated with recombinant, His-tagged hCtBP1 (11.6 nmol) in the presence or absence of 20 or 40 μM nonacetylated (NonAc) or acetylated (Ac) peptides. Bound hCtBP1 was analyzed by Western blotting using a His tag antibody. −, binding in the absence of peptide competitor. (C) K446Q mutation attenuates the RIP140-hCtBP1 interaction in a mammalian two-hybrid assay. Gal4-RIP140 (386-470) and VP16-hCtBP1 were cotransfected with either (Gal4)5-E1b-luc or (Gal4)5-TK-luc reporter constructs into HepG2 cells. Luciferase activity was measured 48 h posttransfection.
To confirm these results, we performed a series of peptide competition assays. Because only the nonacetylated form of RIP140 is able to interact with CtBP, the nonacetylated peptide, but not the acetylated form, should be able to compete with GST-RIP140 (1-495) for CtBP binding. Peptide competition assays were performed using an acetylated RIP140 peptide (SNCVPIDLSCacKHGT) or a nonacetylated peptide (SNCVPIDLSCKHGT). Preincubation of the nonacetylated peptide completely blocked GST-RIP140 (1-495) binding to hCtBP1, while incubation with the acetylated peptide did not, even at the higher concentrations (Fig. 5B). These studies confirm the potent effects of RIP140 acetylation on CtBP binding.
It is not possible to manipulate the level of RIP140 acetylation very precisely in vivo, but evidence for the effect of Lys446 acetylation on CtBP binding can be obtained by examining the Lys446Gln mutant. Mammalian two-hybrid assays were performed using Gal4-RIP140 (386-470) and VP16-hCtBP1. Wild-type Gal4-RIP140 is able to interact with VP16-hCtBP1 in HepG2 cells, a cell line that normally expresses RIP140, and activate the E1b or thymidine kinase promoter (Fig. 5C). Mutation of the PIDL sequence in CtBP to AIAL completely blocked the interaction, while mutation of Lys446 to Gln reduced the interaction by 70 to 80%.
RIP140 represses transcription through its association with CtBP.
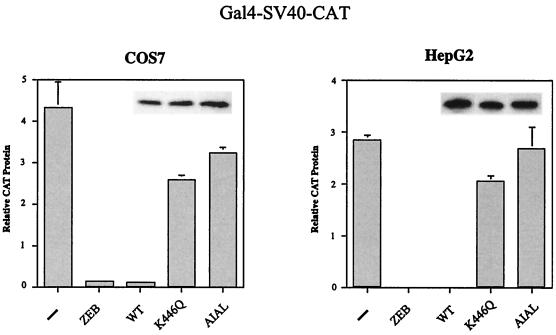
Our studies suggest that one of the mechanisms by which RIP140 represses transcription is through its interaction with the corepressor, CtBP. Disrupting this interaction by acetylation or mutation of Lys446 should result in a loss of repression. To test this hypothesis, we analyzed the ability of wild-type Gal4-RIP140 (386-470), which contains sequences spanning the CtBP interaction domain, as well as the Lys446Gln and AIAL mutants, to repress transcription of a Gal4-SV40-CAT reporter gene. As a control for these experiments, we analyzed Gal4-ZEB (700-776), a known CtBP-dependent transcriptional repressor (18). In both COS7 and HepG2 cells, wild-type Gal4-RIP140 (386-470) represses transcription to a level similar to that of Gal4-ZEB (700-776) (Fig. 6). Alteration of the consensus CtBP interaction motif, either by mutation of Lys446 to Gln or alteration of the PIDL sequence to AIAL, restores transcription levels to those seen in the absence of repressor (Fig. 6). We conclude from these studies that modification of Lys446 contributes to the regulation of RIP140 repressor function.
FIG. 6.
RIP140 represses transcription by binding to hCtBP1. Gal4-RIP140 fusion protein containing the CtBP interaction motif was analyzed for the ability to repress a Gal4-SV40-CAT reporter. The wild type (WT) and the K446Q and AIAL mutants were compared to Gal4-ZEB (700-776). COS7 and HepG2 cells were transiently transfected with 300 ng of reporter, 700 ng of Gal4 fusion constructs, and 20 ng of SV40-lacZ. CAT protein produced in the absence of repressor (−) is indicated. The total amount of DNA was kept constant using empty pcDNA3 vector. Protein levels of the RIP140 constructs are shown in the insets. CAT protein production was measured 48 h after transfection using an enzyme-linked immunosorbent assay system (Roche Molecular Biochemicals), and values are normalized to β-galactosidase activity.
RIP140 represses nuclear hormone receptor-dependent transcription.
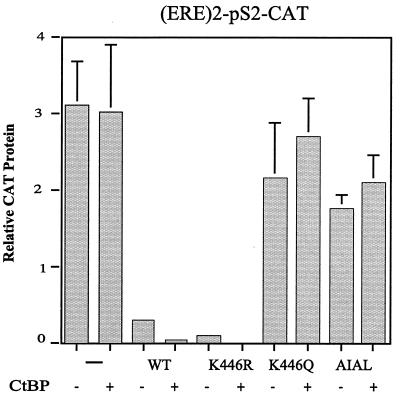
We next examined the ability of full-length RIP140 to repress transcription of an estrogen receptor-dependent reporter gene, (ERE)2-pS2-CAT. This gene contains a duplicated estrogen response element (ERE) upstream from the pS2 promoter and has been used extensively to examine estrogen-responsive transcription (9). In the absence of exogenous RIP140, CtBP did not significantly repress transcription (Fig. 7). However, in the presence of exogenous wild-type RIP140 or a form in which Lys446 had been mutated to Arg, transcription was dramatically repressed. Mutation of Lys446 to Gln or alteration of the PIDL motif to AIAL severely attenuated RIP140-mediated transcription repression. These results support the conclusion that RIP140 represses transcription in large part through its association with CtBP.
FIG. 7.
RIP140 represses estrogen receptor-dependent transcription. The ability of full-length RIP140, wild type and mutants, to repress transcription of an (ERE)2-pS2-CAT reporter gene was analyzed in HepG2 cells treated with 10 nM estradiol. Cells were transfected with 100 ng of CAT reporter, 20 ng of SV40-lacZ, and 100 ng of pEFRIP140 wild type and mutants in the absence or presence of pCMV-hCtBP1. The total concentration of DNA was kept constant by addition of empty pcDNA3 vector. CAT protein production was measured 48 h after transfection, and values are normalized to β-galactosidase activity.
DISCUSSION
RIP140 associates in a ligand-dependent manner with a variety of nuclear hormone receptors, including the estrogen, glucocorticoid, retinoic acid, retinoid X, thyroid, and liver X receptors (4, 11, 13, 28, 30). Unlike other proteins that interact with ligand-bound nuclear hormone receptors, RIP140 is a transcriptional corepressor. In this study, we have analyzed the regulation of RIP140 corepressor function by acetylation. Previous work from our laboratory has demonstrated that the E1A-CtBP interaction is similarly modulated by acetylation (39). In the case of E1A, the acetylation activity of P/CAF was much more efficient than that of p300/CBP. With RIP140, p300/CBP was able to mediate acetylation, whereas P/CAF was completely ineffective. The common theme of both studies was that Lys acetylation regulated the binding of CtBP and, hence, repressor function. Our yeast two-hybrid assays suggest that this mechanism may be a fairly common mode of gene derepression, in that many of the putative CtBP binding sequences were flanked by a Lys residue (Table 1, group I). Whether the repressors in group III can be acetylated and whether this modification blocks CtBP binding have not been addressed. In several instances, however, this residue is an Arg (shown in group II), which is not a known target of the coactivator acetyltransferases. Indeed, it has been shown in the context of E1A that a Lys-to-Arg substitution actually increases CtBP binding in vivo (39), although it is possible that this reflects a basal level of Lys acetylation rather than a true difference in the affinity of Lys- or Arg-containing proteins. Nonetheless, it appears that individual transcriptional repressors may be differentially responsive to coactivator acetyltransferases depending upon the identity of this single amino acid.
Why acetylation of the flanking Lys residue is so critical for CtBP interaction is unknown. Presumably, this issue will be clarified once the structure of one of the CtBP-binding repressors has been solved. Although such information is not yet available, clues to the nature of the CtBP interaction site may possibly be gleaned from other known structures. For example, the prohormone-processing carboxypeptidase Kex1 contains a PXDLTXK sequence that might also be expected to bind CtBP. Indeed, we have shown that CtBP interacts with Kex1 in GST pull-down assays (N. Vo, unpublished observations.) Structural analysis of Kex1 indicates that the motif is located on the surface of the protein and forms a pocket that is filled by the positively charged Lys residue (25). Thus, neutralization of this positively charged Lys residue by acetylation would be predicted to perturb the CtBP-binding interface. In keeping with this model, we have shown that mutation of Lys to Gln prevents CtBP binding. It is possible, therefore, that the flanking Lys is ideally positioned to regulate CtBP binding. In contrast, the results of the two-hybrid assay suggest that Lys is not essential for binding (see group IV [Table 1], where the flanking Lys is not present). We suggest, therefore, that although Lys is not essential, its acetylation prevents the CtBP interaction.
Our model is somewhat reminiscent of that described for the coactivator ACTR, which, when acetylated by p300, disrupts the coactivator complex and terminates transcription (5). Unlike the situation with ACTR, however, acetylation of RIP140 leads to gene activation rather than repression. Coactivator histone acetyltransferases recruited by particular transcription factors are believed to modify specific nucleosomes in the vicinity of activated genes (reviewed in reference 27). Recent studies demonstrate that histone modifications may occur globally as well (33). The idea that the basal state of nucleosomes contains both acetylated and deacetylated histones suggests that chromatin can be primed for transcription. Thus, the ability of a transcription factor such as RIP140 to function as an activator or repressor may depend on the global acetylation state of a given gene. Alternatively, RIP140 could be acetylated locally through recruitment of p300/CBP. Whether nuclear hormone receptors can associate simultaneously with p300/CBP and RIP140 is unknown, however.
In conclusion, the acetylation state of CtBP binding proteins such as E1A and RIP140 directly modulates their ability to mediate transcriptional repression. In the unacetylated state, these proteins can act as transcriptional repressors through their interaction with CtBP, but in the acetylated state, the interaction is prevented. Although the mechanism by which CtBP represses transcription remains unknown, this corepressor has clearly been shown to be critical for the regulation of a large number of developmental and differentiation processes. Potentially, these activities can also be modulated by the acetylation state of the CtBP-interacting transcription factors. It is widely accepted that the level of histone deacetylation is correlated with transcriptional repression while histone acetylation is correlated with transcriptional activation. Our studies suggest that disruption of repressor-corepressor complexes might be an equally plausible mechanism to explain how coactivator acetyltransferases activate selected genes.
ACKNOWLEDGMENTS
We thank G. Chinnadurai, W. L. Kraus, and D. Dean for reagents, H. Yao for help with the two-hybrid assays, and Q. Zhang for helpful comments.
This work was supported by grants from the NIH.
REFERENCES
- 1.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyd J M, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 4.Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 6.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 7.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koipally J, Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J Biol Chem. 2000;275:19594–19602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 9.Kraus W L, Weis K E, Katzenellenbogen B S. Inhibitory cross-talk between steroid hormone receptors: differential targeting of estrogen receptor in the repression of its transcriptional activity by agonist- and antagonist-occupied progestin receptors. Mol Cell Biol. 1995;15:1847–1857. doi: 10.1128/mcb.15.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C H, Chinpaisal C, Wei L N. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol Cell Biol. 1998;18:6745–6755. doi: 10.1128/mcb.18.11.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C H, Wei L N. Characterization of receptor-interacting protein 140 in retinoid receptor activities. J Biol Chem. 1999;274:31320–31326. doi: 10.1074/jbc.274.44.31320. [DOI] [PubMed] [Google Scholar]
- 12.Mannervik M, Nibu Y, Zhang H, Levine M. Transcriptional coregulators in development. Science. 1999;284:606–609. doi: 10.1126/science.284.5414.606. [DOI] [PubMed] [Google Scholar]
- 13.Miyata K S, McCaw S E, Meertens L M, Patel H V, Rachubinski R A, Capone J P. Receptor-interacting protein 140 interacts with and inhibits transactivation by peroxisome proliferator-activated receptor alpha and liver-X-receptor alpha. Mol Cell Endocrinol. 1998;146:69–76. doi: 10.1016/s0303-7207(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 14.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 16.Phippen T M, Sweigart A L, Moniwa M, Krumm A, Davie J R, Parkhurst S M. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J Biol Chem. 2000;275:37628–37637. doi: 10.1074/jbc.M004234200. [DOI] [PubMed] [Google Scholar]
- 17.Postigo A A, Dean D C. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc Natl Acad Sci USA. 2000;97:6391–6396. doi: 10.1073/pnas.97.12.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postigo A A, Dean D C. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postigo A A, Dean D C. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 21.Schaeper U, Boyd J M, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeper U, Subramanian T, Lim L, Boyd J M, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 23.Sewalt R G, Gunster M J, van der Vlag J, Satijn D P, Otte A P. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 25.Shilton B H, Li Y, Tessier D, Thomas D Y, Cygler M. Crystallization of a soluble form of the Kex1p serine carboxypeptidase from Saccharomyces cerevisiae. Protein Sci. 1996;5:395–397. doi: 10.1002/pro.5560050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sollerbrant K, Chinnadurai G, Svensson C. The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res. 1996;24:2578–2584. doi: 10.1093/nar/24.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 28.Subramaniam N, Treuter E, Okret S. Receptor interacting protein RIP140 inhibits both positive and negative gene regulation by glucocorticoids. J Biol Chem. 1999;274:18121–18127. doi: 10.1074/jbc.274.25.18121. [DOI] [PubMed] [Google Scholar]
- 29.Sundqvist A, Sollerbrant K, Svensson C. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 1998;429:183–188. doi: 10.1016/s0014-5793(98)00588-2. [DOI] [PubMed] [Google Scholar]
- 30.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J A. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 31.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vo N, Goodman R H. CREB binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 33.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 34.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 35.Wei L N, Farooqui M, Hu X. Ligand-dependent formation of retinoid receptors, receptor-interacting protein 140 (rip140), and histone deacetylase complex is mediated by a novel receptor-interacting motif of rip140. J Biol Chem. 2001;276:16107–16112. doi: 10.1074/jbc.M010185200. [DOI] [PubMed] [Google Scholar]
- 36.Wei L N, Hu X, Chandra D, Seto E, Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J Biol Chem. 2000;275:40782–40787. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C L, McKinsey T A, Lu J R, Olson E N. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Vo N, Goodman R H. Histone binding protein RbAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/mcb.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Yao H, Vo N, Goodman R H. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]