ABSTRACT.
Chikungunya virus (CHIKV) is a mosquito-transmitted pathogen in family Togaviridae, genus Alphavirus. Although CHIKV is well known for its ability to cause debilitating rheumatoid-like arthritis, it has been also been observed to cause cardiovascular symptoms such as arrhythmias. Here, using samples from a previous study, we sequenced RNA from serum, kidney, skeletal muscle, and cardiac muscle from CHIKV- and mock-infected IFN-αR−/− mice using two sequencing techniques to investigate heart-specific changes in virus mutational profiles and host gene expression. Mutation rates were similar across muscle tissues although heart tissue carried heart-specific CHIKV minority variants, one of which had a coding change in the nsP3 gene and another in the 3′UTR. Importantly, heart-specific transcriptional changes included differential expression of genes critical for ion transport and muscle contraction. These results demonstrate that CHIKV replicates in the hearts of immunodeficient mice and induce heart-specific mutations and host responses with implications for cardiac pathologies.
Chikungunya virus (CHIKV) is a mosquito-vectored alphavirus that, along with other arthritogenic alphaviruses, can cause a severe debilitating rheumatoid-like arthritis that can persist for months to years post infection. The pathophysiology of arthritogenic alphaviruses reflects their musculoskeletal tropism: CHIKV, for instance, has been shown to preferentially infect muscle satellite cells.1 However, a rare but notable complication of CHIKV infection in humans is myocarditis2 characterized by inflammation of cardiac tissue that can result in cardiovascular symptoms including dilated cardiomyopathy and even death.3 Although chikungunya fever (CHIKF) is not typically lethal, cardiovascular complications have been found to be a leading cause of death in several studies.4,5 Further, arrhythmias are a common occurrence in studies reporting electrocardiographic (ECG) data,6,7 and in at least one case of myocarditis detected by magnetic resonance imaging was reported to persist for a year post disease onset.8 This is particularly concerning for areas where preexisting cardiovascular conditions like hypertension are common, such as the United States, as these have also been linked to increased CHIKF disease severity.9 However, despite a demonstrable tropism for skeletal muscle and a known ability to cause or exacerbate heart disease, the ability of CHIKV to infect cardiac muscle and the consequences thereof remain largely unexplored.
As part of our previous study10 comparing defective-RNA (D-RNA) expression in intracellular-derived viral RNA and extracellular-derived RNA, we inoculated four type-1 interferon receptor knock-out mice (129-IFNαR−/−, or A129) mice with 103 plaque-forming units CHIKV-AF15561 derived from an infectious clone and two mice with phosphate buffered saline (mock), and then collected serum, skeletal muscle from the injection site near the left rear footpad (SKMIS), skeletal muscle from the contralateral leg (SKMCL), heart, and kidney 4 days post infection. The RNA was then extracted from these tissues and sequenced using random-primed ClickSeq (CS) approach11 (data available in SRA NCBI archive, study number PRJNA613616). Interestingly, cardiac muscle and skeletal muscle at the site of injection had the highest diversity of viral D-RNA species compared with serum, and a principle component analysis revealed that of all tissues, cardiac muscle clustered the mostly tightly indicating that the sources of variance in these samples were the most similar.10
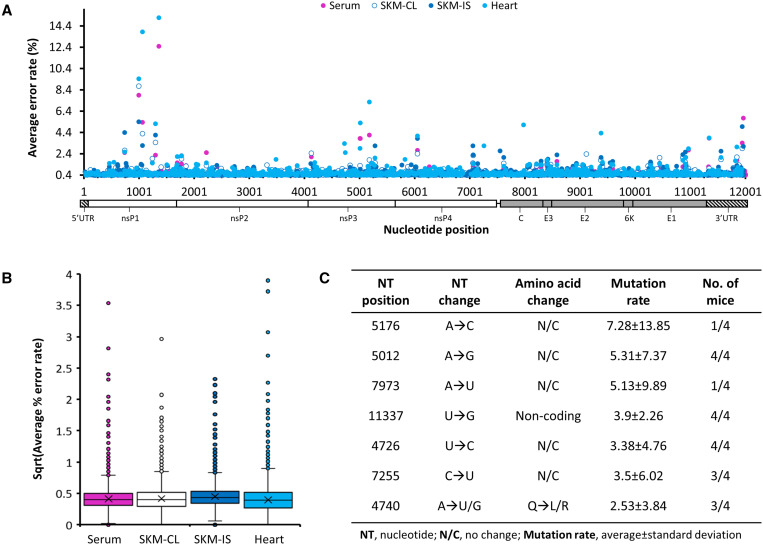
Considering these results, we performed additional bioinformatics analyses to extract nucleotide mutation frequencies from existing random-primed CS pileup files generated using samtools12 to compare virus mutational profiles between heart, serum, and muscle tissue types among the four infected animals. Across all sampled tissues, nsP1 and nsP3 exhibited the highest abundance of single nucleotide mutations, while the average nucleotide mutation rates remained similar (Figure 1A and B). However, tissue-specific mutations were observed in all mice (Supplemental Table S1). Skeletal muscle at the injection site generated the most minority variants (present in at least two mice at an average mutation rate > 1%), mostly in the nsP1 and nsP3 genes. Importantly, we identified several heart-specific minority variants that arose with average nucleotide mutation rates ranging between 1.25% and 5.31% among varying numbers of mice (Figure 1C). Most of these were silent mutations except an A4740(U/G) mutation resulting in either a Q223L or a Q223R amino acid change and a A5151(G/C) resulting in a D359(G/A) amino acid change in the nsP3 gene; however, the former only occurred in three of the four infected mice at an average rate of 2.53% ± 3.84 and the latter occurred in all four mice at an average rate of only 1.25% ± 0.58. There was an additional mutation in the 3′UTR, U11337G, which occurred in all four infected mice specifically in the heart at an average rate of 3.9% ± 2.26.
Figure 1.
Mutation profiles of Chikungunya virus (CHIKV) RNA collected from infected A129 mice. Four type 1 interferon knock-out (A129) mice were inoculated with CHIKV-AF15561, and then serum, skeletal muscle from the injection site near the left rear footpad (SKMIS), skeletal muscle from the contralateral leg (SKMCL), and heart were collected 4 days post infection; RNA derived therefrom was sequenced on a NextSeq550, and mutation rates were extracted from bam files. (A) Mutation rates across the genome of CHIKV by tissue. (B) Average nucleotide mutation rate across the genome by tissue. (C) Heart-specific mutations observed across data sets.
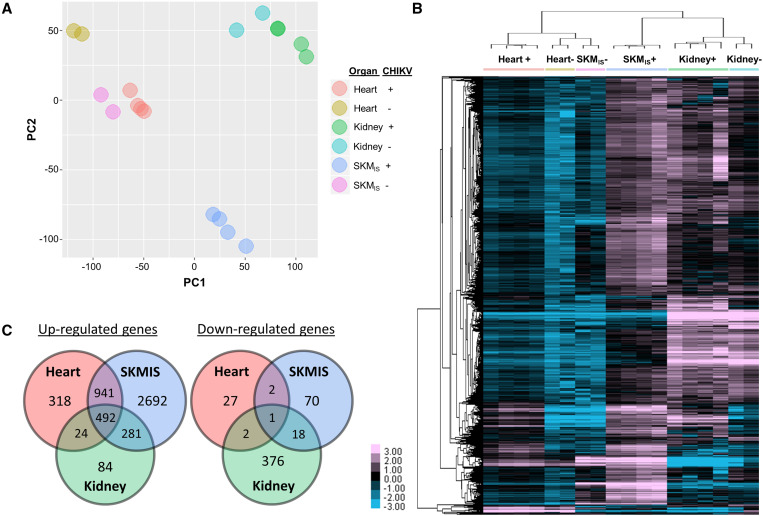
We additionally constructed poly-A primed CS libraries (“PAC-Seq”)13 for the same RNA samples from tissues of infected and mock-infected animals (data available in SRA NCBI archive, study number PRJNA736452) and analyzed sequences using our DPAC14 pipeline followed by DESeq215 to examine changes in host gene expression between infected and uninfected heart and skeletal muscle (represented by SKMIS) (Figure 2). Kidney data were used as a control tissue to identify genes with system-wide regulation changes, such as immune signaling pathways. ENRICHR16,17 was then used to identify GO processes (2018) associated with differentially expressed genes, grouped by either systemic (genes common to all data sets, i.e., present in all three organs), muscle-wide (genes common to both skeletal and heart muscle data sets), and heart (genes exclusively in the heart dataset). In CHIKV-infected mouse hearts, we identified 1,775 significant gene expression changes (fold change ≥ 2, P-adj ≤ 0.1), which showed consistency among replicates and clear distinction from controls by principal component analysis (Figure 2A). Hierarchical clustering showed all samples clustered by both tissue and condition (Figure 2B), further supporting the consistency observed among replicates. Of differentially expressed genes, 493 were differentially expressed systemically, 943 were muscle-wide (e.g., shared overlap between skeletal and heart muscle datasets), and 345 were heart-specific (Figure 2C). Interestingly, both uninfected and infected kidney samples clustered together, indicating that sources of variation between these samples were more similar than between infected muscle tissues. This is likely as a result of lower viral replication in the kidneys, which is also reflected in lower viral read counts in the original CS data,10 and thus kidney expression data function as a good control for system-wide expression changes secondary to infection.
Figure 2.
Differential gene expression profiles for tissues from Chikungunya virus (CHIKV)-infected A129 mice. Poly-A primed ClickSeq libraries were constructed with RNA samples collected from chikungunya virus-infected (CHIKV; N = 4, noted by “+”) or mock-infected (N = 2, noted by “−”) A129 mouse kidney, skeletal muscle (SKMIS), and heart to evaluate changes in gene expression. Significant (fold change ≥ 2, P-adj ≤ 0.1) gene expression changes were identified with DPAC pipeline. (A) Principal component analysis showed clear consistency among replicates and distinctive clustering by both tissue and CHIKV infection status. (B) Hierarchical clustering of the significantly altered genes using Cluster 3.0 followed by TreeView demonstrated the unique cardiac transcriptomic profile upon CHIKV infection (color scale: log2fold change). (C) Significantly up- or downregulated genes as a result of CHIKV infection in specific compartments.
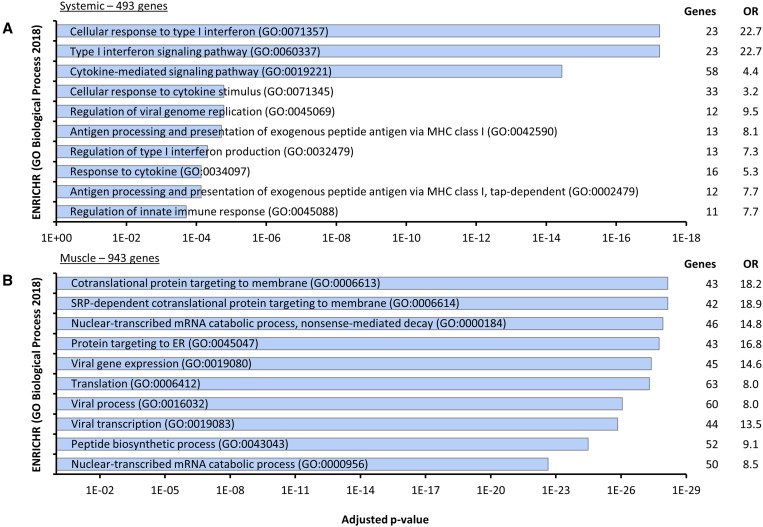
As expected, genes upregulated among all organ groups consisted largely of innate immune signaling pathways and cellular responses to infection (e.g., cytokine-mediated signaling pathways; Figure 3A). Unlike kidney, where CHIKV replication was likely low, muscle tissues (including heart) showed regulatory changes in multiple functional clusters related to viral replication, indicating both these tissues hosted active viral replication (Figure 3B). Finally, we found 318 and 27 genes that are uniquely and significantly up- or downregulated in CHIKV-infected murine heart, respectively (Figure 2C). From these 345 genes, ENRICHR identified 45 processes that were significantly (gene number > 2, P-adj ≤ 0.1) enriched in heart tissue, ranging from processes related to RNA and protein synthesis to apoptosis and cell proliferation; however, genes from these processes overlapped considerably so BiNGO18 was used to generate a network for visualization of related processes in Cytoscape19 (Figure 4A).
Figure 3.
Systemic and muscle-specific murine transcriptional response to Chikungunya virus (CHIKV) infection by biological process. Four type-1 interferon knock-out (A129) mice were inoculated with CHIKV-AF15561 and two mock-infected with PBS, and then skeletal muscle from the injection site near the left rear footpad (SKMIS), kidney, and heart were collected 4 days post infection; RNA was extracted from tissues, and then PACSeq libraries were constructed and sequenced on a Next550. Differential gene expression was analyzed using the DPAC pipeline, and genes were sorted into those significantly differentially expressed systemically (580 genes in kidney, SKMIS, and hear datasets; (A) and in muscle (1,124 genes in SKMIS and heart dayaseys, (B), and genes were analyzed using Enrichr analysis tool (https://maayanlab.cloud/Enrichr/). Shown the top 10 terms by adjusted P value. OR = odds ratio.
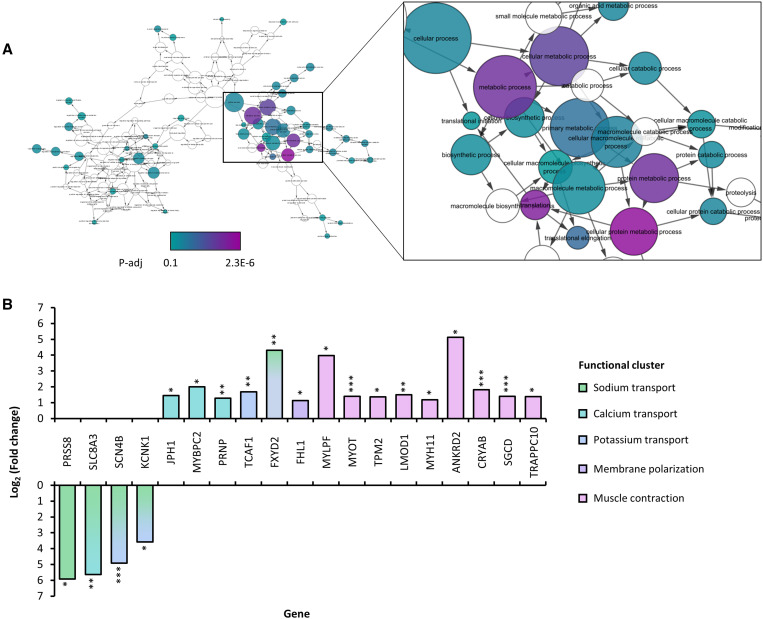
Figure 4.
Heart-specific transcriptional changes in Chikungunya virus (CHIKV)-infected A129 mice. Four type 1 interferon knock-out (A129) mice were inoculated with CHIKV-AF15561 and two mock-infected with PBS, and then skeletal muscle from the injection site near the left rear footpad (SKMIS), kidney, and heart were collected 4 days post infection; RNA was extracted from tissues, and then PAC-Seq libraries were constructed and sequenced on a NextSeq550. Differential gene expression was analyzed using the DPAC pipeline in conjunction with DESeq2 followed by enrichment analysis using ENRICHR analysis tool (https://maayanlab.cloud/Enrichr/) for genes that were significantly differentially regulated (fold change ≥ 2, p-adj ≤ 0.1). BiNGO was then used in conjunction with Cytoscape to generate a network visualization of enriched GO processes. (A) GO 2018 processes significantly enriched in heart tissue (P-adj ≤ 0.1; circles colored by level of significance); (B) genes differentially expressed in CHIKV-infected heart from processes related to ion homeostasis and muscle contraction (*, P-adj ≤ 0.1; **, P-adj ≤ 0.05; ***, P-adj < 0.01). Genes are colored by functional cluster; some ion transport-related genes serve as sym- or antiporters for dual cations and are colored accordingly.
Among 27 downregulated genes in the heart, four out of the top 10 genes with the largest fold changes were solute transporters, two of which were related to sodium transport (SLC8A3, fold change = 49.5, P-adj = 0.0052; SCN4B, fold change = 30, P-adj = 0.0012), while the largest fold change was in a regulator of sodium channel function (PRSS8, fold change = 60.35, P-adj = 0.09). Therefore, we searched enriched GO processes (gene number > 1) for terms related to ion homeostasis (e.g., sodium ion transport, GO0006814) and muscle contraction (e.g., muscle contraction, GO0006936), as these processes are reciprocal and critical for heart function, and then extracted genes from these processes for individual consideration (Figure 4B). Importantly, multiple ion transporters were differentially expressed that have previously been associated with heart disease. For example, SCN4B encodes a critical interacting partner of the Nav1.5 channel, mutations in which have been associated with congenital long-QT syndrome (a condition marked by dysregulation in membrane repolarization and irregular heartbeat),20 whereas KCNK1 encodes a potassium channel, knockdown of which results in bradycardia and atrial dilation in zebrafish.21 In conjunction with differentially expressed genes related to muscle contraction, this would indicate a potential for dysregulated contractile processes in the heart.
These sequencing studies evaluating heart-specific changes in CHIKV mutational profiles and host gene expression collectively provide confirmation that CHIKV can infect the hearts of immunodeficient mice and potentially cause defects in several critical cardiac processes. This is firstly shown by heart-specific mutations in infected cardiac tissue, which demonstrates that CHIKV can replicate in heart tissue and generate cardiac-specific minority variants, although the potential epidemiological significance of such variants is unknown. One mutation occurred in all four infected mice and specifically in the heart, U11337G, which fell in the 3′UTR, a noncoding region that nevertheless has important functions during viral replication.22 Interestingly, the sequence of and secondary RNA structures within the CHIKV 3′UTR have been shown to be important for CHIKV adaptation to its insect vector rather than its mammalian host.23–26 These studies, however, did not use mammalian heart tissue. Because nucleotide changes in an RNA sequence may potentially result in changes to secondary structure and subsequently functions thereof, it is possible that the U11337G variant may affect CHIKV replication in the heart, though has yet to be determined. Furthermore, heart and muscle transcriptional changes overlapped considerably in processes involved in viral replication and muscle function. We additionally showed heart-specific changes in host gene expression. This supports the hypothesis that CHIKV can infect cardiac tissue and subsequently induce cardiac pathologies. Although these results provide important insights into potential effects of alphavirus infection on the cardiovascular system with remarkable consistency between replicates, only two animals were used for mock controls in gene expression studies. Therefore, differential gene expression analyses remain preliminary and should be interpreted with caution. Furthermore, animals in this study were not perfused prior to organ collection, thus both mutation data as well as gene expression data may be contaminated by circulating virus and contaminating host RNAs, respectively. Finally, these studies were performed in an immunodeficient model of CHIKV infection where viral dissemination to nearly all tissues is enhanced, and therefore these results must additionally be validated in an immunocompetent model. Despite these shortcomings, the observations made here support clinical observations of CHIKV-associated heart disease and complications.
Importantly, both muscle-wide and heart-specific gene expression changes clustered functionally into ion channel homeostasis and muscle contraction, two interdependent processes essential for cardiac function; defects in these pathways subsequently may result in heart failure, marked by contractile changes and life-threatening arrhythmias. Ion channels maintain action potentials across the sarcolemma, which guide muscle fibers through the contraction process.27 Calcium ions are additionally important for mediating transactions between contractile proteins, namely myosin, troponin, and actin.28,29 Here, we’ve shown that genes involved in ion transport are significantly differentially regulated in the heart during CHIKV infection, with decreased expression of sodium transport genes (SCN4B) and increased expression of potassium and calcium binding and transport genes. Paired with expression changes in contractile protein genes, this is particularly problematic in the heart where action potentials are carefully orchestrated by specialized cells to induce contractions to specific rhythms, thus potentially leading to arrhythmias. Clinically, arrhythmias are a common feature of human CHIKV infection in studies that collect ECG data.6,7 Thus, these observations may be critical for understanding the pathophysiology of CHIKF-associated heart complications. Additionally, given that preexisting heart conditions are exacerbated by CHIKV infection, these results additionally warrant the investigation of the pathological effects of CHIKV infection in mouse models of heart disease.
Supplemental tables
ACKNOWLEDGMENTS
We would like to thank Scott Weaver for kindly providing containment facilities and virus used for the original animal studies upon which this work is based, as well as Tony Muruato for aiding in the original animal studies.
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.Ozden S. et al. , 2007. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One 2: e527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obeyesekere I, Hermon Y, 1972. Myocarditis and cardiomyopathy after arbovirus infections (dengue and Chikungunya fever). Br Heart J 34: 821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obeyesekere I, Hermon Y, 1973. Arbovirus heart disease: myocarditis and cardiomyopathy following dengue and Chikungunya fever–a follow-up study. Am Heart J 85: 186–194. [DOI] [PubMed] [Google Scholar]
- 4.Lima STS. et al. , 2020. Fatal outcome of Chikungunya virus infection in Brazil. Clin Infect Dis. doi: 10.1093/cid/ciaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitas ARR, Donalisio MR, Alarcón-Elbal PM, 2018. Excess mortality and causes associated with Chikungunya, Puerto Rico, 2014–2015. Emerg Infect Dis 24: 2352–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo-Zambrano DM, Jiménez-Canizales CE, Alzate-Piedrahita JA, Medina-Gaitán DA, Rodriguez-Morales AJ, 2016. Electrocardiographic changes in patients with Chikungunya fever. Rev Panam Infecto 18: 13–15. [Google Scholar]
- 7.Villamil-Gomez WE, Ramirez-Vallejo E, Cardona-Ospina JA, Silvera LA, Rodriguez-Morales AJ, 2016. Electrocardiographic alterations in patients with chikungunya fever from Sucre, Colombia: a 42-case series. Travel Med Infect Dis 14: 510–512. [DOI] [PubMed] [Google Scholar]
- 8.Simon F, Paule P, Oliver M, 2008. Chikungunya virus-induced myopericarditis: toward an increase of dilated cardiomyopathy in countries with epidemics? Am J Trop Med Hyg 78: 212–213. [PubMed] [Google Scholar]
- 9.Economopoulou A, Dominguez M, Helynck B, Sissoko D, Wichmann O, Quenel P, Germonneau P, Quatresous I, 2009. Atypical Chikungunya virus infections: clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Reunion. Epidemiol Infect 137: 534–541. [DOI] [PubMed] [Google Scholar]
- 10.Langsjoen RM, Muruato AE, Kunkel SR, Jaworski E, Routh A, 2020. Differential alphavirus defective RNA diversity between intracellular and extracellular compartments is driven by subgenomic recombination events. MBio 11: e00731–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Routh A, Head SR, Ordoukhanian P, Johnson JE, 2015. ClickSeq: fragmentation-free next-generation sequencing via click ligation of adaptors to stochastically terminated 3′-Azido cDNAs. J Mol Biol 427: 2610–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP, 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Routh A, Ji P, Jaworski E, Xia Z, Li W, Wagner EJ, 2017. Poly(A)-ClickSeq: click-chemistry for next-generation 3′-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res 45: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Routh A, 2019. DPAC: a tool for differential poly(A) cluster usage from poly(A)-targeted RNAseq data. G3 (Bethesda) 9: 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuleshov MV. et al. , 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maere S, Heymans K, Kuiper M, 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medeiros-Domingo A. et al. , 2007. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation 116: 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen AH. et al. , 2016. The two-pore domain potassium channel, TWIK-1, has a role in the regulation of heart rate and atrial size. J Mol Cell Cardiol 97: 24–35. [DOI] [PubMed] [Google Scholar]
- 22.Hyde JL, Chen R, Trobaugh DW, Diamond MS, Weaver SC, Klimstra WB, Wilusz J, 2015. The 5′ and 3′ ends of alphavirus RNAs–non-coding is not non-functional. Virus Res 206: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, Wang E, Tsetsarkin KA, Weaver SC, 2013. Chikungunya virus 3′ untranslated region: adaptation to mosquitoes and a population bottleneck as major evolutionary forces. PLoS Pathog 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stapleford KA. et al. , 2016. Whole-genome sequencing analysis from the Chikungunya virus caribbean outbreak reveals novel evolutionary genomic elements. PLoS Negl Trop Dis 10: e0004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R, Puri V, Fedorova N, Lin D, Hari KL, Jain R, Rodas JD, Das SR, Shabman RS, Weaver SC, 2016. Comprehensive genome scale phylogenetic study provides new insights on the global expansion of Chikungunya virus. J Virol 90: 10600–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filomatori CV, Bardossy ES, Merwaiss F, Suzuki Y, Henrion A, Saleh MC, Alvarez DE, 2019. RNA recombination at Chikungunya virus 3′UTR as an evolutionary mechanism that provides adaptability. PLoS Pathog 15: e1007706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant AO, 2009. Cardiac ion channels. Circ Arrhythm Electrophysiol 2: 185–194. [DOI] [PubMed] [Google Scholar]
- 28.Lehman W, Craig R, Vibert P, 1994. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature 368: 65–67. [DOI] [PubMed] [Google Scholar]
- 29.Affolter H, Chiesi M, Dabrowska R, Carafoli E, 1976. Calcium regulation in heart cells. The interaction of mitochondrial and sarcoplasmic reticulum with troponin-bound calcium. Eur J Biochem 67: 389–396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.