Abstract
There is emerging evidence of a gut microbiome–skeletal muscle axis. The purpose of this study was to determine if an intact gut microbiome was necessary for skeletal muscle adaptation to exercise. Forty-two 4-month-old female C57BL/6J mice were randomly assigned to untreated (U) or antibiotic-treated (T) non-running controls (CU or CT, respectively) or progressive weighted wheel running (PoWeR, P) untreated (PU) or antibiotic-treated (PT) groups. Antibiotic treatment resulted in disruption of the gut microbiome as indicated by a significant depletion of gut microbiome bacterial species in both CT and PT groups. The training stimulus was the same between PU and PT groups as assessed by weekly (12.35 ± 2.06 vs. 11.09 ± 1.76 km/week, respectively) and total (778.9 ± 130.5 vs. 703.8 ± 112.9 km, respectively) running activity. In response to PoWeR, PT showed less hypertrophy of soleus type 1 and 2a fibres and plantaris type 2b/x fibres compared to PU. The higher satellite cell and myonuclei abundance of PU plantaris muscle after PoWeR was not observed in PT. The fibre-type shift of PU plantaris muscle to a more oxidative type 2a fibre composition following PoWeR was blunted in PT. There was no difference in serum cytokine levels among all groups suggesting disruption of the gut microbiome did not induce systemic inflammation. The results of this study provide the first evidence that an intact gut microbiome is necessary for skeletal muscle adaptation to exercise.
Keywords: dysbiosis, exercise, gut microbiome, hypertrophy, skeletal muscle
Graphical Abstract
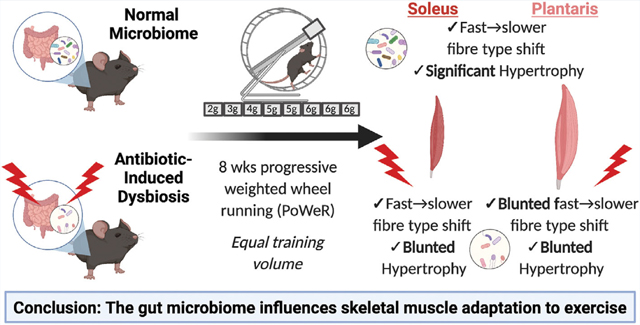
Adult female mice were continuously treated with antibiotics to induce dysbiosis of the gut microbiome and then underwent progressive weighted wheel running (PoWeR) for 8 weeks. In control, untreated mice, PoWeR induced hypertrophy and a fiber-type shift in both the soleus and plantaris muscles that was significantly diminished in mice with dysbiosis. These findings suggest a healthy gut microbiome is required for skeletal muscle to fully adapt to exercise.
Introduction
Skeletal muscle is one of the largest organs of the body, representing close to 50% of total body mass. Skeletal muscle functions as a biological motor that allows for the production of work, heat generation, regulation of blood glucose and acts as a reservoir of amino acids (DeFronzo, 1988; Gibala, 2001; Lecker et al. 2006; Rowland et al. 2015; Lieber et al. 2017). A remarkable quality of skeletal muscle is the ability to alter its physical characteristics in a specific manner in response to regular bouts of exercise. The cellular and molecular mechanisms that underlie this adaptive ability are an area of intense research given the importance of skeletal muscle to overall health.
There is exciting evidence to suggest the gut microbiome may play a role in regulating skeletal muscle mass and function. In a pioneering study, Bäckhed and co-workers reported skeletal muscle of germ free (GF) mice (completely devoid of any commensal bacteria) had significantly higher levels of phosphorylated AMPactivated protein kinase and acetyl-CoA carboxylase compared to specific pathogen free (SPF) mice, indicating GF mice rely more heavily on fatty acid oxidation as an energy source and possibly explaining why GF mice are more resistant to obesity (Backhed et al. 2007). Additionally, skeletal muscle of GF mice is atrophic compared to SPF mice and inoculating GF mice with microbiota was able to restore muscle mass (Lahiri et al. 2019). Exercise has been shown to modulate the composition of the microbiome with specific microbes associated with enhanced exercise capacity (Clarke et al. 2014; Allen et al. 2015; Grosicki et al. 2019; Scheiman et al. 2019). Exercise-induced changes in the gut microbiome were reported to have systemic benefits, indicating the microbiome of an exercise trained host is protective against a range of different pathologies (Bleau et al. 2015; Campbell et al. 2016; Allen et al. 2018; Codella et al. 2018). These findings have generated great excitement in the exercise physiology field as well as other fields of research see king to utilize the microbiome to treat diseases such as cancer, depression, obesity, Parkinson’s and autism (Backhed et al. 2007; Sampson et al. 2016; Routy et al. 2018; Sharon et al. 2019; Bastiaanssen et al. 2020). A better understanding of the role of the gut microbiome in skeletal muscle plasticity could ultimately provide new interventions to treat conditions such as cachexia and sarcopenia, the age-related loss in skeletal muscle mass and function.
Although there is emerging evidence to the concept of a gut microbiome–skeletal muscle axis (Grosicki et al. 2018; Ni Lochlainn et al. 2018; Lustgarten, 2019; Ticinesi et al. 2019a,b), there remains a scarcity of studies investigating the potential role of the gut microbiome in skeletal muscle adaptation to exercis purpose of this study was to test the hypothesis that the gut microbiome is required for skeletal muscle adaptations to exercise. To test this hypothesis, mice were treated with or without antibiotics throughout a 9-week progressive weighted wheel running (PoWeR) protocol which has been shown to induce hypertrophy and a fibre-type shift to a more oxidative phenotype (Dungan et al. 2019). Despite the same exercise stimulus, antibiotic-treated mice had a blunted hypertrophic response and fibre-type shift compared to the untreated mice. These findings provide the first evidence showing that disruption of the gut microbiome impairs skeletal muscle adaptation to exercise and suggest the exciting possibility that microbially derived metabolites have an important role in skeletal muscle plasticity.
Methods
Ethical approval
All experiments involving C57BL/6J mice were approved by the University of Kentucky Institutional Animal Care and Use Committee (2018–3005) and all steps to minimize pain and suffering were taken throughout the study.
Animals
Adult (4 months of age), female C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Upon arrival, mice were housed four to five per cage and acclimated for 1 month to allow for the gut microbiome to become stabilized to the new housing facility (Montonye et al. 2018). Mice were then randomly assigned to one of four groups (n = 10–11/group): (1) non-running control untreated (CU), (2) PoWeR untreated (PU), (3) non-running control treated with antibiotics (CT), and (4) PoWeR treated with antibiotics (PT). All non-running control mice were housed individually in a running wheel cage as for the PoWeR mice, except the running wheel remained locked for the duration of the study. One mouse in the CT group died unexpectedly 3 days before completion of the study, and thus the CT group had n = 9. Animals were kept on a 10:14 light cycle with irradiated chow (Teklad 2918 protein rodent diet; Envigo, Indianapolis, IN) and water ad libitum. Fresh, autoclaved water with or with antibiotics was provided each week. Food and water consumption were measured weekly. Cage and bedding were changed each week with dirty bedding from the previous week pooled for each group and then added to clean bedding in a ratio of 2/3 clean bedding and 1/3 dirty bedding in an effort to reduce microbial drift during the course of the study (Ericsson et al. 2018).
Antibiotic treatment
Upon completion of the acclimation period, mice were randomly assigned to a group (CU, CT, PU or PT), then placed singly into a running wheel cage with antibiotic treatment initiated for CT and PT groups. Antibiotic treatment was started 1 week prior to PoWeR training, designated week 0, to allow for depletion of the microbiome prior to exercise (Nay et al. 2019). Antibiotics were continuously administered via the drinking water and prepared fresh twice per week. Each time new antibiotics were administered, drinking volume was collected resulting in fluid consumption measurements taken twice per week. In the untreated groups, mice received the same autoclaved water, which was changed at the same time as for the antibiotic-treated groups. The antibiotic cocktail was modified from Suarez-Zamorano et al. (2015) and Koh et al. (2018) and consisted of 100 μg/ml each of metronidazole (Sigma-Aldrich, St. Louis, MO, USA), neomycin (J&K Scientific, San Jose, CA, USA), and ampicillin (Sigma-Aldrich, St. Louis, MO, USA) and 50 μg/ml each of vancomycin (Sigma-Aldrich, St. Louis, MO, USA) and streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). This antibiotic cocktail was chosen because it was shown to not cause weight loss and lethargy (Koh et al. 2018).
Progressive weighted wheel running
Mice underwent 8 weeks of the PoWeR protocol as previously described by our group; PoWeR training was shown to induce hypertrophy and a fibre-type shift in muscles of the lower hind limbs (Dungan et al. 2019). Mice were singly housed in running wheel cages with free access to the running wheel. Individual running data (km/day) and total running volume (km) was recorded ClockLab software (Actimetrics, Wilmette, IL, USA). During the first week of training, mice ran with an unloaded wheel to allow for acclimation to the running wheel. After the acclimation period (week 1), weight was loaded onto the wheel for a training intensity that consisted of 2 g during week 2, 3 g during week 3, 4 g during week 4, 5 g during week 5 and 6 and 6 g during weeks 7–9. The weight consisted of 1 g magnets (product no. B661, K&J Magnetics, Pipersville, PA, USA), which were attached on one side of the running wheel.
Immunohistochemistry
Immunohistochemistry (IHC) analysis was performed as previously described by us (Fry et al. 2017; Dungan et al. 2019) on the soleus and plantaris muscles to determine skeletal muscle fibre cross-sectional area (CSA), fibre-type composition and myonuclei abundance. Excised soleus and plantaris muscles were immediately weighed, with one limb snap-frozen in O.C.T. compound using liquid nitrogen-cooled isopentane and muscles from the other limb snap-frozen in liquid nitrogen and then stored in −80°C for biochemical and molecular analyses. Muscle samples were mounted and cut into 7 μm sections. Muscle sections were air-dried overnight at room temperature. After drying, the sections were incubated in a cocktail of iso-type specific anti-mouse primary antibodies against myosin heavy chain (MyHC) 1, MyHC 2a (Developmental Studies Hybridoma Bank, Iowa City, IA, USA) in addition to an antibody against dystrophin (cat. no. ab15277, Abcam, Cambridge, MA, USA). Type 2b and type 2x were not stained and counted based on the lack of staining. For muscle fibre-type composition, we combined type 2b and type 2x fibres based on their similar phenotype (Murach et al. 2020b). Primary antibodies were diluted 1:100 in phosphate-buffered saline (PBS) with slides incubated for 2 h at room temperature, then washed three times in PBS and incubated for 90 min at room temperature with the appropriate secondary antibodies at a dilution of 1:250. During the final 10 min of the secondary incubation, 4',6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA, USA, 1:10,000) was added to counterstain nuclei. Sections were then washed three times in PBS and mounted using PBS–glycerol solution at a 1:1 ratio. IHC for Pax7+ was performed according to the protocol established by our laboratory (McCarthy et al. 2011; Fry et al. 2017).
Image capture and analysis
IHC sections were captured at ×20 magnification using an upright fluorescence microscope (AxioImager M1, Carl Zeiss Microscopy, Oberkochen, Germany). Quantification of skeletal muscle CSA, fibre type-specific CSA, fibre-type distribution and myonuclei abundance was quantified using MyoVision automated analysis software (Wen et al. 2018).
Faecal DNA extraction
Faecal samples were collected upon transferring co-housed mice to individual wheel cages, prior to antibiotic administration and 48 h prior to euthanasia at week 10. Faecal samples were collected by placing a single mouse in a clean, sterile plastic cage and allowing the mouse to defecate at will. Fresh faeces were placed in a sterile Eppendorf tube and immediately placed in dry ice. Samples were then stored at −80°C until further analysis. DNA was isolated using the PureLink™ Microbiome DNA Purification kit (cat. no. A29790, Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, samples were weighed and then subjected to mechanical (bead beating), chemical and heat lysis, followed by washing and then final elution in 25 μl of DNase-free water.
Metagenomic sequencing
A subset of samples (n = 6 for CU, PU, CT and PT (pre) and n = 6 for CU, PT, n = 5 for PU and CT (post) were sent to CosmosID Inc. (Rockville, MD, USA) for metagenomics sequencing as previously described (Hourigan et al. 2018). Faecal DNA concentration was quantified via Qubit (Thermo Fisher Scientific) and DNA libraries were prepared using the IonXpress Plus Fragment Library Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Library quantity was assessed with Qubit and sequenced on a Ion S5 XL sequencer (Thermo Fisher Scientific). Single read sequences, without adapters generated k-mers between 150 and 300 base pairs. The short sequence reads were first inputed into the reference genome curated by ComosID, which generated variable length k-mer fingerprints that were associated with distinct phylogeny of microbes. Next, the short reads were aligned against a known set of variable length k-mers in order to yield a precise taxonomic classification and relative abundance estimates for microbial next generation sequencing datasets. False positives were excluded by using a filtering threshold that was determined by analysing large numbers of diverse metagenomes. Sequences were referenced with CosmosID’s GenBook® database and analysed using the CosmosID cloud app (CosmosID Metagenomics Cloud, app.cosmosid.com). The curated database allows for high resolution of millions of short reads corresponding to discrete microorganisms which are void of contamination from the host. The Genbook® contains over 150,000 bacteria, viruses, fungi and protist genomes, including both coding and non-coding sequences (Newman et al. 2021).
Cytokine assay
Serum cytokines (tumour necrosis factor α (TNF-α), interferon γ (INF-γ), interleukin (IL)-6, IL-10 and IL-17a) were measured using a U-PLEX customized multiplex immunoassay (Meso Scale Diagnostics, Rockville, MD, USA) according to the manufacturer’s directions. We have chosen to measure the serum concentration of the aforementioned cytokines because they are accepted markers of systemic inflam (Cani et al. 2007, 2008; Schirmer et al. 2016; Douzandeh-Mobarrez & Kariminik, 2019). Following collection, blood was centrifuged at 1500 g for 15 min with the resulting serum centrifuged at 3000 g for 15 min. The cleared serum was stored at -80°C until analysis. To measure serum cytokine levels, the stored serum was thawed in duplicate, 25 μl aliquots added per well to MSD plate coated with capture antibodies for TNF-α, INF-γ, IL-6, IL-10 and IL-17a.
Euthanasia
Twenty-four hours prior to euthanasia, all running wheels were locked for the PoWeR-trained mice. In addition, mice were fasted for 5–6 h prior to euthanasia. Mice were anaesthetized by isoflurane (1–2%) inhalation, and once fully sedated, blood was collected followed by excision of the heart (to ensure death) and then skeletal muscles.
Statistical analysis
Data are presented as means ± standard deviation (SD) with significance set at P-value <0.050. A normality check was performed using a Shapiro–Wilk test. In the event the data were not normally distributed, we performed a log transformation and reassessed for normality in order to obtain a normal distribution. Weekly food and water consumption, body weight and running volume were compared using a repeated measures two-factor ANOVA (Group × Time). Skeletal muscle fibre CSA, fibre type-specific CSA, fibre-type distribution, myonuclei and Pax7+ abundance, end point body weight, caeca weight, number of reads and number of bacterial species were analysed by a two-factor ANOVA (Antibiotic treatment × PoWeR training). In the case of significance, post hoc analysis was performed using Tukey’s HSD test to correct for multiple comparisons. During the IHC analysis, one soleus in the CU group was damaged beyond repair and was not included in any of the soleus comparisons. Outliers were detected first by subtracting 1.5 × interquartile range (IQR) from the first quartile and adding 1.5 × IQR to the third quartile. We then cross checked using Grubb’s test (Prism, GraphPad Software Inc., La Jolla, CA, USA).
Results
The study design is presented in Fig. 1. During the 4-week acclimation period there were no reported complications with the mice. Upon completion of the acclimation period, mice were randomly separated into singly housed running wheel cages with a pre-faecal (pre) sample immediately collected. Once pre samples were collected, mice in the antibiotic assigned groups began antibiotic treatment via drinking water for 1 week, during which time running wheels remained locked. After the first week of antibiotic treatment, running wheels were unlocked for those mice in the PoWeR groups. Upon completion of PoWeR training, faeces were collected 48 h prior to euthanasia from all mice to obtain a post-faecal (post) sample. No complications were reported at any time during the study including when the mice were singly housed in wheel running cages, faecal collections or during the training period. One mouse in the CT group died 3 days prior to the completion of the study with the cause of death unknown.
Figure 1. Study design.
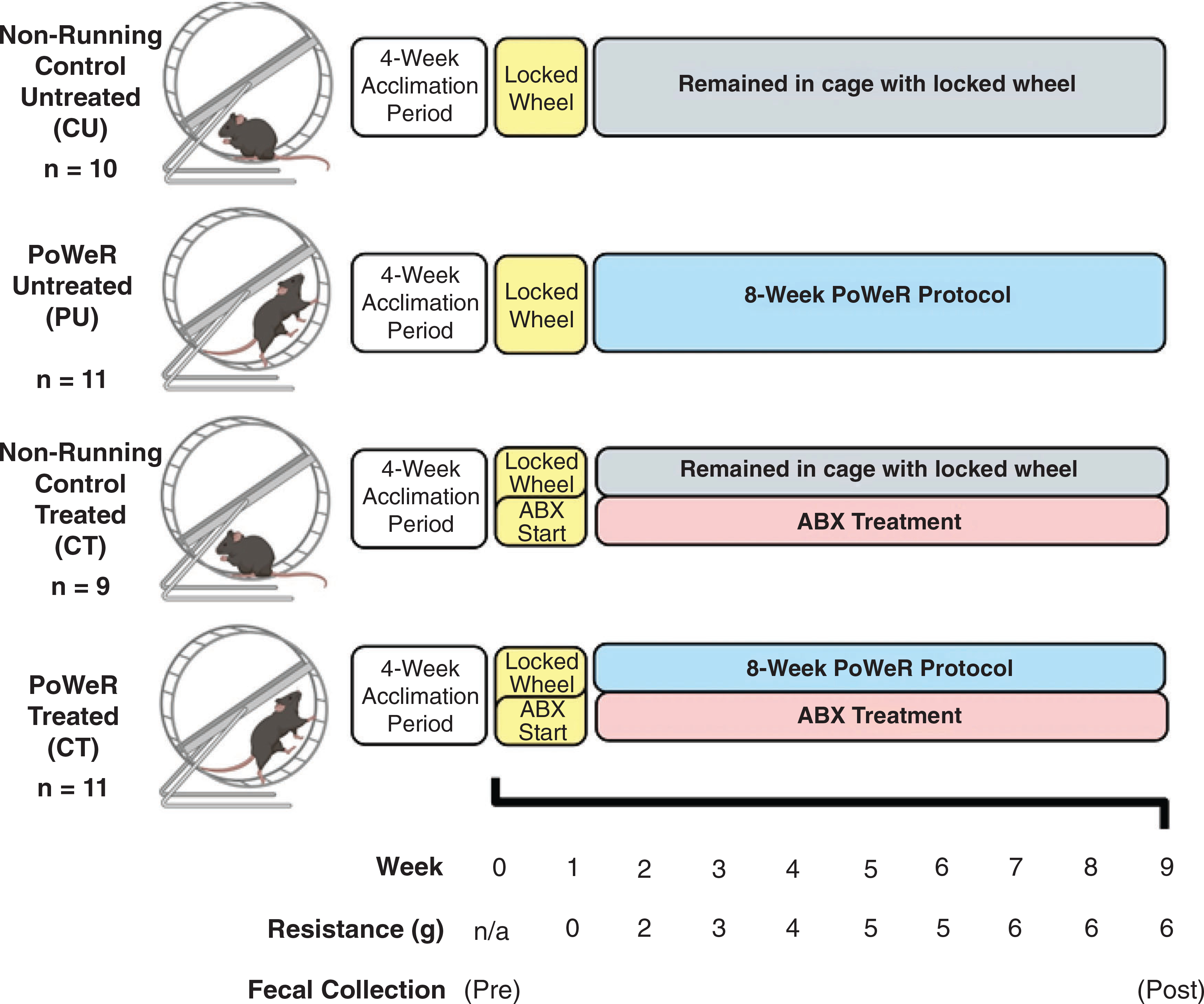
Animals were co-housed in groups of four to five for 4 weeks in order to allow for acclimation to the animal facility at the University of Kentucky. Upon the completion of the acclimation period, mice were randomly split into four different groups and singly housed in running wheel cages. Immediately after randomization into study groups, the first faecal samples were collected (pre), prior to antibiotic administration. During the first week of being singly housed, all wheels were locked, and the antibiotic treatment began. One week after, the wheels were unlocked for those mice in the PoWeR groups, initiating the acclimation week. After the first week of acclimation with an unloaded wheel, 2 g was placed on one side of the running wheel to add resistance Each week thereafter, an additional 1 g was added to the wheel until the load reached a total of 6 g for the final 3 weeks of training. A final faecal sample was collected roughly 48 h prior to euthanasia. After completion of 8 weeks of PoWeR training, mice were euthanized, and tissues were collected for analysis. Image created with BioRender.
Dysbiosis of the gut microbiome by antibiotic treatment
As shown in Fig. 2A, there was no difference in the number of bacterial species detected in the gut microbiome across all groups prior to antibiotic treatment. Antibiotic treatment significantly (main effect of antibiotic treatment, P < 0.0001) reduced the number of bacterial species detected within the gut microbiome (Fig. 2B); furthermore, PoWeR training did not change the number of detected bacterial species compared to their respective non-running control counterparts (Fig. 2B). Importantly, the loss of bacterial species with antibiotic trea was not caused by a change in the depth of reads as there was no difference in the number of reads pre- and post-treatment between all groups (Fig. 2C and D). For the reads at the post time point, one mouse in the CU group was determined to be an outlier (Fig. 2D). This mouse also had the highest number of species at the post time point (Fig. 2B), which could be contributing to the higher number of reads. Antibiotic treatment caused a significantly higher caecum weight (main effect of PoWeR training, P = 0.0087; main effect of antibiotic treatment, P < 0.0001; interaction, P = 0.0352) (Fig. 2E and F). An overview of the microbiome at the genus taxonomic level revealed the antibiotic treatment was effective in reducing the composition of the microbiota (Fig. 3). Two of the five CT samples had undetectable levels of bacteria and could not be included in the post lanes of the heat map (Fig. 3). Together, these findings illustrate that antibiotic treatment effectively induced dysbiosis of the gut microbiome which was not affected by PoWeR training.
Figure 2. Gut microbial dysbiosis with antibiotics.
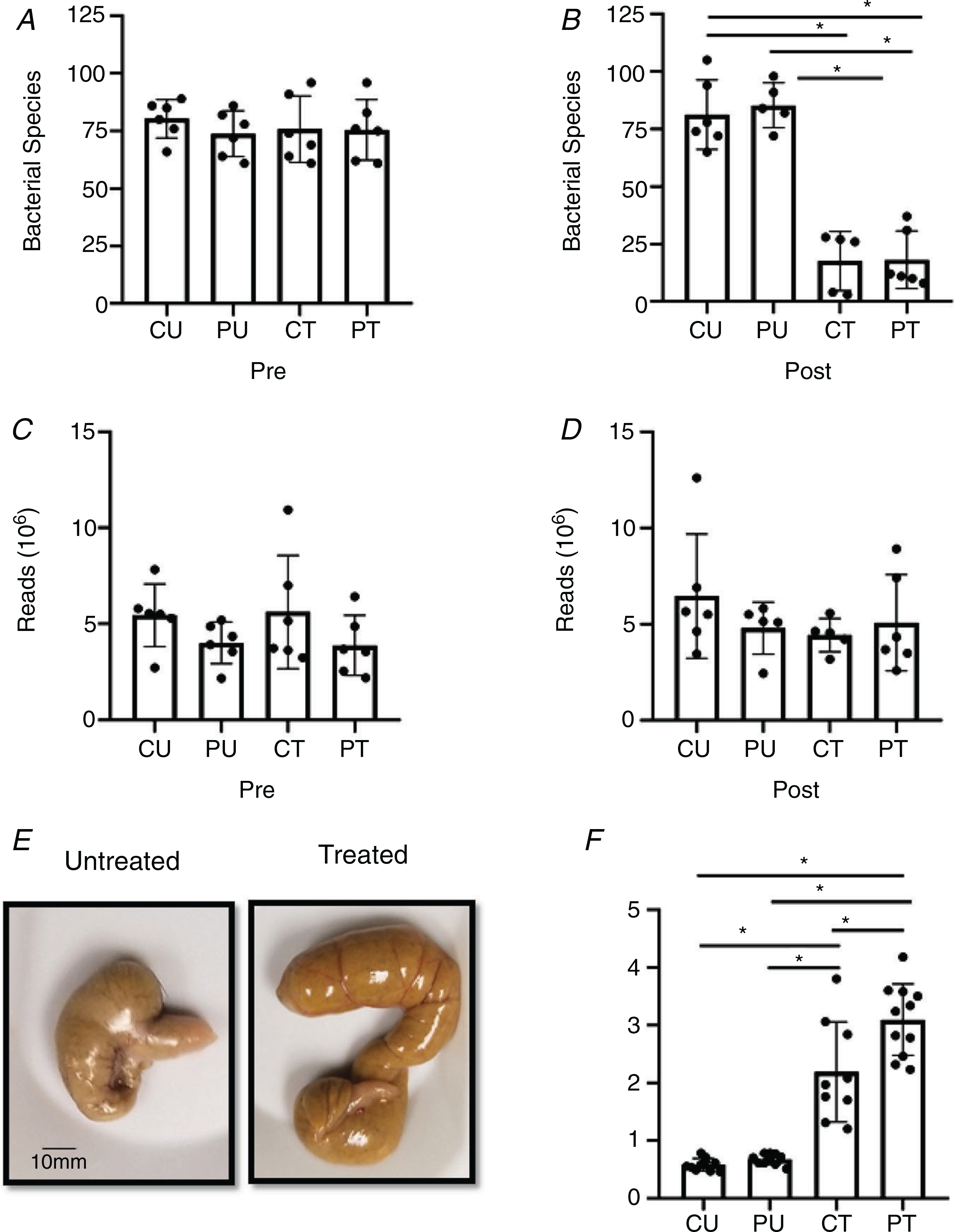
Antibiotic induced dysbiosis of the gut microbiome. A and B, number of individual bacterial species before (pre) (A) and after (post) (B) antibiotic treatment and PoWeR training n = 5–6 per group. C and D, number of reads during sequencing corresponding to the pre (C) and post (D) time points; n = 5–6 per group. E, representative image of a caecum harvested from an untreated and treated mouse. F, differences in caeca weight between the groups; n = 9–11 per group. Bars are mean values; circles represent individual values. Errors bars show standard deviation. *P < 0.05.
Figure 3. PoWeR- and antibiotic-induced changes to the microbiome composition.
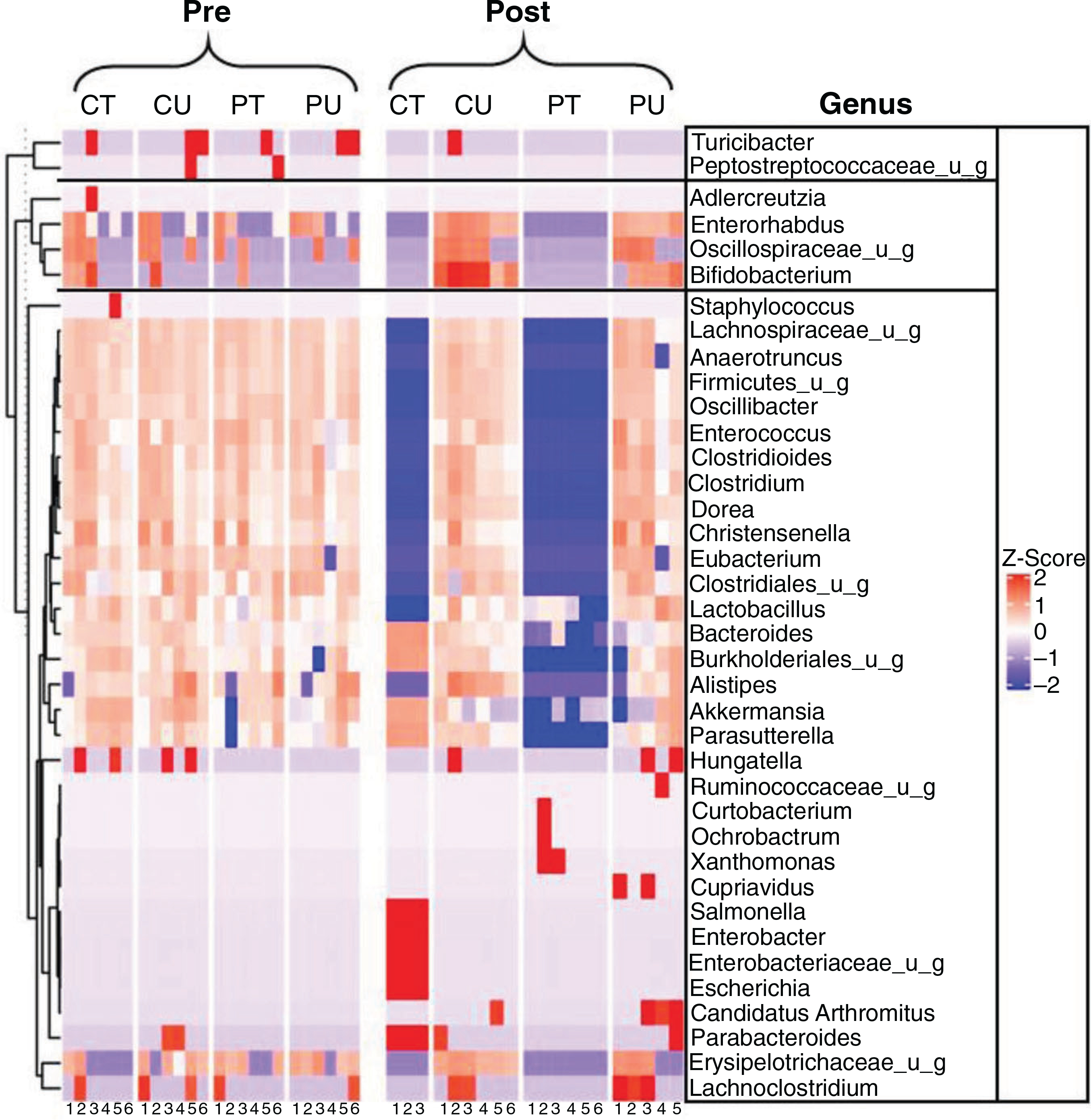
Heat map indicating the microbial composition at the genus levels between groups at the pre and post time points. n = 3–6 per group.
PoWeR increased food and water consumption
Time course analysis revealed higher food consumption, starting at weeks 1 and 2, and water consumption, starting at weeks 3 and 5, and remained significantly higher in the PoWeR groups compared to their respective non-running control counterparts for the duration of the study (Fig. 4A and B).
Figure 4. PoWeR training, not dysbiosis of the gut microbiome, leads to increased food and water consumption but not body weight changes.
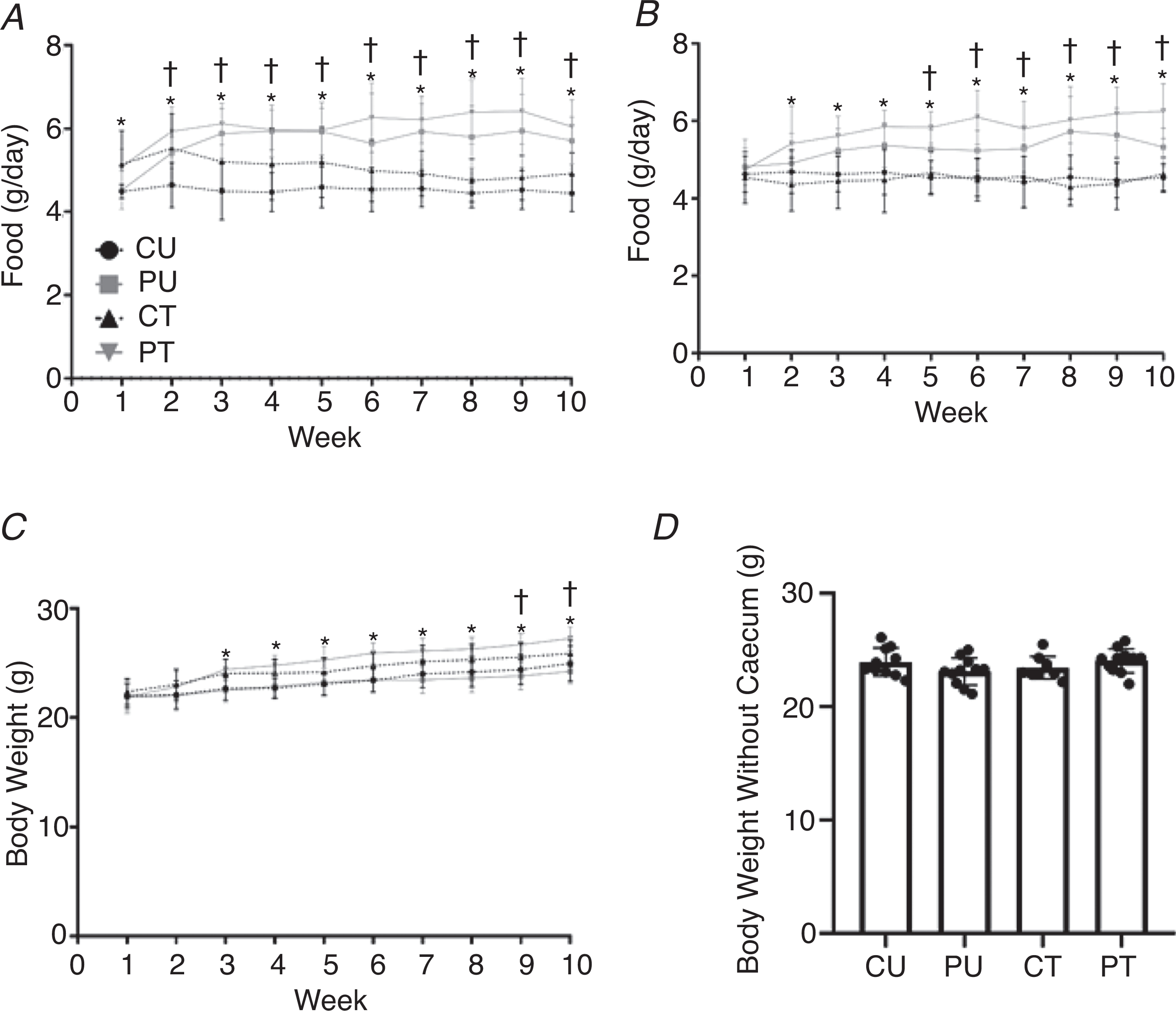
Food consumption, water intake and body weight changes during PoWeR training. Weekly food consumption during training (A), weekly water intake during study (B), weekly body weight during study (C), and body weight at the point of sacrifice with the caecum removed (D). Bars are means and circles represent individual values. Points in A–C represent group averages for that week, errors bars show standard deviation. n = 9–11 per group. * Significantly different (P < 0.05) PT vs. CT and †significantly (P < 0.05) different PU vs. CU for food and water intake. *Significantly (P < 0.01) different PT vs. CU and PU and †significantly (P < 0.05) different CT vs. PU for body weights.
Body weight
There was a significant main effect of time (P < 0.0001) and group (P = 0.0004) with a significant interaction (P < 0.0001) for body weight throughout the study. The body weight of the PT group was significantly higher than those of the PU and CU groups from weeks 3 through 10. In addition, the body weight of the CT group was significantly higher than that of the PU group for weeks 9 and 10 (Fig. 4C). The higher final body weight of the antibiotic-treated groups was caused by enlargement of the caecum. When the caecum weight was subtracted from the final body weight, there was no difference in body weight between any groups (Fig. 4D).
Running volume
As shown in Fig. 5A and B, there was no difference in the weekly or total running volume between the PoWeR groups despite a progressive increase in the amount of weight (2 to 6 g) added to the running wheel over the course of the 8-week PoWeR training period.
Figure 5. Antibiotic induced dysbiosis of the gut microbiome does not impair exercise activity.
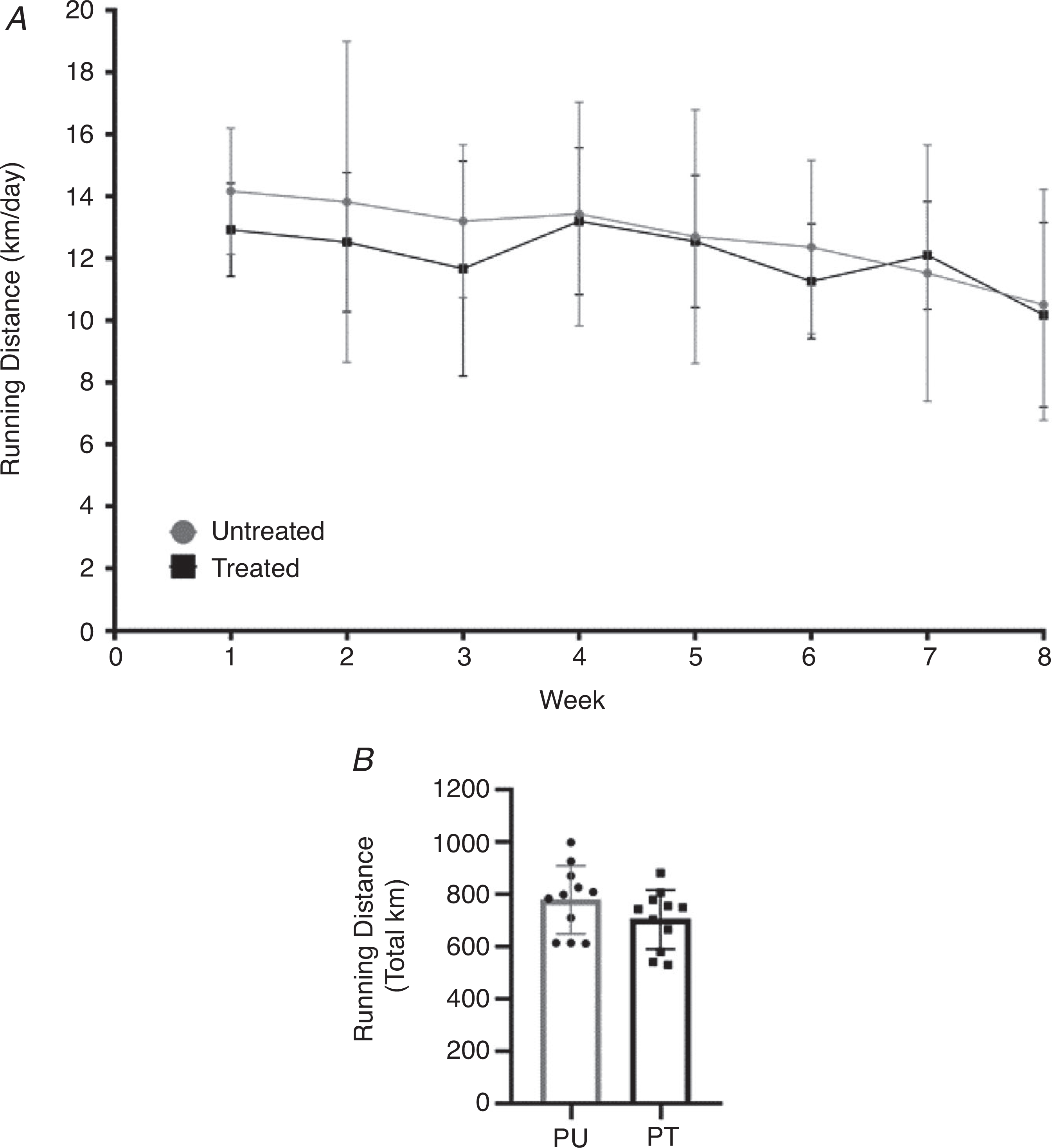
Running volume during PoWeR training. A, weekly running volume (km/day) of the PoWeR untreated and treated groups. B, mean total distance run during PoWeR training between PoWeR untreated and treated groups. n = 11 per group. Bars represent means, circles represent individual mice, filled circles in A represent group averages for the corresponding week and error bars show standard deviation.
Soleus
Muscle wet weight
In response to PoWeR training, there was a significant main effect of PoWeR training (P = 0.0002) and interaction (P = 0.0005) with a trend (P = 0.0540) for a significant effect of antibiotic treatment for normalized (to body weight) soleus wet weight. Normalized soleus wet weight was significantly larger in the PU group compared to the CU (P < 0.0001), CT (P < 0.0008) and PT (P < 0.0007) groups. The PT showed no increase in normalized soleus wet weight. These results were not affected by caecum weight as normalizing soleus muscle wet weight to body weight without the caecum weight included still showed the normalized soleus wet weight was significantly higher in the PU group compared to the CU (P < 0.0001), CT (P = 0.0008) and PT (P = 0.0007) groups (Fig. 6B).
Figure 6. Antibiotic-induced dysbiosis of the gut microbiome results in a blunted hypertrophic response in the soleus.
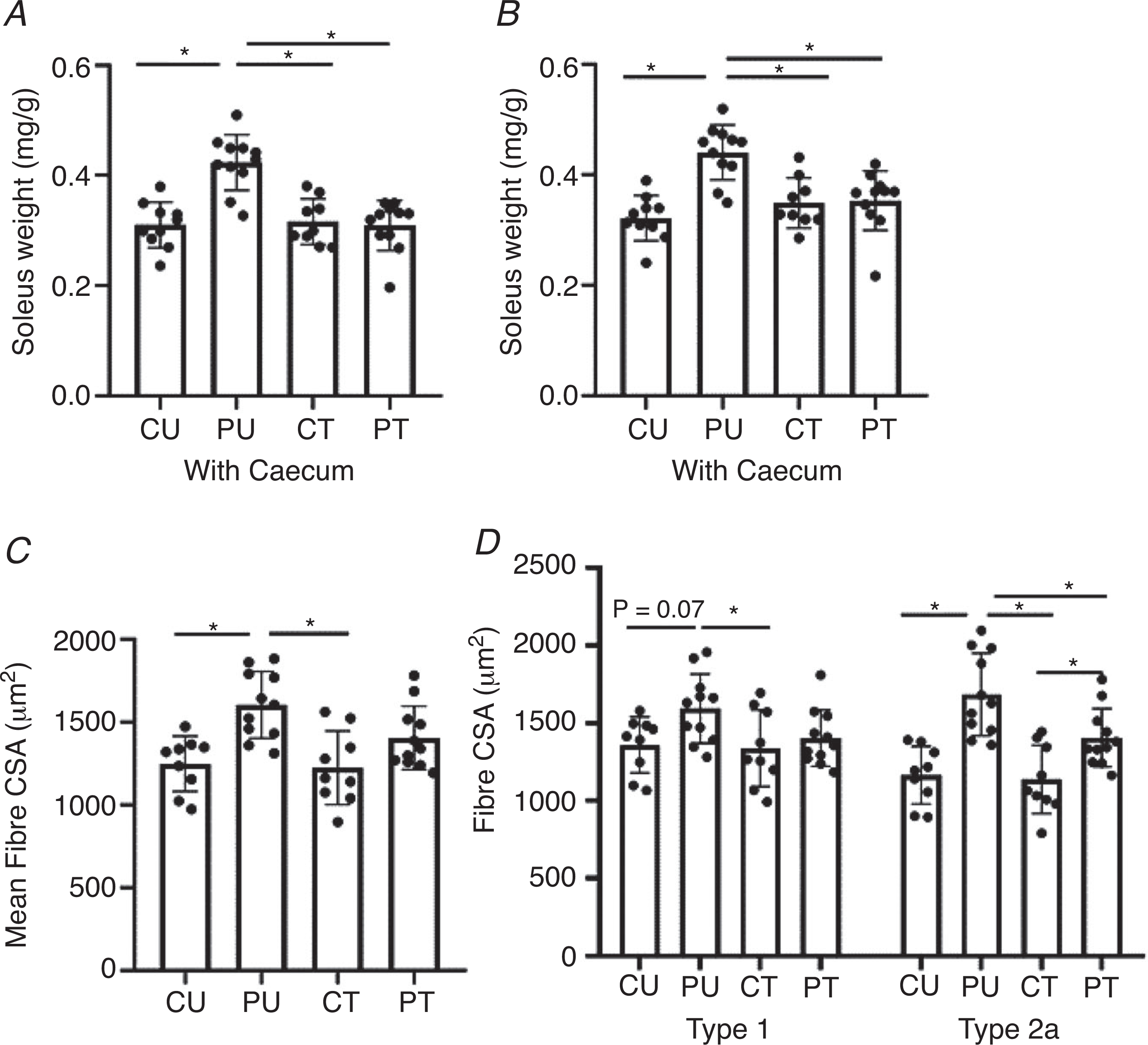
Analysis of the soleus muscle after PoWeR training with or without dysbiosis. A, normalized soleus wet weight to body weight including the caecum. B, normalized soleus wet weight to body weight excluding the caecum. C, soleus mean cross-sectional area (7 μm sections). D, type 1 and type 2a skeletal muscle fibre cross-sectional area (7 μm sections). n = 8–11 per group. Bars represent means, circles represent individual mice and error bars show standard deviation. *P < 0.05.
Fibre cross-sectional area.
In agreement with normalized soleus muscle weight, there was a significant main effect of PoWeR training (P = 0.0001) on soleus mean fibre CSA (Fig. 6C). Post hoc analysis revealed the mean fibre CSA of the PU soleus was 29% larger than that of the CU group (P = 0.0012) and 31% larger compared to the CT group (P = 0.0007) while there was no difference in mean fibre CSA between the CU, CT and PT groups. When comparing type 1 fibre CSA, there was a significant (P = 0.0302) main effect of PoWeR training on type 1 fibre CSA. Type 1 fibre CSA was trending towards significance (P = 0.0778) between the PU and CU groups and was significantly larger (P = 0.0464) between the PU and CT groups (Fig. 6D). Fibre type-specific CSA analysis showed the larger mean fibre CSA of the PU soleus was primarily a result of larger type 2a fibre CSA (main effect of antibiotic treatment, P = 0.0330; main effect of PoWeR training, P < 0.0001; interaction, P = 0.0779) (Fig. 6D). The PU group had significantly larger type 2a fibre CSA in comparison to the fibre CSA of the CU (P < 0.0001), CT (P < 0.0001) and PT (P = 0.0238) groups. The PT group had significantly larger type 2a fibre CSA compared to the CT group (P = 0.0463).
Myonuclei abundance.
There was a significant (P = 0.0330) main effect of PoWeR training on myonuclei abundance in the soleus muscle; however, post hoc analysis revealed no significant difference between groups in response to PoWeR training.
Fibre-type composition.
One mouse in the CT group was classified as an outlier (see Methods) for fibre-type distribution and subsequently removed from all fibre-type distribution analyses. In response to PoWeR training there was a main effect (P < 0.0001) for type 1 fibre abundance. PU had 17% more type 1 fibres compared to CU (P = 0.0058) and 20% more compared to CT (P = 0.0026) while PT had 21% more type 1 fibres compared to CU (P = 0.0008) and 23% more compared to CT (P = 0.0004) (Fig. 7B). Comparing the abundance of type 2a fibres, there was a significant main effect of PoWeR training (P < 0.0001). PU had 20% fewer type 2a fibres compared to CU (P = 0.0220) and 15% fewer compared to CT (P = 0.0116). PT had 25% fewer type 2a fibres compared to CU (P = 0.0074) and 26% fewer compared to CT (P = 0.0039) (Fig. 7B). In addition, there was a significant (P < 0.0001) main effect of PoWeR training on type 2b/x fibre abundance. PU had 88% fewer type 2b+x fibres compared to CU (P = 0.0003) and 87% fewer compared to CT (P = 0.0009). PT had 55% fewer type 2b+x fibres than CU (P = 0.0312) and a trend towards lower type 2b/x fibre abundance compared to CT (P = 0.0613). There was no difference in the abundance of fibres that co-expressed myosin heavy chain isoforms type 1+2a (Fig. 7B).
Figure 7. Antibiotic-induced dysbiosis of the gut microbiome does not impair skeletal muscle fibre-type shift in response to PoWeR training in the soleus.
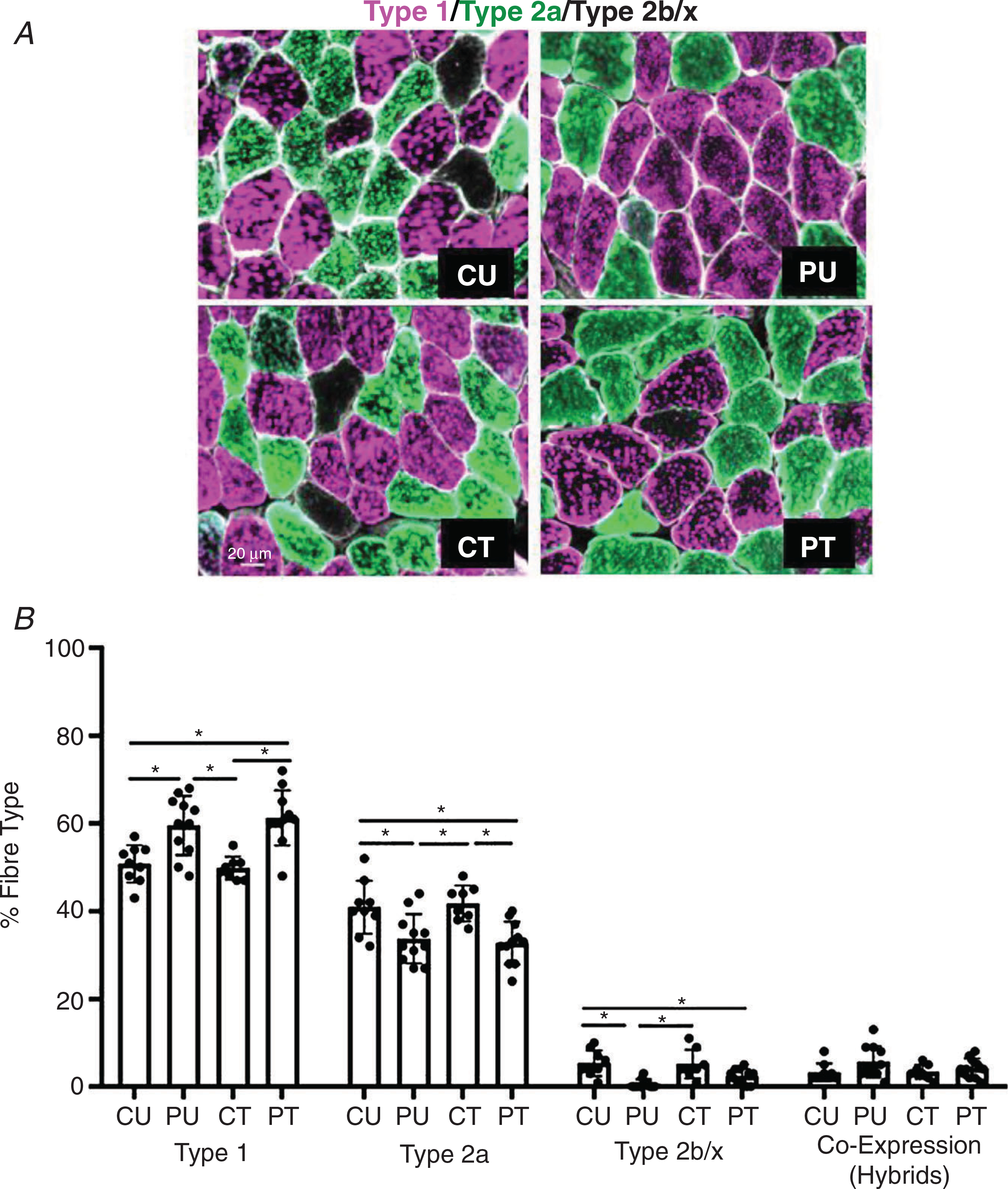
Analysis of the fibre-type distribution in the soleus muscle after PoWeR training. A, representative image of soleus cross section. B, fibre-type distribution for type 1, type 2a, type 2b/x and co-expression (hybrid) skeletal muscle fibres. n = 7–11 per group. Bars represent means, circles represent individual mice and error bars show standard deviation. *P < 0.05.
Plantaris
Muscle wet weight.
Analysis of normalized (to body weight) plantaris muscle wet weight with the caecum included showed a significant (P = 0.0001) main effect of antibiotic treatment. Post hoc analysis revealed that the PU group was significantly heavier compare the CT (P = 0.0108) and PT (P = 0.0014) groups. Additionally, the CU group was significantly (P = 0.0467) heavier compared to the PT group. In contrast to the soleus muscle, these differences did not persist when plantaris muscle wet weight was normalized to weight excluding the caecum (Fig. 8A and B).
Figure 8. Antibiotic-induced dysbiosis of the gut microbiome results in blunted hypertrophy, myonuclei accretion and altered satellite cell abundance in the plantaris muscle.
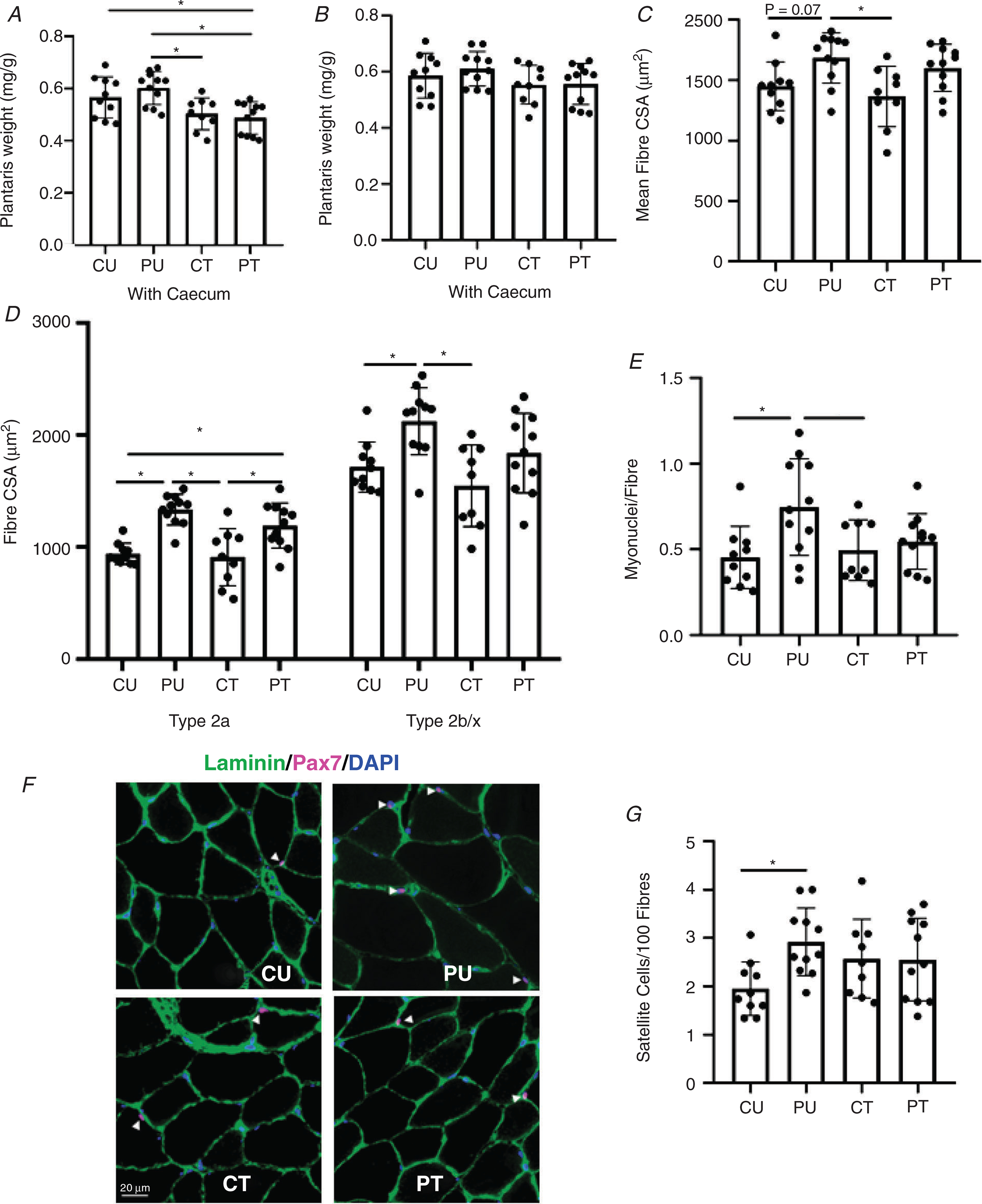
Analysis of the plantaris muscle after PoWeR training. A, normalized plantaris wet weight to body weight including the caecum. B, normalized plantaris wet weight to body weight excluding the caecum. C, plantaris mean fibre cross-sectional area (7 μm sections). D, type 2a and type 2b/x skeletal muscle fibre cross-sectional area (7 μm sections). E, number of myonuclei per fibre. F, representative image of Pax 7+ myonuclei. G, satellite cells per 100 muscle fibres. n = 9–11 per group. Bars represent means, circles represent individual mice and error bars show standard deviation. *P < 0.05.
Fibre cross-sectional area.
In response to PoWeR training, there was a significant (P = 0.0012) main effect of training on mean fibre CSA (Fig. 8C). PU mean fibre CSA was 23% larger compared to CT (P = 0.0109). There was a trend for significantly larger mean fibre CSA between PU and CU (P = 0.0767) and between PT and CT groups (P = 0.0831) (Fig. 8C). In regards to fibre type-specific CSA, there was a significant (P < 0.0001) main effect of PoWeR training on type 2a fibre CSA. PU had 42% and 47% larger type 2a fibre CSA compared to CU(P < 0.0001) and CT (P < 0.0001), respectively (Fig. 8D). PT had 27% and 31% larger type 2a fibre CSA compared to CU (P = 0.0137) and CT (P = 0.0064), respectively (Fig. 8D). There was a significant main effect of PoWeR training (P = 0.0011) and main effect of antibiotic treatment (P = 0.0282) for type 2b/x fibre CSA (Fig. 8D). Type 2b/x fibres of PU were 24% larger compared to CU (P = 0.0257) and 37% larger compared to CT (P = 0.0013). Type 2b/x fibre CSA was not different when comparing PT to CU (P = 0.8000) and CT (P = 0.1827) groups (Fig. 8D).
Myonuclei abundance.
There was a significant (P = 0.0120) main effect of PoWeR training for myonuclei abundance. Myonuclei abundance of PU was 65% higher compared to CU (P = 0.0136) and 51% compared to CT (P = 0.0494) (Fig. 8E). There was no difference in body myonuclei abundance when comparing PT and CT groups (P = 0.9443).
Satellite cell abundance.
Satellite cell abundance was determined using Pax7 immunohistochemistry. There was a significant main effect of PoWeR training (P = 0.0467) and interaction (P = 0.0401) for the abundance of Pax7+ nuclei/100 fibres. Post hoc analysis revealed the PU group had 50% more Pax7+ nuclei compared to the CU group (P = 0.0236). There was no difference (P = 0.2782) in satellite cell abundance between the CU and CT groups. Finally, there was no difference (P = 0.6546) when comparing the PU and PT groups in satellite cell abundance (Fig. 8G).
Fibre-type composition.
There was a significant (P < 0.0001) main effect of PoWeR training on the percentage of type 2a fibres (Fig. 9B). PU had 89% more type 2a fibres compared to CU (P < 0.0001), 94% more compared to CT (P < 0.0001) and 24% more compared to PT (P = 0.0467) (Fig. 9B). PT had 52% more type 2a fibre compared to CU (P = 0.0031) and 57% compared to CT (P = 0.0025) (Fig. 9B). We also observed a significant main effect of PoWeR training (P < 0.0001) and interaction (P = 0.0127) for type 2b/x fibre abundance (Fig. 9C). PU had 66% fewer type 2b/x fibres compared to CU (P < 0.0001), 60% fewer than CT (P < 0.0001) and 24% fewer than PT (P = 0.0186) (Fig. 9B). In addition, PT had 34% fewer type 2b/x fibres compared to CU (P = 0.0004) and 29% fewer compared to CT (P = 0.0040) (Fig. 9C). As shown in Fig. 9B, there was no difference in the abundance of type 1 and hybrid fibres that co-expressed myosin heavy chain isoforms (type 2a+type2b/x) across all groups.
Figure 9. Antibiotic-induced dysbiosis of the gut microbiome results in a blunted fibre-type shift in the plantaris.

Analysis of the fibre-type distribution in the plantaris muscle after PoWeR training. A, representative image of plantaris cross section. B, fibre-type distribution for type 1, type 2a, type 2b/x and co-expression (hybrid) skeletal muscle fibres. n = 9–11 per group. Bars represent means, circles represent individual mice and error bars show standard deviation. *P < 0.05.
Serum cytokine levels.
IL-6 concentrations of one mouse from the PU group were found to be an outlier. Additionally, IL-17a from another mouse in the PU group was also determined to be an outlier. We moved these values from the specific cytokine analysis. There was no change in the serum concentration of IL-10, IFN-γ, TNF-α or IL-17a in response to antibiotic treatment and/or PoWeR training (Fig. 10). When looking at IL-6, there was a significant (P = 0.0236) main effect of PoWeR training. Post hoc analysis revealed a trend for IL-6 levels to be elevated when comparing PU to CU (P = 0.0590).
Figure 10. Ten weeks of antibiotic administration and PoWeR training did not augment inflammation.
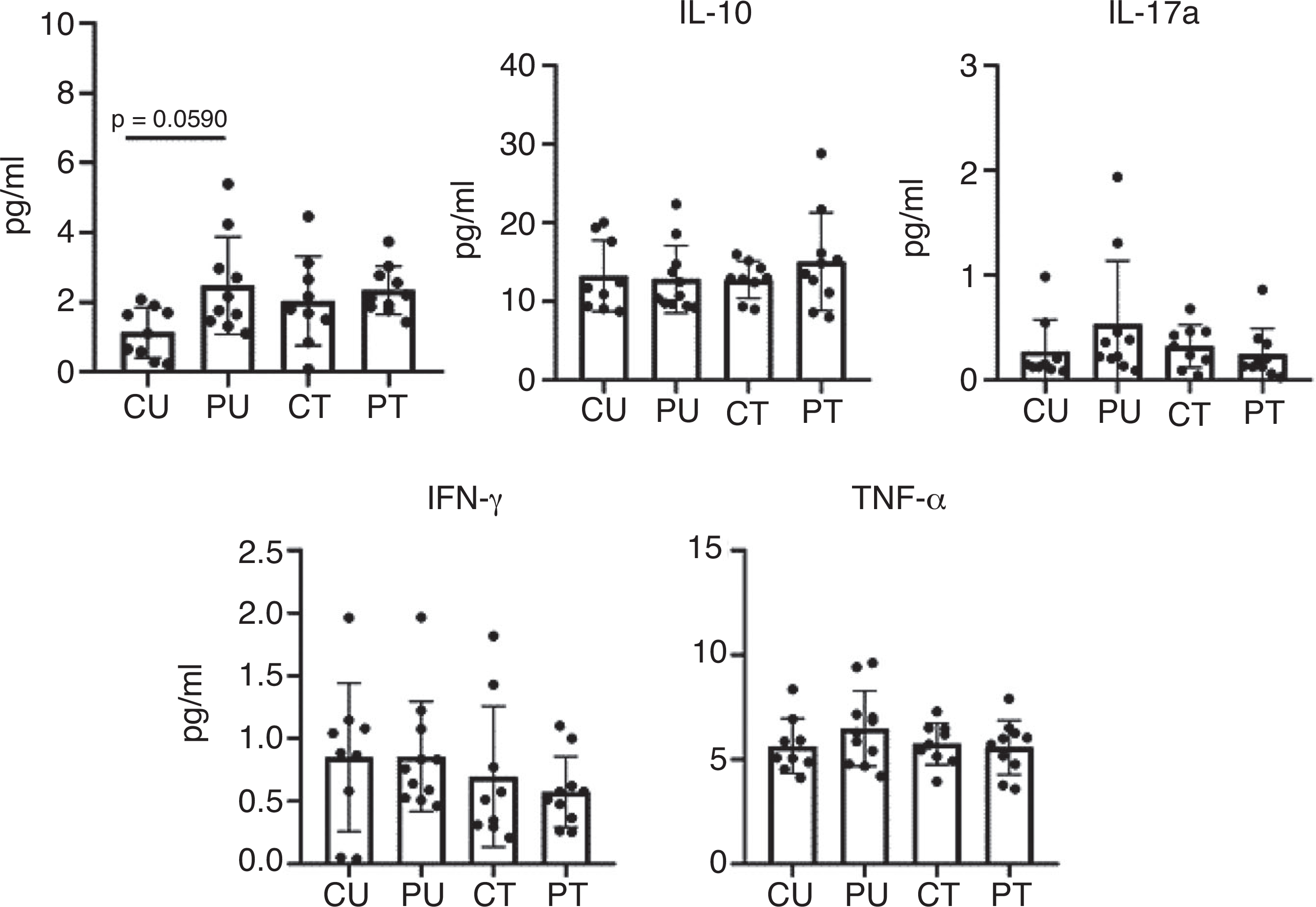
Serum concentrations of inflammatory markers taken after 9 weeks of training and antibiotic administration. n = 9–11 per group. Bars represent means, circles represent individual mice and error bars show standard deviation. IL-6, IL-10 and TNF-α were log transformed.
Discussion
The major finding of the study is that antibiotic-induced dysbiosis of the gut microbiome impaired the ability of skeletal muscle to adapt to exercise training. Despite a similar training stimulus between untreated and antibiotic-treated groups, dysbiosis resulted in blunted hypertrophy in both the soleus and plantaris muscles following PoWeR training. In the soleus muscle of mice with a disrupted gut microbiome, skeletal muscle hypertrophy of type 1 fibres was blunted and the trend for larger type 2a fibres observed in untreated mice did not occur. Similarly, in the plantaris muscle, dysbiosis of the microbiome prevented the greater type 2b/x fibre CSA following PoWeR training observed in mice with an intact gut microbiome. In untreated mice, the greater abundance of both satellite cells and myonuclei in the plantaris muscle in response to PoWeR training was absent in dysbiotic mice, which likely contributed to their blunted hypertrophic response. We did not find an increase in myonuclei accretion in the soleus muscle and therefore decided to not quantify satellite cell abundance in the soleus muscle. Lastly, the major fibre-type shifts induced by PoWeR training was unaffected by antibiotic-induced dysbiosis of the gut microbiome in the soleus muscle. However, type 2a fibre abundance nearly doubled in the plantaris muscle, which was significantly less in mice with a suppressed microbiome. These findings agree with previous studies showing the gut microbiome influences skeletal muscle mass and fibre-type composition and provide the first detailed evidence that an intact gut microbiome is necessary for skeletal muscle to fully adapt to exercise training (Yan et al. 2016; Lahiri et al. 2019).
An important finding from our study was that antibiotic-induced dysbiosis of the microbiome did not affect running activity which was critical to ensure a similar training stimulus between the two PoWeR groups. In fact, it might be argued that the training stimulus was greater in the antibiotic-treated mice given their heavier body weight caused by an enlarged caecum. In contrast, previous studies found that both antibiotic-treated mice and GF mice have reduced exercise performance, suggesting a role for the gut microbiome in exercise activity (Hsu et al. 2015; Nay et al. 2019; Okamoto et al. 2019). The discrepancy in exercise activity between these studies and our own could be explained by the lower dose of antibiotics used in the current study or that the mice in this study were given free access to a running wheel while the other studies used time to exhaustion with untrained mice. The antibiotic treatment itself may have negatively impacted exercise time to exhaustion as has been reported in athletes (Fayock et al. 2014). Alternatively, it could be plausible that an intact gut microbiome is needed for maximal exercise capacity as compared to sub-maximal wheel running exercise activity.
The use of antibiotics in lieu of GF mice is a common method to assess the role of the gut microbi health and disease (Ericsson & Franklin, 2015). GF mice are costly, labour intensive and, as shown previously, display muscle atrophy associated with higher expression of atrogenes (Lahiri et al. 2019), which creates an issue when trying to investigate the role of the gut microbiome in skeletal muscle adaptation to exercise. We decided to use antibiotics to suppress the microbiome in order to allow us to use mice in which skeletal muscles, as well as other systems like the immune system, had under-gone normal development and maturation with an intact gut microbiome (Kennedy et al. 2018; Bayer et al. 2019). The lack of an established antibiotic dosing regimen that effectively depletes the gut microbiome with minimal side effects, however, leaves open the possibility that the antibiotics themselves may be directly interfering with cellular and/or molecular mechanisms involved in skeletal muscle adaptation to exercise.
Dysbiosis of the gut microbiome by antibiotics is known to alter immune cell activation, which causes a thinning of the mucosal layer with a concomitant increase in intestinal epithelial cell permeability (Willing et al. 2011; Tulstrup et al. 2015; Song et al. 2020). The increase in intestinal permeability with dysbiosis is known to induce low-level systemic inflammation, which might induce a state of anabolic resistance leading to the impaired hypertrophic growth observed in antibiotic-treated PoWeR-trained mice. While this possible scenario awaits further investigation, we found no difference in the serum levels of inflammatory cytokines across all groups indicating the absence of low-level systemic inflammation induced by antibiotic treatment. However, these measurements were taken upon completion of PoWeR training allowing for the possibility that systemic inflammation was different at some earlier time point of the study. This concern is offset by the finding that non-running control muscle weights and running activity were not different between antibiotic-treated mice and mice with an intact gut microbiome, thus indicating antibiotic treatment had minimal effect on skeletal muscle maintenance and function.
What evidence is there to suggest antibiotics might interfere with skeletal muscle adaptation to exercise? A study reported high doses of the antibiotic metronidazole can cause skeletal muscle atrophy; however, this study used a dose 10-fold greater than the dose administered in the current study (Manickam et al. 2018). Other studies have shown that high doses of aminoglycosides such as gentamycin, neomycin and streptomycin, can affect calcium signalling, but these studies used a dose 100-fold greater than the dose used in the current study. Hayo and colleagues utilized eccentric exercise to induce muscle damage in rats that were administered streptomycin, which can reduce intracellular calcium concentration by blocking stretch-activated channels. Although these authors found that streptomycin reduced muscle membrane permeability, it did not affect the amount of myofibre swelling using a dose 80-fold greater than that used in the current study (Hayao et al. 2018). Most recently, Qiu and colleagues reported antibiotic-induced dysbiosis caused muscle atrophy which was shown to be mediated by a suppression of bile acid signalling to fibroblast growth factor 15; however, as with the aforementioned studies, these authors used antibiotic doses 100 time greater than those used in the current study (Qiu et al. 2021). Finally, intraperitoneal injection of the antibiotic imipenem for five consecutive days did not affect muscle specific force (Owen et al. 2019). Collectively, these studies demonstrate that antibiotic doses 10–100 times greater than the dose we used can negatively affect muscle size but not muscle function. Given that we observed no difference in muscle phenotype (weight, fibre size and composition) between non-running control groups as well as wheel running activity, we think the relatively low dose of antibiotics administered was able to effectively induce dysbiosis while minimizing any side effects that might have interfered with skeletal muscle adaptation to exercise.
The blunted hypertrophic growth response observed in both slow- and fast-twitch myofibres could be due to a common gut-derived microbial metabolite(s) which is necessary for maximal muscle growth. In support of such a mechanism, Chen and colleagues discovered that Bacteroides thetaiotaomicron (B. theta C34) bacteria produce L-phenylalanine, an agonist of G-coupled protein receptor 56, which was previously shown to be sufficient for skeletal muscle hypertrophic growth as well as being required for mechanical overload-induced hypertrophy (White et al. 2014; Chen et al. 2019). Similarly, in a series of studies led by Sato and Sasaki, activation of the G-coupled protein bile acid receptor TRG5 was shown to promote skeletal muscle hyper and improve glucose metabolism (Thomas et al. 2009; Sasaki et al. 2018). In particular, these authors reported administration of the gut microbially derived bile acid metabolite taurolithocholic acid induced expression of pro-hypertrophy genes, Nr4a1 and Pgc-1α4, in transgenic mice that overexpressed TGR5 (Sasaki et al. 2018). Future studies will need to mechanistically determine if gut microbially derived metabolites play a role in skeletal muscle adaptation to exercise.
Antibiotic dysbiosis of the gut microbiome did not impair the exercise-induced fibre-type shifts in the soleus muscle but did in the plantaris muscle. In the soleus, the loss of type 2a fibres was similar bet PoWeR-trained groups whereas in the plantaris, the higher abundance of type 2a fibres was significantly less in mice with a suppressed microbiome. This finding suggests there is some microbially derived factor(s) that may be necessary to promote the transition to a more oxidative phenotype. In support of this idea, Yan and colleagues demonstrated that the microbiome from two different types of pigs (the obese Rongchang and the lean Yorkshire) could give rise to their distinct muscle fibre-type composition when transferred to GF mice (Yan et al. 2016). Based on succinate dehydrogenase staining and gene expression, GF mice were found to have a loss of oxidative capacity across different hind limb muscles that was partially restored following microbial transfer (Lahiri et al. 2019). Collectively, these findings along with the results from the current study support the idea that the gut microbiome facilitate the transition to a more oxidative phenotype in skeletal muscle.
Satellite cell and myonuclei abundance of the plantaris muscle were higher in response to PoWeR training in mice with an intact gut microbiome. The effect of dysbiosis on satellite cells was more ambiguous because some mice in both groups, sedentary and runners, showed higher abundance of satellite cells while other mice appeared to be unaffected by dysbiosis. This variability led to there being no difference in satellite cell abundance between the two PoWeR-trained groups. This variable response to dysbiosis may reflect the inherent molecular heterogeneity of satellite cells as revealed by single-cell RNA-sequencing to changes in microbially derived metabolites, though confirmation of such a mechanism awaits further study (Cho & Doles, 2017).
The current study has some important limitations that need to be acknowledged. For this initial study, we chose to use C57BL/6J female mice because they are known to be better runners than their male counterparts (Murach et al. 2020a) and we were concerned the disruption of the gut microbiome might negatively impact exercise activity as previously reported (Hsu et al. 2015; Nay et al. and 2019). Since the running activity was the same between the antibiotic-treated and untreated groups, thus having an equivalent training stimulus, we were able to draw stronger conclusions about the influence of the gut microbiome on skeletal muscle adaptation to exercise. Given the reported sex differences in host-gut–microbiome interactions, it will be important for a future study to determine if dysbiosis in males also blunts muscle adaptation to exercise training as observed in females (Rizzetto et al. 2018; Razavi et al. 2019). Beyond wheel running activity, we did not perform any muscle function analyses to determine if the observed phenotypic differences between the PoWeR-trained groups was associated with any change in functional characteristics of the muscle. Thus, future work will need to determine if the changes in fibre size and composition induced by dysbiosis have an impact on maximum and specific force and fatigability of the muscle. Finally, there is a wide variety of rodent diets used in pre-clinical research which have been reported to significantly alter the composition of the gut microbiome and the production of short-chained fatty acids (Tuck et al. 2020). Importantly, all mice in the current study consumed exactly the same high protein diet for the duration of the study to ensure adequate protein intake to support skeletal muscle growth in response to PoWeR training.
The findings of this study demonstrate that an intact gut microbiome is required for skeletal muscle to fully adapt to exercise training. Additionally, the findings from this study add to the growing body of evidence supporting a gut microbiome–skeletal muscle axis (Bindels et al. 2012; Fielding et al. 2019; Lahiri et al. 2019; Whon et al. 2021). Future studies will seek to identify the bacterial species and associated metabolites that play a critical role in facilitating skeletal muscle hypertrophy and the fibre-type shift that occur in response to exercise with the expectation they will be unique for each of these processes.
Supplementary Material
Key points.
Dysbiosis of the gut microbiome caused by continuous antibiotic treatment did not affect running activity.
Continuous treatment with antibiotics did not result in systemic inflammation as indicated by serum cytokine levels.
Gut microbiome dysbiosis was associated with blunted fibre type-specific hypertrophy in the soleus and plantaris muscles in response to progressive weighted wheel running (PoWeR).
Gut microbiome dysbiosis was associated with impaired PoWeR-induced fibre-type shift in the plantaris muscle.
Gut microbiome dysbiosis was associated with a loss of PoWeR-induced myonuclei accretion in the plantaris muscle.
Acknowledgements
The authors would like to thank Dr Jennifer Moylan, of the University of Kentucky for her assistance for the cytokine analysis. We would also like to thank Dr Kevin Murach, of the University of Arkansas for his assistance with the graphical abstract. We also thank Brian Fanelli and CosmosID® for their assistance with the metagenomic sequencing. The graphical abstract and figure one was generated using BioRender (https://biorender.com/).
Funding
This study was funded by an NIH T-32 training grant (GM118292) awarded to T.R.V. and an NIH R21 (AG071888) awarded to J.J.M.
Biography
Taylor Valentino is a PhD candidate in the Department of Physiology at the University of Kentucky. He received a BS in Kinesiology and MS in Exercise Physiology from San Francisco State University where he studied thermoregulation, gastro-intestinal distress and muscle soreness during ultramarathon events. He joined the lab of John McCarthy PhD in 2017 to investigate the role of the gut microbiome in the regulation of skeletal muscle adaptation to exercise. His long-term goal is to better understand how metabolites regulate skeletal muscle plasticity.

Footnotes
Competing interests
All authors declare no competing interests.
Supporting information
Additional supporting information can be found online in the Supporting Information section at the end of the HTML view of the article. Supporting information files available:
The peer review history is available in the Supporting Information section of this article (https://doi.org/10.1113/JP281788#support-information-section).
Data availability statement
All data supporting the results of this study are included in the manuscript. The datasets generated during the current study are available from the corresponding author on reasonable request.
References
- Allen JM, Berg Miller ME, Pence BD, Whitlock K, Nehra V, Gaskins HR, White BA, Fryer JD & Woods JA (2015). Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol 118, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Allen JM, Mailing LJ, Cohrs J, Salmonson C, Fryer JD, Nehra V, Hale VL, Kashyap P, White BA & Woods JA (2018). Exercise training-induced modification of the gut microbiota persists after microbiota colonization and attenuates the response to chemically-induced colitis in gnotobiotic mice. Gut Microbes 9, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF & Gordon JI (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 104, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaanssen TFS, Cussotto S, Claesson MJ, Clarke G, Dinan TG & Cryan JF (2020). Gutted! Unraveling the role of the microbiome in major depressive disorder. Harv Rev Psychiatry 28, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer F, Ascher S, Pontarollo G & Reinhardt C (2019). Antibiotic treatment protocols and germ-free mouse models in vascular research. Front Immunol 10, 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels LB, Beck R, Schakman O, Martin JC, De Backer F, Sohet FM, Dewulf EM, Pachikian BD, Neyrinck AM, Thissen JP, Verrax J, Calderon PB, Pot B, Grangette C, Cani PD, Scott KP & Delzenne NM (2012). Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS One 7, e37971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau C, Karelis AD, St-Pierre DH & Lamontagne L (2015). Crosstalk between intestinal microbiota, adipose tissue and skeletal muscle as an early event in systemic low-grade inflammation and the development of obesity and diabetes. Diabetes Metab Res Rev 31, 545–561. [DOI] [PubMed] [Google Scholar]
- Campbell SC, Wisniewski PJ, Noji M, McGuinness LR, Haggblom MM, Lightfoot SA, Joseph LB & Kerkhof LJ (2016). The effect of diet and exercise on intestinal integrity and microbial diversity in mice. PLoS One 11, e0150502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmee E, Cousin B, Sulpice T, Chamontin B, Ferrieres J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC & Burcelin R (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM & Burcelin R (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481. [DOI] [PubMed] [Google Scholar]
- Chen H, Nwe PK, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, Cline GW, Kruse AC, Ring AM, Crawford JM & Palm NW (2019). A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 177, 1217–1231.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DS & Doles JD (2017). Single cell transcriptome analysis of muscle satellite cells reveals widespread transcriptional heterogeneity. Gene 636, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, O’Sullivan O, Lucey AJ, Humphreys M, Hogan A, Hayes P, O’Reilly M, Jeffery IB, Wood-Martin R, Kerins DM, Quigley E, Ross RP, O’Toole PW, Molloy MG, Falvey E, Shanahan F & Cotter PD (2014). Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920. [DOI] [PubMed] [Google Scholar]
- Codella R, Luzi L & Terruzzi I (2018). Exercise has the guts: how physical activity may positively modulate gut microbiota in chronic and immune-based diseases. Dig Liver Dis 50, 331–341. [DOI] [PubMed] [Google Scholar]
- DeFronzo RA (1988). Lilly lecture 1987. The triumvirate: beta-cell, muscle, liver. A collusion responsible for NIDDM. Diabetes 37, 667–687. [DOI] [PubMed] [Google Scholar]
- Douzandeh-Mobarrez B & Kariminik A (2019). Gut microbiota and IL-17A: physiological and pathological responses. Probiotics Antimicrob Proteins 11, 1–10. [DOI] [PubMed] [Google Scholar]
- Dungan CM, Murach KA, Frick KK, Jones SR, Crow SE, Englund DA, Vechetti IJ Jr, Figueiredo VC, Levitan BM, Satin J, McCarthy JJ & Peterson CA (2019). Elevated myonuclear density during skeletal muscle hypertrophy in response to training is reversed during detraining. Am J Physiol Cell Physiol 316, C649–C654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC & Franklin CL (2015). Manipulating the gut microbiota: methods and challenges. ILAR J 56, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Gagliardi J, Bouhan D, Spollen WG, Givan SA & Franklin CL (2018). The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci Rep 8, 4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayock K, Voltz M, Sandella B, Close J, Lunser M & Okon J (2014). Antibiotic precautions in athletes. Sports Health 6, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding RA, Reeves AR, Jasuja R, Liu C, Barrett BB & Lustgarten MS (2019). Muscle strength is increased in mice that are colonized with microbiota from high-functioning older adults. Exp Gerontol 127, 110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Kirby TJ, Kosmac K, McCarthy JJ & Peterson CA (2017). Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 20, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibala MJ (2001). Regulation of skeletal muscle amino acid metabolism during exercise. Int J Sport Nutr Exerc Metab 11, 87–108. [DOI] [PubMed] [Google Scholar]
- Grosicki GJ, Durk RP & Bagley JR (2019). Rapid gut microbiome changes in a world-class ultramarathon runner. Physiol Rep 7, e14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosicki GJ, Fielding RA & Lustgarten MS (2018). Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle axis. Calcif Tissue Int 102, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayao K, Tamaki H, Nakagawa K, Tamakoshi K, Takahashi H, Yotani K, Ogita F, Yamamoto N & Onishi H (2018). Effects of streptomycin administration on increases in skeletal muscle fiber permeability and size following eccentric muscle contractions. Anat Rec 301, 1096–1102. [DOI] [PubMed] [Google Scholar]
- Hourigan SK, Subramanian P, Hasan NA, Ta A, Klein E, Chettout N, Huddleston K, Deopujari V, Levy S, Baveja R, Clemency NC, Baker RL, Niederhuber JE & Colwell RR (2018). Comparison of infant gut and skin microbiota, resistome and virulome between neonatal intensive care unit (NICU) environments. Front Microbiol 9, 1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YJ, Chiu CC, Li YP, Huang WC, Huang YT, Huang CC & Chuang HL (2015). Effect of intestinal microbiota on exercise performance in mice. J Strength Cond Res 29, 552–558. [DOI] [PubMed] [Google Scholar]
- Kennedy EA, King KY & Baldridge MT (2018). Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol 9, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, Bergh PO, Gerdes V, Hartstra A, de Brauw M, Perkins R, Nieuwdorp M, Bergstrom G & Backhed F (2018). Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175, 947–961.e17. [DOI] [PubMed] [Google Scholar]
- Lahiri S, Kim H, Garcia-Perez I, Reza MM, Martin KA, Kundu P, Cox LM, Selkrig J, Posma JM, Zhang H, Padmanabhan P, Moret C, Gulyas B, Blaser MJ, Auwerx J, Holmes E, Nicholson J, Wahli W & Pettersson S (2019). The gut microbiota influences skeletal muscle mass and function in mice. Sci Transl Med 11, eaan5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL & Mitch WE (2006). Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. JAmSocNephrol 17, 1807–1819. [DOI] [PubMed] [Google Scholar]
- Lieber RL, Roberts TJ, Blemker SS, Lee SSM & Herzog W (2017). Skeletal muscle mechanics, energetics and plasticity. J Neuroeng Rehabil 14, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustgarten MS (2019). The role of the gut microbiome on skeletal muscle mass and physical function: 2019 update. Front Physiol 10, 1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam R, Oh HYP, Tan CK, Paramalingam E & Wahli W (2018). Metronidazole causes skeletal muscle atrophy and modulates muscle chronometabolism. Int J Mol Sci 19, 2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, Van Zant G, Campbell KS, Esser KA, Dupont-Versteegden EE & Peterson CA (2011). Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138, 3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montonye DR, Ericsson AC, Busi SB, Lutz C, Wardwell K & Franklin CL (2018). Acclimation and institutionalization of the mouse microbiota following transportation. Front Microbiol 9, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach KA, McCarthy JJ, Peterson CA & Dungan CM (2020. a). Making mice mighty: recent advances in translational models of load-induced muscle hypertrophy. J Appl Physiol 129, 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach KA, Mobley CB, Zdunek CJ, Frick KK, Jones SR, McCarthy JJ, Peterson CA & Dungan CM (2020b). Muscle memory: myonuclear accretion, maintenance, morphology, and miRNA levels with training and detraining in adult mice. J Cachexia Sarcopenia Muscle 11, 1705–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nay K, Jollet M, Goustard B, Baati N, Vernus B, Pontones M, Lefeuvre-Orfila L, Bendavid C, Rue O, Mariadassou M, Bonnieu A, Ollendorff V, Lepage P, Derbre F & Koechlin-Ramonatxo C (2019). Gut bacteria are critical for optimal muscle function: a potential link with glucose homeostasis. Am J Physiol Endocrinol Metab 317, E158–E171. [DOI] [PubMed] [Google Scholar]
- Newman TM, Shively CA, Register TC, Appt SE, Yadav H, Colwell RR, Fanelli B, Dadlani M, Graubics K, Nguyen UT, Ramamoorthy S, Uberseder B, Clear KYJ, Wilson AS, Reeves KD, Chappell MC, Tooze JA & Cook KL (2021). Diet, obesity, and the gut microbiome as determinants modulating metabolic outcomes in a non-human primate model. Microbiome 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Lochlainn M, Bowyer RCE & Steves CJ (2018). Dietary protein and muscle in aging people: the potential role of the gut microbiome. Nutrients 10, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Morino K, Ugi S, Nakagawa F, Lemecha M, Ida S, Ohashi N, Sato D, Fujita Y & Maegawa H (2019). Microbiome potentiates endurance exercise through intestinal acetate production. Am J Physiol Endocrinol Metab 316, E956–E966. [DOI] [PubMed] [Google Scholar]
- Owen AM, Patel SP, Smith JD, Balasuriya BK, Mori SF, Hawk GS, Stromberg AJ, Kuriyama N, Kaneki M, Rabchevsky AG, Butterfield TA, Esser KA, Peterson CA, Starr ME & Saito H (2019). Chronic muscle weakness and mitochondrial dysfunction in the absence of sustained atrophy in a pre-clinical sepsis model. Elife 8, e49920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Yu J, Li Y, Yang F, Yu H, Xue M, Zhang F, Jiang X, Ji X & Bao Z (2021). Depletion of gut microbiota induces skeletal muscle atrophy by FXR-FGF15/19 signalling. Ann Med 53, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi AC, Potts KS, Kelly TN & Bazzano LA (2019). Sex, gut microbiome, and cardiovascular disease risk. Biol Sex Differ 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzetto L, Fava F, Tuohy KM & Selmi C (2018). Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex. J Autoimmun 92, 12–34. [DOI] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G & Zitvogel L (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Rowland LA, Bal NC & Periasamy M (2015). The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol Rev Camb Philos Soc 90, 1279–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R & Mazmanian SK (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Kuboyama A, Mita M, Murata S, Shimizu M, Inoue J, Mori K & Sato R (2018). The exercise-inducible bile acid receptor Tgr5 improves skeletal muscle function in mice. J Biol Chem 293, 10322–10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, Wibowo MC, Wurth RC, Punthambaker S, Tierney BT, Yang Z, Hattab MW, Avila-Pacheco J, Clish CB, Lessard S, Church GM & Kostic AD (2019). Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med 25, 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Ter Horst R, Jansen T, JacobsL MJ, Kurilshikov A, Fu J, Joosten LAB, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG&Xavier RJ (2016). Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167, 1125–1136.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Cruz NJ, Kang DW, Gandal MJ, Wang B, Kim YM, Zink EM, Casey CP, Taylor BC, Lane CJ, Bramer LM, Isern NG, Hoyt D, Noecker C, Sweredoski MJ, Moradian A, Borenstein E, Jansson JK, Knight R, Metz TO, Lois C, Geschwind DH, Krajmalnik-Brown R & Mazmanian SK (2019). Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 177, 1600–1618.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C & Kasper DL (2020). Microbial bile acid metabolites modulate gut ROR? + regulatory T cell homeostasis. Nature 577, 410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Zamorano N, Fabbiano S, Chevalier C, Stojanovic O, Colin DJ, Stevanovic A, Veyrat-Durebex C, Tarallo V, Rigo D, Germain S, Ilievska M, Montet X, Seimbille Y, Hapfelmeier S & Trajkovski M (2015). Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med 21, 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J & Schoonjans K (2009). TGR5-mediated bile acid sensing controls glucose homeostasis. Cell etab 10, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticinesi A, Lauretani F, Tana C, Nouvenne A, Ridolo E & Meschi T (2019a). Exercise and immune system as modulators of intestinal microbiome: implications for the gut-muscle axis hypothesis. Exerc Immunol Rev 25, 84–95. [PubMed] [Google Scholar]
- Ticinesi A, Nouvenne A, Cerundolo N, Catania P, Prati B, Tana C & Meschi T (2019b). Gut microbiota, muscle mass and function in aging: a focus on physical frailty and sarcopenia. Nutrients 11, 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck CJ, De Palma G, Takami K, Brant B, Caminero A, Reed DE, Muir JG, Gibson PR, Winterborn A, Verdu EF, Bercik P & Vanner S (2020). Nutritional profile of rodent diets impacts experimental reproducibility in microbiome pre-clinical research. Sci Rep 10, 17784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulstrup MV, Christensen EG, Carvalho V, Linninge C, Ahrne S, Hojberg O, Licht TR & Bahl MI (2015). Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS One 10, e0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Murach KA, Vechetti IJ Jr, Fry CS, Vickery C, Peterson CA, McCarthy JJ & Campbell KS (2018). MyoVision: software for automated high-content analysis of skeletal muscle immunohistochemistry. J Appl Physiol 124, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Wrann CD, Rao RR, Nair SK, Jedrychowski MP, You JS, Martinez-Redondo V, Gygi SP, Ruas JL, Hornberger TA, Wu Z, Glass DJ, Piao X & Spiegelman BM (2014). G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci USA 111, 15756–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon TW, Kim HS, Shin NR, Jung ES, Tak EJ, Sung H, Jung MJ, Jeong YS, Hyun DW, Kim PS, Jang YK, Lee CH & Bae JW (2021). Male castration increases adiposity via small intestinal microbial alterations. EMBO Rep 22, e50663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Russell SL & Finlay BB (2011). Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 9, 233–243. [DOI] [PubMed] [Google Scholar]
- Yan H, Diao H, Xiao Y, Li W, Yu B, He J, Yu J, Zheng P, Mao X, Luo Y, Zeng B, Wei H & Chen D (2016). Gut microbiota can transfer fiber characteristics and lipid metabolic profiles of skeletal muscle from pigs to germ-free mice. Sci Rep 6, 31786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results of this study are included in the manuscript. The datasets generated during the current study are available from the corresponding author on reasonable request.


