Abstract
Sphingolipids are major components of the plasma membrane of eukaryotic cells and were once thought of merely as structural components of the membrane. We have investigated effects of inhibiting sphingolipid biosynthesis, both in germinating spores and growing hyphae of Aspergillus nidulans. In germinating spores, genetic or pharmacological inactivation of inositol phosphorylceramide (IPC) synthase arrests the cell cycle in G1 and also prevents polarized growth during spore germination. However, inactivation of IPC synthase not only eliminates sphingolipid biosynthesis but also leads to a marked accumulation of ceramide, its upstream intermediate. We therefore inactivated serine palmitoyltransferase, the first enzyme in the sphingolipid biosynthesis pathway, to determine effects of inhibiting sphingolipid biosynthesis without an accumulation of ceramide. This inactivation also prevented polarized growth but did not affect nuclear division of germinating spores. To see if sphingolipid biosynthesis is required to maintain polarized growth, and not just to establish polarity, we inhibited sphingolipid biosynthesis in cells in which polarity was already established. This inhibition rapidly abolished normal cell polarity and promoted cell tip branching, which normally never occurs. Cell tip branching was closely associated with dramatic changes in the normally highly polarized actin cytoskeleton and found to be dependent on actin function. The results indicate that sphingolipids are essential for the establishment and maintenance of cell polarity via control of the actin cytoskeleton and that accumulation of ceramide is likely responsible for arresting the cell cycle in G1.
Sphingolipids are ubiquitous components of eukaryotic cell membranes and are particularly enriched in the plasma membrane. In Saccharomyces cerevisiae, sphingolipids account for 30% of total phospholipids of the plasma membrane (32). The sphingolipid biosynthesis pathway has been well characterized in many organisms, and many genes in this pathway have also been cloned (7). Sphingolipids are composed of a long-chain sphingoid base normally 18 carbons long with an amide linkage to a fatty acid at the 2-amino group and with various polar additions to the 1-hydroxyl group. The synthesis of the long-chain base component of sphingolipids begins with the condensation of serine and palmitoyl coenzyme A (CoA) to yield 3-ketosphinganine. The 3-ketosphinganine is reduced to form the long-chain base sphinganine, which is N-fatty-acid acylated to yield dihydroceramide. In animals, dihydroceramide is oxidized to ceramide by the introduction of a trans-4,5 double bond. In fungi sphinganine is hydroxylated on C-4 to form phytosphinosine before N-fatty-acid acylation to form phytoceramide. Ceramide is then rapidly converted to sphingomyelin by the addition of phosphocholine in animals or converted to inositol phosphorylceramide (IPC) by the addition of myo-inositol phosphate in fungi. Sphingolipids are subsequently further modified by addition of various sugars and sometimes sulfates to form a large number of complex sphingolipids. Three major species of sphingolipids are found in S. cerevisiae, that is, IPC, mannose-inositol-P-ceramide, and mannose-(inositol-P)-2-ceramide (7).
Previously considered to play mainly a structural role in membranes, the sphingolipid metabolic pathway is now recognized as an important signaling system conserved from fungi to humans. Metabolites derived from the breakdown of complex sphingolipids, or sometimes from de novo synthesis, are found to be highly bioactive molecules that are implicated as second messengers mediating diverse cellular functions. The metabolite that has been studied most extensively is ceramide, a central component of the sphingolipid pathway. Ceramide is not only a building block for sphingolipid synthesis but also a source for other bioactive molecules, such as sphingosine and sphingosine-1-phosphate (11, 22). A role for ceramide in various stress responses is now well established in many biological systems, including the heat shock response of S. cerevisiae (11, 17, 22, 39). A large number of stress agents are shown to transiently regulate sphingolipid metabolism and cause ceramide to accumulate. Moreover, increasing cellular levels of ceramide experimentally has been shown to be sufficient to induce many of the stress responses, including cell cycle arrest and apoptosis, that are normally associated with the treatment of stress agents (11, 22).
In addition to being a particularly rich source of highly bioactive metabolites, sphingolipids, as major components of membranes with so many distinct species in any given cell type, may have many, still unknown, physiological and cellular functions. Indeed, a genetic study of S. cerevisiae showed that sphingolipids are in fact essential for growth, even under nonstressful conditions (27). Furthermore, no significant turnover of complex sphingolipids has been observed in S. cerevisiae in response to any stress. Rather, accumulation of ceramide normally comes from de novo synthesis (17, 39). Thus, complex sphingolipids are directly required for growth in S. cerevisiae. How sphingolipids are required for yeast growth is not understood.
Aspergillus nidulans, a filamentous fungus, is a genetically tractable model organism well suited and widely used to study cell cycle regulation, polarized hyphal cell growth, and development in fungi (2, 26). To study the biological functions of sphingolipids, we analyzed the cellular consequences of inactivation of serine palmitoyltransferase (SPT) and IPC synthase using A. nidulans as a model system. SPT and IPC synthase are two rate-limiting enzymes in the sphingolipid biosynthesis pathway (7). SPT catalyzes the first committed step of sphingolipid biosynthesis, the formation of 3-ketosphinganine through condensation of serine and palmitoyl-CoA. IPC synthase catalyzes the addition of myo-inositol phosphate to the 1-hydroxy group of ceramide to produce IPC. In this study we uncovered a novel role of sphingolipids in cell polarity by regulating polarized organization of the actin cytoskeleton. In addition, we provide evidence demonstrating that IPC synthase plays an important role in mediating the level of cellular ceramide and that ceramide regulates cell cycle progression through G1.
MATERIALS AND METHODS
Strains and general techniques.
A. nidulans strains used in this study were the A773 strain (pyrG89 pyroA4 wA1); SO182 (nimT23 pyrG89 pabaA1 chaA1); a strain containing an aurA gene with a mutation producing a G275V change (the aurAG275V-10 strain) (pyrG89 pyroA4 aurAG275V aurA+ pyrG+ wA1); the pyrG strain (pyrG89 pyroA4 pyrG+ wA1); the aurA3Δ-15, -18, and -30 strains (pyrG89 pyroA4 alcA::aurA aurA3′ΔpyrG+ wA1); JCC152-19 (pyrG89 pyroA4 nimT23 alcA::aurA aurA3′ΔpyrG+ wA1); and the lcbA3Δ-263, -326, and -327 strains (pyrG89 pyroA4 alcA::lcbA pyrG+ lcbA3′Δ wA1). Media and general techniques for culture, transformation, and genetic crossing of Aspergillus strains were as previously described (41). The nimT23 block-release experiment, 4′,6′-diamidino-2-phenylindole (DAPI; Sigma, St. Louis, Mo.) staining, a hydroxyurea (HU) S-phase block, and nocodazole (Sigma) treatment for chromosome mitotic index (CMI) determination were also carried out as previously described (41). Cytochalasin A (CcA; Sigma) treatment to depolymerize the actin cytoskeleton was as described previously (36). Aureobasidin A (AbA) and myriocin (Sigma) were dissolved in ethanol at 10 and 5 mg per ml, respectively, and added into medium to the final concentrations indicated in Results. The S. cerevisiae strain used in this study was YPH499 (MATa ura3-52 lys2-801amber ade2-101ochre trp1Δ63 his3Δ200 leu2Δ1) from Stratagene (La Jolla, Calif.), and the yeast cells were grown in inositol-free synthetic complete medium.
Labeling and analysis of sphingolipids.
Pulse-labeling of sphingolipids with [2-3H]inositol and [4,5-3H] dihydrosphingosine ([4,5-3H]DHS) were performed as described previously (12). Briefly, cells were grown to early exponential phase (107/ml) and pulse-labeled with [3H]inositol (5 μCi/ml) for 60 min or with 10 μM [3H]DHS (2 μCi/ml) for 60 min in 10-ml cultures of inositol-free medium. Cells were harvested by centrifugation (3,000 × g) and treated with 5% cold trichloroacetic acid on ice for 60 min. Lipids were extracted using the 95% ethanol-water-diethylether-pyridine-ammonium hydroxide (15:15:5:1:0.018 [vol/vol/vol/vol]) solvent system. Extracted lipids were dried under nitrogen. Monomethylamine reagent (methanol-water-butanol-methylamine, 4:3:1:5 [vol/vol]) was added to dried extract, which was then incubated at 52°C for 30 min to deacylate the lipids. The mixture was dried under nitrogen and resuspended in 0.2 ml of chloroform-methanol-H2O (16:16:5). Radiolabeled lipids were then separated on Silica Gel 60 thin-layer chromatography (TLC) plates (EM Merck) using chloroform–methanol–4.2 N ammonium hydroxide (9:7:2 [vol/vol/vol]) and visualized on a Molecular Dynamics Typhoon PhosphorImager by using a tritium-sensitive phosphor screen.
Determination of IPC synthase activity.
IPC synthase activity was measured by monitoring the incorporation of N-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl) aminocaproyl] (NBD)–C6–ceramide (Molecular Probes, Eugene, Oreg.) into chloroform soluble NBD-IPC in a mixed micellar assay system (9). Microsomes were prepared from homogenized A. nidulans cells (10 g [wet weight]) by differential centrifugation. Chloroform-soluble NBD-IPC was analyzed by TLC on Silica Gel 60 plates (EM Science) using the chloroform-methanol-water (65:24:4 [vol/vol/vol]) solvent system. NBD-IPC was quantified by direct fluorescence using a Molecular Dynamics Typhoon imager.
aurA disruption.
An aurA disruption construct was made by three-step cloning using pBluescript II SK(+) (Stratagene). A 2,018-bp 5′-end-flanking sequence was amplified from A. nidulans genomic DNA by PCR using a pair of primers, ATAAGAATGCGGCCGCTCTGTGGCTTCCGGTTGGCTAC and GCTCTAGACCAGGGTTGAGTCGGCAGCATG, and cloned into the NotI and XbaI sites of pBluescript II sk(+). The 3,130-bp 3′-end-flanking sequence was amplified in the same manner using the primers CGGGATCCTGAAGCCCGTCTTCGTGACC and GAGCAGATATCGGTGGTCCAAATACAGGTACC and cloned into the BamHI and EcoRV sites. Finally, a 1,892-bp A. fumigatus pyrG fragment (38) was cloned into the XbaI and BamHI sites as an XbaI and BglII fragment to give rise to pAURAΔ, in which the aurA open reading frame (ORF) was disrupted and the sequence from Ala297 to Met308 was replaced by pyrG of A. fumigatus.
A linear NotI-EcoRV fragment from pAURAΔ was used to replace aurA by transformation of the A773 strain. Transformants were screened for heterokaryons on selective yeast extract-glucose-agar plates as previously described (42). Suspected heterokaryons were maintained as mycelial colonies on the selective medium. PCR analysis of the selected heterokaryons was carried out to confirm aurA disruption using a primer specifically derived from pyrG of A. fumigatus and a primer derived from sequences from the aurA locus further upstream or downstream of the sequences used to construct pAURAΔ.
lcbA cloning.
The lcbA gene of A. nidulans was cloned by rapid amplification of cDNA ends (RACE)-PCR based on a 300-bp expressed sequence tag (g4a07a1.r1) from the Fungal Genetic Stock Center A. nidulans cDNA database (http://www.genome.ou.edu/asper_blast.html), which was identified by homology search using LCB1 of S. cerevisiae. Aspergillus mycelia grown in yeast extract-glucose to early log phase were harvested. After snap-freezing in liquid nitrogen, the mycelia were lyophilized. The lyophilized mycelia were ground to a fine powder using a mortar and pestle, and total RNA was isolated using TRIzol reagent (GIBCO BRL, Rockville, Md.). The mRNA was then purified from the total RNA using the PolyATtract mRNA isolation system (Promega, Madison, Wis.). Adapter-ligated cDNA was synthesized, and the subsequent RACE-PCR was carried out using the Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.) according to the manufacturer's instructions. The 5′ and 3′ RACE primers used were CGAAAGCTGGACGACACCAGGCTTGG and GCCTGCGCAGCGTGTGCGCTATTGCTG, respectively. After the first round of PCR, the DNA was further amplified by a second round of PCR using nested primers: TACTTCGGGGCGAGCAAATAGCGG for the 5′ RACE and TATGTGAAGTCTAGCTACCAGAATG for the 3′ RACE. The specific PCR products were separated by electrophoresis on low-melting-temperature gels, excised, cloned into the vector pCR2.1 (Invitrogen), and sequenced. The assembled full-length cDNA contains a 497-amino-acid (aa) ORF whose deduced amino acid sequence shows high homology to LCB1. This gene is thus designated lcbA according to A. nidulans gene naming conventions.
Generation of alcA::aurA- and alcA::lcbA-dependent strains.
To create an alcohol-dependent aurA strain, we cloned a 1,290-bp fragment of aurA with a 3′ 159-aa truncation under the control of the alcA promoter, which is alcohol inducible and glucose repressible (37), into pBluescript II sk(+), with the pyrG gene of A. fumigatus as a selection marker. The resulting plasmid was designated pAURA3′Δ. The circular pAURA3′Δ DNA was used to transform the A773 strain. A single homologous recombination of pAURA3′Δ at the endogenous aurA locus would result in 3′ truncation of the endogenous aurA gene and, at the same time, a full-length aurA gene under the control of the alcA promoter. The transformants were first screened for heterokaryons on selective YAG plates. Selected heterokaryons were then further tested for dependence on medium containing alcohol as the sole carbon source for growth. Alcohol-dependent strains were then analyzed by PCR as described above for aurA deletion. An alcohol-dependent lcbA strain was also generated in the same manner with a construct containing a 3′ truncation of 215 aa of the product of lcbA.
Indirect immunofluorescence staining of microtubules, actins, and nucleoli.
The procedures for indirect immunofluorescence staining of microtubules, actins, and nucleoli in A. nidulans were as described previously (41). A mouse monoclonal antitubulin antibody, B5-1-2, (Sigma) was used at a 1:200 dilution to stain microtubules. A mouse monoclonal antiactin antibody, Ab-1 (Oncogene), was used at a 1:5,000 dilution to stain actin, and the human autoantibody ANA-N (Sigma) was used to stain nucleoli at a 1:4 dilution. The secondary antibody, goat anti-mouse immunoglobulin G conjugated with fluorescein isothiocyanate (F2653; Sigma), was used at a 1:200 dilution in actin and tubulin staining, and the secondary antibody, goat anti-human immunoglobulin G conjugated with fluorescein isothiocyanate (F3512; Sigma), was used at a 1:64 dilution in nucleolar staining.
Nucleotide sequence accession number.
The nucleotide sequence of the lcbA gene has been deposited in GenBank under accession number AY032867.
RESULTS
aurA function is essential for sphingolipid biosynthesis and growth in A. nidulans
The aurA gene of A. nidulans was cloned with a dominant mutation producing resistance to the antifungal compound AbA (18). Sequence homology suggests that aurA is a homolog of AUR1 of S. cerevisiae, which was also cloned originally as a mutated gene that produces resistance to AbA (14, 16). AUR1 has been shown to be required for IPC synthase activity, and thus AUR1 is thought to encode IPC synthase or an essential subunit of IPC synthase in S. cerevisiae (27). To see if aurA is also required for IPC synthase activity in A. nidulans, we first assayed IPC synthase activity in early-log-phase A. nidulans cells. Exponentially growing yeast cells were used as a positive control. Compared to yeast cells, surprisingly, actively growing A. nidulans cells contain very little IPC synthase activity (data not shown).
It is possible that A. nidulans IPC synthase requires some unknown factors for activity or that the assay conditions optimized for the yeast IPC synthase are not suitable for the A. nidulans IPC synthase. To circumvent these potential problems, we detected IPC synthase activity by monitoring the incorporation of [3H]myo-inositol into sphingolipids in the presence and absence of the IPC synthase inhibitor AbA. As shown in Fig. 1A, both A. nidulans and yeast cells efficiently incorporated the labeled inositol into sphingolipids. Although A. nidulans produces different sphingolipid species, sphingolipid synthesis in both yeast and A. nidulans was equally inhibited by AbA. To determine whether inhibition of sphingolipid synthesis by AbA is specific to inactivation of AURA function, we recreated the dominant resistance mutation (G275V) by in vitro mutagenesis and then introduced the mutant aurA gene into A. nidulans cells by transformation. A. nidulans cells carrying aurA with the mutation producing the G275V change became highly resistant to AbA (Fig. 1B). Furthermore, sphingolipid synthesis in the AbA resistance cells was not significantly inhibited by AbA (Fig. 1A). The results thus indicate that the aurA gene of A. nidulans, like the AUR1 gene of S. cerevisiae, is required for IPC synthase activity.
FIG. 1.
Requirement of aurA for IPC synthase activity and growth in A. nidulans. (A) Autoradiogram of base stable sphingolipids labeled with [3H]inositol and separated by chromatography on a Silica Gel 60 TLC plate. Cells were pretreated with AbA for 15 min before addition of [3H]inositol. The bands for sphingolipid species, IPC, mannose-inositol-P-ceramide (MIPC), and mannose-(inositol-P)-2-ceramide [M(IP)2C] of S. cerevisiae are indicated. Note the inhibition of [3H]inositol incorporation into sphingolipids in both S. cerevisiae and A. nidulans cells by AbA and the resistance to this inhibition conferred by the mutation in aurA in A. nidulans producing the G275V change. (B) Colonies of the wt and aurAG275V mutant strains grown on MAG plates containing AbA at the concentrations indicated. The strains were spot inoculated with toothpicks and were allowed to grow for 2 days at 32°C. (C) Dependence of aurA3Δ strains on the expression of functional AURA off the alcA promoter. Shown are colonies of a wt and three aurA3Δ alcA::aurA strains grown on media containing glycerol, ethanol, or glucose as the sole carbon source.
To directly determine if aurA is essential for A. nidulans growth, we inactivated AURA function by gene deletion using a linear DNA construct in which the sequence encoding A297 to M308 of the product of the aurA ORF was replaced with pyrG from A. fumigatus as a nutritional selection marker. A. fumigatus pyrG encodes orotidine-5′-phosphate decarboxylase and complements the pyrG89 mutation in A. nidulans. If deletion of aurA is lethal, the deleted allele would then be maintained in heterokaryons (29, 31). The ability of A. nidulans to maintain nuclei containing a deletion of an essential gene in a heterokaryotic state under selective conditions allows the analysis of the null phenotype in germinating spores, because conidiation (asexual sporulation) breaks down the heterokaryotic state to form uninucleate spores. Thus, 50% of the spores derived from a heterokaryon, as further confirmed by PCR, are expected to carry the parental nuclei with the pyrG89 mutation and another 50% are expected to carry nuclei with the mutated aurA gene and pyrG+. As expected, when germinated in the presence of uridine and uracil, which complement the pyrG89 mutation, about 50% of the spores (parental) grew normally whereas another 50% of the germinating spores failed to initiate polarized growth (data not shown and Fig. 2). The results therefore demonstrate that aurA is essential for A. nidulans growth.
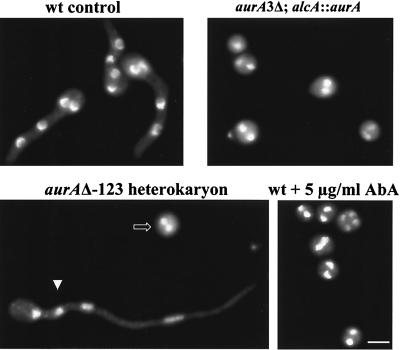
FIG. 2.
Terminal phenotype of germinating spores in the absence of aurA function. Spores of the indicated strains were germinated on coverslips in YG medium containing uridine and uracil for 7 h and then fixed and stained with DAPI to visualize DNA. The alcA promoter is repressed in YG medium, and inclusion of uridine and uracil allows the parental spores (arrowhead) derived from the aurAΔ heterokaryon to germinate, thus clearly distinguishing them from the spores with the aurA deletion (open arrow). Bar, 5 μm.
As aurA is essential, to further analyze its function, we generated a conditional mutant strain in which the only functional copy of aurA is under the control of the alcA promoter (see Materials and Methods for details). The alcA promoter is induced by alcohol as the sole carbon source and was tightly repressed by glucose, whereas glycerol serves as a noninducing and nonrepressing carbon source. The wild-type (wt) strain grew well on all three carbon sources, whereas the mutant strain grew as well as the wt strain only on media containing glycerol or alcohol (Fig. 1C). The mutant strain failed to grow on medium containing glucose, which tightly represses the alcA promoter, thus further confirming the essentiality of aurA in A. nidulans.
To see how AURA is required for A. nidulans growth, we examined terminal phenotypes of germinating spores lacking AURA function. During germination, spores with inactive AURA underwent the initial isotropic growth in a manner similar to that of the wt spores but failed to initiate polarized hyphal growth. DNA staining with DAPI revealed that the cell cycle was arrested after one round of nuclear division (Fig. 2). Inactivation of AURA by aurA deletion, alcA promoter repression, or treatment with AbA generated the same phenotypes (Fig. 2). Lack of polarized growth and uniform cell cycle arrest suggest that AURA may normally have a role in cell cycle progression and cell polarity.
Inactivation of AURA causes cell cycle arrest in G1.
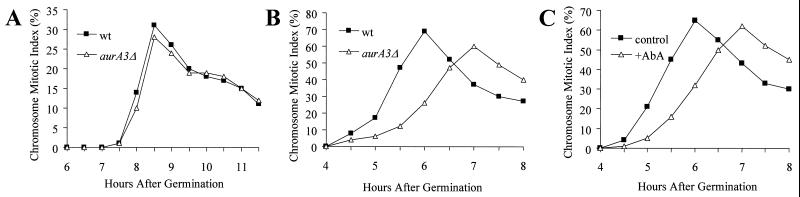
To determine whether AURA indeed has a role in cell cycle progression, we analyzed the kinetics of entry into the first mitosis in germinating spores in the presence or absence of AURA as spores entered the cell cycle uniformly from G1. To better monitor the kinetics of entry into mitosis, we included nocodazole, a microtubule poison, in the germinating medium to trap cells in a mitotic state as cells progressed into mitosis. In glycerol-containing medium, both alcA::aurA-dependent and wt strains entered into the first mitosis with similar kinetics, with the mitotic index peaking 8.5 h after germination (Fig. 3A). By contrast, in the glucose-containing (repressing) medium, the alcA::aurA-dependent strain showed a marked delay in entry into mitosis, as its mitotic index did not peak until 7 h, while the wt strain had a peak mitotic index 6 h after germination (Fig. 3B). Similarly, addition of 5 μg of AbA per ml in the medium to inactivate AURA also markedly delayed entry into mitosis (Fig. 3C), thus implicating an important role for AURA in cell cycle progression.
FIG. 3.
Inactivation of AURA significantly delays cell cycle progression in germinating spores. Spores with or without AURA function were germinated on coverslips in the presence of 5 μg of nocodazole per ml, fixed, and stained with DAPI. The CMI was determined as the percentage of cells with condensed DNA. Shown are CMIs of wt and aurA3Δ alcA::aurA spores germinated in medium containing glycerol as the sole carbon source (A), of wt and aurA3Δ alcA::aurA spores germinated in medium containing glucose (B), and of wt spores germinated in the presence or absence of AbA (C).
To determine at which stage of the cell cycle AURA is required, we first determined whether AURA has a role in the G2/M transition and subsequent progression through mitosis. To do that, we employed a G2-specific temperature-sensitive mutant gene, nimT23cdc25, which encodes a homolog of fission yeast Cdc25 tyrosine phosphatase, to synchronize cells at G2 at the restrictive temperature of 42°C before inactivating AURA. We then compared the kinetics of entry into and progression through mitosis upon release to the permissive temperature of 32°C in the presence and absence of AURA. nimTcdc25 is required for p34cdc2-cyclin B activation by dephosphorylation to promote entry into mitosis (30).
When germinated at 42°C, the nimT23cdc25 mutant cells were blocked in G2. AbA was added to the G2-arrested cells to inactivate AURA 2 h before release to 32°C. Upon release to 32°C, cells rapidly entered and then progressed through mitosis synchronously, with the mitotic index peaking at 10 min either with or without AbA treatment (Fig. 4A).
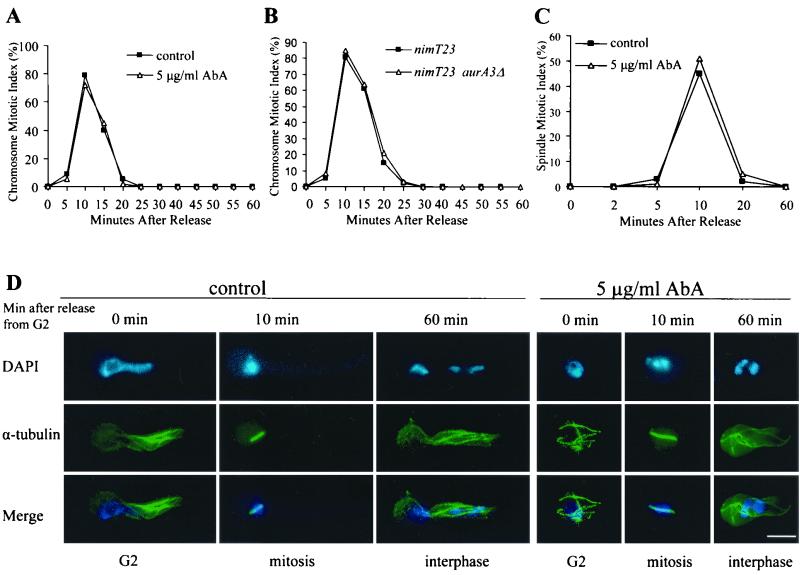
FIG. 4.
AURA function is not required for G2/M transition and progression through mitosis. (A) CMIs during nimT23cdc25 block and release in the presence or absence of AbA. The nimT23cdc25 mutant spores were germinated on coverslips at 42°C for 5 h to allow G2 arrest before AbA addition to inactivate AURA. Cells remained at 42°C for an additional 2 h after AbA addition and were then released to 32°C. (B) CMI during nimT23cdc25 block and release in nimT23cdc25 single and nimT23cdc25 aurA3Δ (alcA::aurA) double mutant cells. (C) Spindle mitotic index during nimT23cdc25 block and release in the presence or absence of AbA. nimT23cdc25 block and release and AbA treatment were exactly as described for panel A. (D) Representative cells showing microtubules and DNA staining during nimT23cdc25 blocking and release in the presence or absence of AbA. Bar, 5 μm.
To ensure that AbA addition had effectively inactivated AURA, we repeated the nimT23cdc25 block-release experiment using an nimT23cdc25 alcA::aurA double mutant strain. In glycerol-containing medium, spores usually enter into the first mitosis 8 to 9 h after germination (Fig. 3A). To minimize undesirable effects of prolonged incubation at the restrictive temperature, we thus first allowed cells to germinate at 32°C for 10 h before upshifting them to 42°C for 2 h to inactivate NIMTcdc25 and cause G2 arrest. Then we turned off the expression of aurA from the alcA promoter by transferring the G2-arrested cells to glucose-containing medium prewarmed to 42°C and continued to incubate at 42°C for an additional 2.5 h to eliminate AURA. Upon return to the permissive temperature, still in glucose-containing medium, both the single and double mutant cells entered and progressed through mitosis with identical kinetics (Fig. 4B).
Although DAPI staining showed that cells lacking AURA function underwent an apparently normal nuclear division during the nimT23cdc25 block and release, we noticed that the cell cycle was arrested immediately after mitosis, giving rise to a nuclear phenotype similar to that shown in Fig. 2. To further investigate if cells could complete normal mitosis successfully in the absence of AURA function, we observed microtubule morphologies and measured the spindle mitotic index during nimT23cdc25 block and release in the presence or absence of AbA as described for the experiment shown in Fig. 4A. At the G2 arrest point, both AbA-treated and control cells had very similar interphase microtubule arrays, although polarized growth in the treated cells was severely inhibited (Fig. 4D). Upon release from the G2 block, short mitotic spindles were rapidly formed as DNA became highly condensed. As cells rapidly progressed through mitosis, we observed normal spindle elongation as DNA segregation occurred. By 20 min after release from G2 arrest, interphase microtubule arrays were reestablished in >90% of the cells. We did not observe any obvious difference in either microtubule morphologies or the kinetics of the spindle mitotic index in the presence or absence of AbA treatment (Fig. 4C and D). Taken together, we conclude that AURA function is not required for cell cycle progression from G2 and through mitosis.
We next asked whether AURA is required for cell cycle progression through S phase. HU is a potent inhibitor of DNA synthesis, and addition of HU to germinating A. nidulans spores arrests the cell cycle in early S phase (3). We thus included HU in the medium to synchronize the cell cycle of germinating spores at early S phase. We then inactivated AURA by adding AbA to HU S-phase-arrested cells and analyzed the consequences of AURA inactivation on cell cycle progression upon removal of HU from the medium to release the S-phase block. Again, nocodazole was included in the medium to facilitate determination of kinetics of mitotic indices. Cells progressed into mitosis from S phase with similar kinetics in the presence and absence of AbA and reached peak CMI 2 h after release from the HU S-phase block (data not shown). The results thus indicate that AURA does not have a role in S-phase progression. Therefore, the marked delay in cell cycle progression from G1 observed with germinating spores lacking AURA function, as demonstrated in Fig. 3, must occur prior to the HU arresting point at early S phase, most likely in G1.
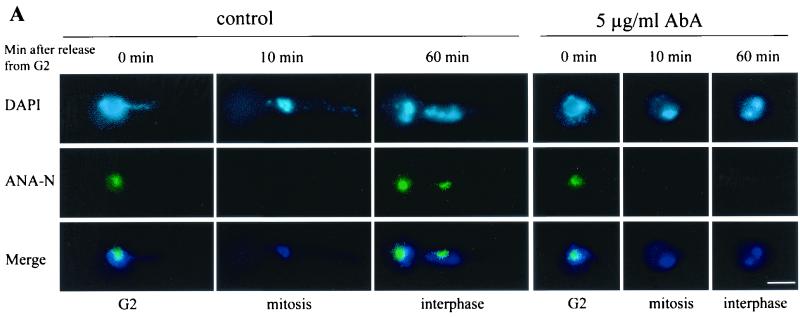
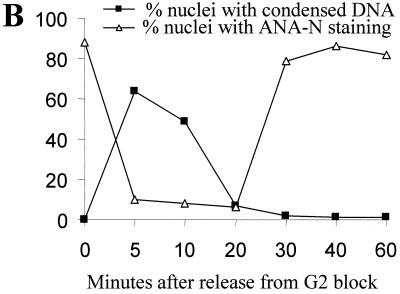
DAPI staining showed that nuclei in the arrested cells lacking AURA had no discernible nucleoli (Fig. 2 and 4D). To detect the presence of nucleoli with a more sensitive and specific probe, we double stained nuclei with a nucleolus-specific antibody, ANA-N (33), and DAPI during a cell cycle synchrony generated by a nimT23cdc25 block and release in the presence or absence of AbA as described for Fig. 4. At the G2 arrest point, the nuclei of both AbA-treated and untreated control cells contained large nucleoli (Fig. 5A). Upon release from the G2 block, nucleoli were disassembled and became ANA-N negative as cells entered into mitosis (Fig. 5). Then, as cells progressed through mitosis and entered the next cell cycle, nucleolar staining reappeared in the control cells. However, nucleolar staining was not observed in cells lacking AURA function for the entire duration of the experiment up to 2 h after release from the nimT23cdc25 G2 block, although these cells had completed mitosis successfully (Fig. 5). The results further indicate that inactivation of AURA causes cell cycle arrest at G1 before reassembly of the nucleolus during G1/S.
FIG. 5.
Nuclei in AbA-arrested cells lack nucleolus staining. The blocking and release of nimT23cdc25 mutant cells, treatment with AbA, and immunostaining with ANA-N antibody for nucleoli and DAPI for DNA were carried out exactly as described for Fig. 4. (A) Representative cells showing ANA-N nucleolar staining during nimT23 blocking and release in the presence or absence of AbA; (B) inverse relationship between mitotic DNA condensation and ANA-A nucleolar staining during nimT23 blocking and release. Bar, 5 μm.
AURA is required for polarized hyphal growth via regulation of the actin cytoskeleton.
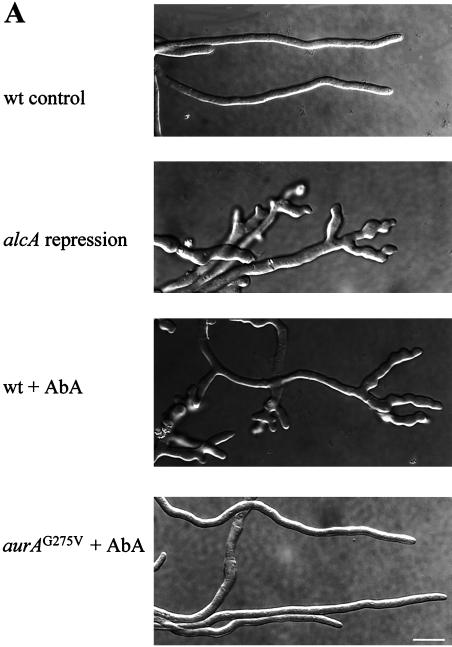
As shown in Fig. 2, inactivation of AURA inhibited polarized growth of germinating spores, suggesting a potential role of AURA in cell polarity. Upon germination, A. nidulans spores undergo highly polarized growth through tip extension, giving rise to long tubular structures called hyphae. Hyphal branching occurs usually several cells posterior to the hyphal tip and never occurs at the tip. To better understand how AURA may regulate polarized hyphal growth, we first let spores germinate to form small hyphae before inactivating AURA. As expected, control hyphae grow in a highly polarized fashion, and no hyphal tip branching was observed (Fig. 6A). By contrast, inactivation of AURA by either shutting off aurA expression from alcA or by addition of AbA rapidly inhibited polarized hyphal growth and subsequently promoted multiple branching at or near the hyphal tips (Fig. 6A). The treated hyphae became abnormally wide compared to those of the control. These dramatic morphogenic changes were specifically caused by inactivation of AURA, as the AbA-resistant mutant cells continued polarized growth in the presence of AbA.
FIG. 6.
Hyphal tip branching and reorganization of the actin cytoskeleton in A. nidulans germlings lacking AURA function. (A) DIC micrographs showing hyphal tip branching in germlings lacking AURA function. For repression of alcA::aurA, spores of a wt and of an alcA::aurA-dependent strain (aurA3Δ alcA::aurA) were first germinated on coverslips in medium containing glycerol as the sole carbon source for 12 h, and then the germlings were transferred to glucose-containing medium to shut off aurA expression from alcA. The micrographs were taken 6 h after transfer to the glucose medium. For AbA treatment, spores of a wt and an aurAG275V strain were germinated in YG on coverslips for 9 h before addition of AbA. Micrographs were taken 4 h after AbA addition. (B) Immunostaining of the actin cytoskeleton at various times after the addition of AbA. Spores were germinated and treated with AbA as described for panel A. CcA was added alone (not shown) or together with AbA (f and g). The solid arrows indicate the intensely stained actin band near the hyphal tip of control cells (a), and the arrowheads indicate the emergence of hyphal branches and the associated actin aggregates (in panels b and c at 1 h and in panels d and e at 2 and 4 h after AbA treatment, respectively). The open arrow points to the actin ring associated with septation (e). Panel g is the differential interference-contrast micrograph of cells shown in panel f. Bar, 5 μm.
The function of the actin cytoskeleton is known to be required for polarized growth in A. nidulans (36). Immunofluorescence staining of microtubules, as shown for the nimT23cdc25 block-release experiments, demonstrated that AURA does not have a role in microtubule function. We thus suspected that AURA, likely through its role in sphingolipid synthesis, is required for the normal organization of the actin cytoskeleton to maintain hyphal tip polarity.
The actin cytoskeleton was visualized by immunofluorescence staining. In control hyphae, actin located in patches is gradually enriched towards hyphal tips, culminating in an intensely stained band of ∼2 to ∼4 μm wide, about 2 μm behind the growing tip (Fig. 6B, panel a). This staining pattern is specific to actin, as depolymerization of actin with CcA prevented any specific actin staining (data not shown; also see Fig. 6B, panel f). Upon inactivation of AURA with AbA, we observed a dramatic remodeling of the actin cytoskeleton at the hyphal tips clearly preceding the initiation of branches. A time course showed that the first observable change in the actin cytoskeleton was the collapse of the intensely stained actin band near the hyphal tip into aggregates (Fig. 6B, panel b). These aggregates then migrated to the hyphal tip, where each aggregate was associated with the initiation of a hyphal branch (Fig. 6B, panel c). Then by 2 h after AURA inactivation, the actin patches in the more distal part of the hyphae were also rearranged into more intensely stained aggregates to promote the initiation of more branches (Fig. 6B, panels d and e). The initiation and extension of hyphal branches require actin function, as addition of CcA to depolymerize the actin cytoskeleton completely abolished the emergence of branches (Fig. 6B, panels f and g).
Interestingly, although inactivation of AURA completely disrupted the normal organization of the actin cytoskeleton required for polarized hyphal growth, the formation of actin rings associated with septation appeared not to be affected. Furthermore, septation continued several hours after inactivation of AURA, when the normal actin cytoskeleton had been completely disorganized (Fig. 6B, panel e).
Sphingolipids are required for hyphal polarity, and accumulation of ceramide is associated with G1 arrest.
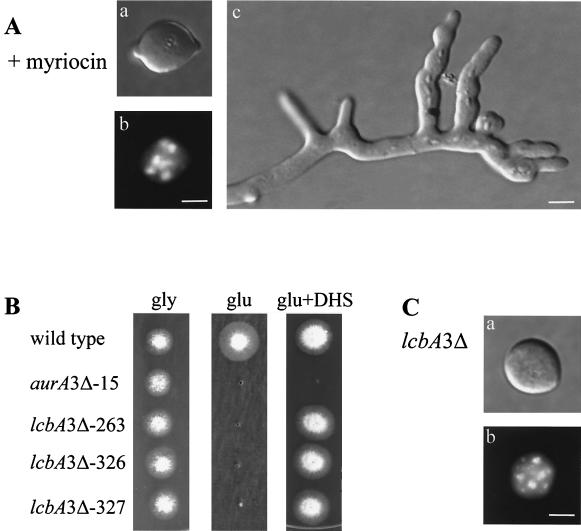
In S. cerevisiae, inactivation of IPC synthase activity not only inhibits sphingolipid synthesis but also leads to accumulation of the upstream intermediate, ceramide (27), which is thought to play a regulatory role in a wide range of cellular activities (11, 22). To differentiate whether defects in the cell cycle and hyphal polarity of cells lacking AURA are caused by inhibition of sphingolipid synthesis or by accumulation of ceramide, we analyzed the effects of myriocin, a specific inhibitor of SPT (24). SPT catalyzes the first committed step in sphingolipid biosynthesis. We reasoned that if defects in the cell cycle and cell polarity in the absence of AURA function are caused by the inhibition of sphingolipid synthesis, but not by the accumulation of ceramide, then inactivation of SPT, which abolishes the entire sphingolipid biosynthesis pathway, would exactly phenocopy AURA inactivation. Indeed, pharmacological inactivation of SPT with myriocin prevented polarized growth of germinating spores (Fig. 7A, panel a). Moreover, addition of myriocin to germlings, as in the experiment described in the legend to Fig. 6, promoted hyphal tip branching exactly as occurred after AbA treatment (Fig. 7A, panel c). The data thus show that polarity defects in cells lacking AURA function were caused by inhibition of sphingolipid biosynthesis, indicating an important role of sphingolipids in hyphal polarity.
FIG. 7.
SPT activity is required for cell polarity but has no role in cell cycle regulation. (A) Myriocin treatment. (a and b) DIC (a) and DAPI (b) fluorescence micrographs of one representative spore germinated in the presence of 80 μg of myriocin per ml for 10 h; (c) DIC micrograph of a germling 4 h after myriocin addition. (B) The lcbA3Δ strains are dependent on alcA::lcbA or DHS supplementation for growth. Shown are colonies of a wt, an alcA::aurA, and three alcA::lcbA strains grown in glycerol medium or glucose medium with or without 4 μg of DHS supplement per ml for 2 days. (C) Repression of alcA::lcbA. (a and b) DIC (a) and DAPI (b) fluorescence micrographs of one representative spore of an alcA::lcbA-dependent strain 10 h after germination in glucose-containing (repressing) medium. Bar, 5 μm.
Unlike AbA treatment, however, myriocin did not cause cell cycle arrest (Fig. 7A, panel b). In fact, several rounds of nuclear division continued in the presence of myriocin with kinetics similar to those shown by the control cells. As myriocin is less potent than AbA against A. nidulans, it was possible that the lack of cell cycle arrest was due to an incomplete inhibition of SPT. To eliminate this possibility, we cloned lcbA from A. nidulans, based on sequence homology to S. cerevisiae LCB1, which encodes a subunit of SPT and is required for SPT activity (5). We generated a conditional, alcohol-dependent mutant strain in which the endogenous lcbA gene was disrupted and a functional copy of lcbA was brought under the control of the alcA promoter (Fig. 7B). Analysis of sphingolipid synthesis in this mutant strain showed that repression of lcbA expression by glucose completely inhibited sphingolipid synthesis (data not shown). If LCBA were a subunit of SPT, as is Lcb1p in S. cerevisiae, then addition of DHS, a downstream intermediate, to the growth medium should complement the lack of LCBA function. As shown in Fig. 7B, the alcA::lcbA mutant strains were able to grow in glucose (repressing) medium containing DHS, whereas the alcA::aurA strain was not able to grow, thus indicating that LCBA of A. nidulans is indeed a functional homolog of Lcb1p, a subunit of SPT, and is essential.
Similar to myriocin treatment, repression of lcbA expression off the alcA promoter inhibited polarized growth but again did not cause cell cycle arrest (Fig. 7C). This suggests that cell cycle arrest and cell polarity defects caused by AURA inactivation are independent events, with cell cycle arrest in G1 likely being attributable to an accumulation of ceramide.
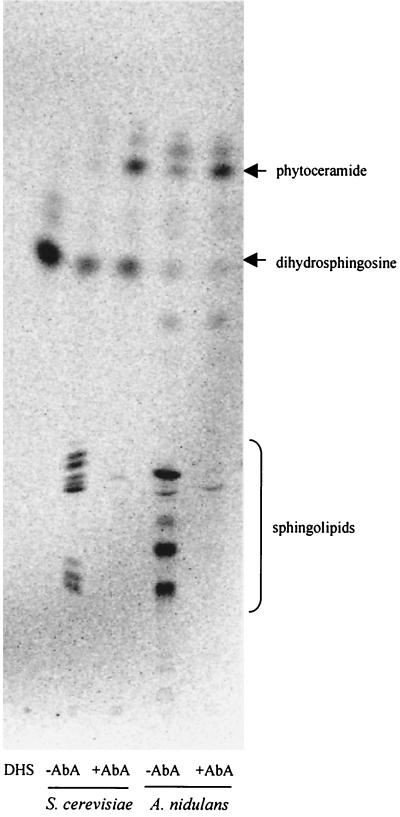
To directly determine whether inactivation of AURA indeed leads to the accumulation of ceramide in A. nidulans as in S. cerevisiae, we labeled S. cerevisiae and A. nidulans cells with [3H]DHS, the upstream intermediate of ceramide, in the presence or absence of AbA treatment. Yeast cells were again used as a positive control. In the absence of AbA, both yeast and A. nidulans cells efficiently incorporated DHS into complex sphingolipids and the level of ceramide was very low (Fig. 8). As expected, AbA treatment completely inhibited the incorporation of DHS into complex sphingolipids, and indeed it also caused a marked elevation of the cellular ceramide levels (Fig. 8).
FIG. 8.
Inactivation of AURA causes accumulation of ceramide. An autoradiogram of base stable sphingolipids labeled with [3H]DHS is shown. The culture of S. cerevisiae and A. nidulans cells, AbA treatment, [3H]DHS labeling, sphingolipid extraction, separation, and detection were as described for Fig. 1A.
DISCUSSION
Sphingolipids are major components of the plasma membranes of eukaryotic cells and are shown to be essential for growth in S. cerevisiae (27). Although much is known about the sphingolipid biosynthetic pathway and the importance of sphingolipid metabolites as second messengers in signal transduction, little work has been directed towards investigating the direct biological functions of sphingolipids. In this study we characterized the function of aurA and lcbA in A. nidulans and showed that they are functional homologs of S. cerevisiae AUR1 and LCB1, respectively. AUR1 is required for IPC synthase activity, and LCB1 encodes a subunit of serine palmitoyl-CoA transferase (5). The present study provides strong evidence for an important role of IPC synthase in cell cycle progression through G1 and for the essentiality of sphingolipids for hyphal cell polarity in A. nidulans.
Several lines of evidence indicate that IPC synthase activity is required for cell cycle progression through G1 phase. First, inactivation of IPC synthase in germinating spores, which enter the cell cycle from a natural resting state in G1, caused a marked delay in progression of the first cell cycle. Cells were then terminally arrested after one round of nuclear division with two small nuclei, most likely in G1. However, when germinating spores were first allowed to progress through G1 and then arrested in G2 or early S phase prior to inactivation of IPC synthase, then this delay in progression of the first cell cycle was abolished. Second, results from nimT23cdc25 block-release experiments showed that, upon release from nimT23cdc25 G2 arrest, the cell cycles of cells lacking IPC synthase activity were arrested immediately after progression through a normal mitosis. These arrested cells contained interphase microtubule arrays and two small nuclei devoid of nucleoli, consistent with G1 arrest. Third, cell cycle arrest caused by inactivation of IPC synthase was associated with a marked increase in levels of cellular ceramide. In fact, accumulation of cellular ceramide is likely responsible for the G1 arrest. This supposition is supported by the observation that inactivation of SPT activity, which eliminated the entire sphingolipid synthesis pathway, including ceramide synthesis, produced all the phenotypes associated with IPC synthase inactivation, except for cell cycle arrest. Additionally, in S. cerevisiae an increase in the level of cellular ceramide caused by overexpression of YSR2, a DHS-1-phosphate phosphatase, is associated with a G1 delay of the cell cycle (21). Furthermore, treatment of S. cerevisiae cells with a synthetic ceramide causes cell cycle arrest in G1, which is further shown to be mediated through a ceramide-activated protein phosphatase (28).
Our present results strongly suggest that IPC synthase plays a pivotal role in mediating the level of cellular ceramide. Ceramide is implicated as a second messenger in stress responses (11, 22). In normal growing cells the level of cellular ceramide remains very low. Upon stress treatments such as heat shock, the level of cellular ceramide is rapidly and transiently increased. In S. cerevisiae, the increase in cellular ceramide upon heat shock is due to de novo synthesis (17, 39). In A. nidulans, heat shock rapidly and transiently inhibits both cell growth and nuclear division (43). Progression of the cell cycle needs to be tightly coupled to cell growth. Perhaps IPC synthase helps couple the cell cycle to growth during stress responses in fungi, as sphingolipid biosynthesis is required for cell growth and increases in amounts of cellular ceramide cause G1 arrest. In this scenario, heat shock or other stresses would rapidly and transiently inhibit IPC synthase activity, which stops sphingolipid biosynthesis and simultaneously leads to the accumulation of the upstream intermediate, ceramide. Inhibition of sphingolipid biosynthesis would prevent cell growth, and accumulation of ceramide would coordinately inhibit cell cycle progression, thus coupling the cell cycle to growth to allow successful adaptation of cells to stress conditions.
A. nidulans produces uninucleate conidiospores that germinate to form hyphal cells. Germinating spores employ two distinct modes of growth. They first grow isotropically, by adding new cell wall material uniformly in all directions. Following the first nuclear division, germinating spores then switch to polarized growth to form a germ tube. Subsequent hyphal growth is highly polarized, occurring exclusively at the tip of the germ tube and thus giving rise to a tubular hyphal filament. Here we showed that sphingolipids are required for both the establishment and the maintenance of hyphal cell polarity. Inhibition of sphingolipid biosynthesis by inactivation of IPC synthase or SPT in germinating spores allowed isotropic spore cell expansion but prevented subsequent polarized growth. This phenotype was particularly striking in germinating spores lacking SPT function. In the absence of SPT function, spores continued isotropic growth and nuclear division before eventually collapsing due to plasmolysis. Plasmolysis is likely to be caused by defects in the plasma membrane, as sphingolipids are its major components. Unlike spores lacking SPT function, germinating spores with inactive IPC synthase do not continue isotropic growth and nuclear division after an initial isotropic expansion and one round of nuclear division. The phenotypic differences between spores lacking SPT function and those lacking IPC synthase function can be explained by the fact that inactivation of IPC synthase causes marked accumulation of ceramide. Ceramide is highly bioactive and known to cause rapid cell cycle arrest or cell death in many organisms (11, 22).
Sphingolipids are also required for the maintenance of hyphal cell polarity, as inhibition of sphingolipid biosynthesis in germlings rapidly inhibited the normal polarized hyphal growth and consequently hyphal cells became abnormally wide. Interestingly, multiple short branches were subsequently initiated at or near the hyphal tip, further indicating that the normal hyphal cell polarity was abolished in the absence of sphingolipid biosynthesis. Hyphal tip branching never occurs in wt cells, and hyphal branches are initiated invariably several cells posterior to the hyphal tip.
Several genes required for the establishment of hyphal cell polarity have been identified recently from a collection of temperature-sensitive mutants of A. nidulans (13, 25). Temperature shift experiments further show that some of the genes are also required for the maintenance of hyphal cell polarity. Although these genes have not been cloned, judging from the phenotypes suppressible by high-level-osmosis medium (25), most of them may function in the Rho1-Pkc1-mediated cell wall integrity signaling pathway. This assumption is in agreement with the report that in A. nidulans deletion of mpkA, a component of the cell wall integrity pathway, results in similar defects in hyphal cell polarity and cell lysis, both suppressible by high-level-osmosis growth medium (6). As defects in cell polarity caused by inhibition of sphingolipid biosynthesis are not suppressible by high-level osmolarity (J. Cheng and X. Ye, unpublished observation), this indicates that sphingolipids do not mediate hyphal cell polarity through the cell wall integrity signaling pathway.
Polarized cell growth requires the polarized organization of the actin cytoskeleton in fungi. Actin immuno-staining showed that the actin cytoskeleton of the growing hyphae is normally organized in a highly polarized fashion. The actin patches are highly enriched towards the hyphal tip and form a discrete, intensely stained band right behind the growing tip. Inhibition of sphingolipid biosynthesis rapidly disrupts this pattern of actin organization, concomitant with the cessation of the normal polarized hyphal growth and clearly preceding the emergence of multiple short hyphal branches at or near the hyphal tip. These observations clearly demonstrate that sphingolipids are required for the polarized organization of the actin cytoskeleton, thus providing a molecular basis for their role in hyphal cell polarity.
Sphingolipids do not appear to be required for all functions of the actin cytoskeleton, however. For instance, in the absence of sphingolipid biosynthesis, the actin ring associated with septation formed normally, even several hours after the polarized organization of the actin cytoskeleton had been abolished. Additionally, the emergence of each hyphal branch at or near the hyphal tip in the absence of sphingolipid biosynthesis was always associated with an actin aggregate and required the actin function, as addition of CcA completely prevented branch emergence. This indicates that sphingolipids do not play a general role in actin function but are specifically required for the polarized organization of the actin cytoskeleton.
A role for sphingolipids in cell morphogenesis appears to be conserved in fungi. It is shown that inhibition of sphingolipid synthesis also causes marked changes in the morphology of S. cerevisiae and Schizosaccharomyces pombe cells, consistent with defects in polarized growth (8, 10, 14, 15, 44). However, whether the morphological defects of the yeast cells in the absence of sphingolipid synthesis also result from defects in the polarized organization of their actin cytoskeletons, like in A. nidulans shown in this study, remains to be established. While the effect of inhibiting sphingolipid synthesis on actin organization in S. pombe was not investigated (15), conflicting results on the effect of inhibiting sphingolipid synthesis on actin organization in S. cerevisiae were reported by the same group in two different studies (8, 14). Inactivating IPC synthase by promoter rundown from GAL1 promoter-controlled AUR1 showed no effect on the actin cytoskeleton organization, although cellular microtubules were completely depolymerized in the cells (14). On the other hand, treatment of S. cerevisiae cells with AbA to inactivate IPC synthase caused marked changes in the polarized distribution of cortical actin patches and the depolymerization of actin cables (8). Recently it was reported that the loss of LCB1 activity in S. cerevisiae also causes marked defects in the polarized organization of the actin cytoskeleton, which results in the loss of endocytosis activity of the cells (10, 44). Interestingly, the defects in actin organization and endocytosis caused by the loss of LCB1 activity in S. cerevisiae can be corrected by the addition of sphingoid bases, even in the absence of sphingolipid synthesis (44), or by increased protein phosphorylation (10). However, as shown in this study, sphingolipids but not sphingoid bases are required for polarized organization of the actin cytoskeleton in A. nidulans. Inactivation of IPC synthase, which does not affect the synthesis of sphingoid bases, rapidly promoted a dramatic reorganization of the normally highly polarized actin cytoskeleton in the growing hyphae of A. nidulans.
The mechanism by which sphingolipids regulate polarized organization of the actin cytoskeleton in the growing hyphae of A. nidulans is not understood at present. Mounting evidence shows that eukaryotic cells contain sphingolipid and cholesterol-rich membrane domains, called lipid rafts. The function of lipid rafts is currently of tremendous interest to cell biologists (4, 35). The concept of lipid rafts originated from a study of epithelial cell polarity to explain how lipids and lipid-anchored proteins are selectively directed to different surfaces of polarized cells (34). Sphingolipids differ from other phospholipids in that they contain long and saturated acyl chains that readily pack tightly together. One of the most important properties of lipid rafts is the selective inclusion or exclusion of certain proteins (4, 35). Perhaps similar sphingolipid-rich plasma membrane domains exist at the hyphal tip regions in filamentous fungi and they have a high affinity for anchoring proteins of the actin cytoskeleton, hence the polarized organization of the actin cytoskeleton. It is also interesting that inactivation of myoA, which encodes an essential type I myosin, generates polarity defects in A. nidulans similar to those caused by inhibition of sphingolipid biosynthesis (23). The MYOA protein is also localized as patches in the hyphal tip region (40), as with actin localization. However, it has not been determined if MYOA and the actin cytoskeleton colocalize with each other. Recent studies of both budding and fission yeasts show that type I myosins stimulate Cdc42-dependent actin assembly through interactions with the Arp2-Arp3 complex (19, 20). Type I myosin has a lipid-binding domain in the tail region (1). Conceivably, the lipid-binding domain may help localize MYOA to the sphingolipid-rich hyphal tip region, where it promotes actin assembly.
In summary, we show here that IPC synthase is required for cell cycle progression through G1. We further show that IPC synthase plays an important role in mediating the level of cellular ceramide and that accumulation of ceramide is likely responsible for G1 arrest of cells lacking IPC synthase activity. Additionally, we demonstrate that sphingolipids are essential for cell polarity in A. nidulans through polarized organization of the actin cytoskeleton. Future studies will be aimed at elucidating the molecular mechanisms of ceramide-mediated G1 arrest and the requirement for sphingolipids in the polarized organization of the actin cytoskeleton.
ACKNOWLEDGMENTS
We thank Robert Dean for performing the initial IPC synthase activity assays. We also thank Jeff Radding and members of X. S. Ye's lab for valuable discussions during the course of this work and Sheng-bin Peng and Donald LeBlanc for critically reading the manuscript.
REFERENCES
- 1.Adams R J, Pollard T D. Binding of myosin I to membrane lipids. Nature. 1989;340:565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- 2.Adams T H, Wieser J K, Yu J H. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergen L G, Upshall A, Morris N R. S-phase, G2, and nuclear division mutants of Aspergillus nidulans. J Bacteriol. 1984;159:114–119. doi: 10.1128/jb.159.1.114-119.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown D B, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 5.Buede R, Rinker-Schaffer C, Pinto W J, Lester R L, Dickson R C. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol. 1991;173:4325–4332. doi: 10.1128/jb.173.14.4325-4332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussink H J, Osmani S A. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus Aspergillus nidulans. FEMS Microbiol Lett. 1999;173:117–125. doi: 10.1111/j.1574-6968.1999.tb13492.x. [DOI] [PubMed] [Google Scholar]
- 7.Dickson R C. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- 8.Eno M, Takesako K, Kato I, Yamaguchi H. Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimcriob Agents Chemother. 1997;41:672–676. doi: 10.1128/aac.41.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischl A S, Liu Y, Browdy A, Cremesti A E. Inositolphosphoryl ceramide synthase from yeast. Methods Enzymol. 2000;311:123–130. doi: 10.1016/s0076-6879(00)11073-0. [DOI] [PubMed] [Google Scholar]
- 10.Friant S, Zanolari B, Riezman H. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 2000;19:2834–2844. doi: 10.1093/emboj/19.12.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannun Y A. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 12.Hanson B A, Lester R L. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J Lipid Res. 1980;21:309–315. [PubMed] [Google Scholar]
- 13.Harris S D, Hofmann A F, Tedford H W, Lee M P. Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics. 1999;151:1015–1025. doi: 10.1093/genetics/151.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashida-Okado T, Ogawa A, Endo M, Yasumoto R, Takesako K, Kato I. AUR1, a novel gene conferring aureobasidin resistance on Saccharomyces cerevisiae: a study of defective morphologies in Aur1p-depleted cells. Mol Gen Genet. 1996;251:236–244. doi: 10.1007/BF02172923. [DOI] [PubMed] [Google Scholar]
- 15.Hashida-Okado T, Yasumoto R, Endo M, Takesako K, Kato I. Isolation and characterization of the aureobasidin A-resistant gene, aur1R, on Schizosaccharomyces pombe: role of Aur1p+ in cell morphogenesis. Curr Genet. 1998;33:38–45. doi: 10.1007/s002940050306. [DOI] [PubMed] [Google Scholar]
- 16.Heidler S A, Radding J A. The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A ( LY295337) Antimicrob Agents Chemother. 1995;39:2765–2769. doi: 10.1128/aac.39.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins G M, Richards A, Wahl T, Mao G, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda M, Hashida-Okado T, Yasumoto R, Gomi K, Kato I, Takesako K. An aureobasidin A resistance gene isolated from Aspergillus is a homolog of yeast AUR1, a gene responsible for inositol phosphorylceramide (IPC) synthase activity. Mol Gen Genet. 1999;261:290–296. doi: 10.1007/s004380050969. [DOI] [PubMed] [Google Scholar]
- 19.Lechler T, Shevchenko A, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee W L, Bezanilla M, Pollard T D. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares function with WASp. J Cell Biol. 2000;151:789–799. doi: 10.1083/jcb.151.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao C, Saba J D, Obeid L M. The dihydosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- 22.Mathias S, Pena L A, Kolesnick R N. Signal transduction of stress via ceramide. Biochem J. 1998;335:465–480. doi: 10.1042/bj3350465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGoldrick C A, Gruver C, May G S. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 25.Momany M, Westfall P J, Abramowsky G. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics. 1999;151:557–567. doi: 10.1093/genetics/151.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris N R. The study of cytoskeletal proteins and mitosis using Aspergillus molecular genetics. Cell Motil Cytoskeleton. 1989;14:58–61. doi: 10.1002/cm.970140112. [DOI] [PubMed] [Google Scholar]
- 27.Nagiec M M, Nagiec E E, Baltisberger J A, Wells G B, Lester R L, Dickson R C. Sphingolipid synthesis as a target for antifungal drugs: complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 28.Nickels J T, Broach J R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 29.Oakley B R, Osmani S A. Cell-cycle analysis using the filamentous fungus Aspergillus nidulans. In: Fantes P, Brooks R, editors. The cell cycle, a practical approach. Oxford, United Kingdom: IRL Press; 1993. pp. 127–142. [Google Scholar]
- 30.O'Connell M J, Osmani A H, Morris N R, Osmani S A. An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO J. 1992;11:2139–2149. doi: 10.1002/j.1460-2075.1992.tb05273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osmani S A, Engle D B, Doonan J H, Morris N R. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell. 1988;52:241–252. doi: 10.1016/0092-8674(88)90513-2. [DOI] [PubMed] [Google Scholar]
- 32.Patton J L, Lester R L. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J Bacteriol. 1991;173:3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowland R R, Kervin R, Kuckleburg C, Sperlich A, Benfield D A. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 34.Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 35.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 36.Torralba S, Raudaskoski M, Pedregosa A M, Laborda F. Effect of cytochalasin A on apical growth, actin cytoskeleton organization and enzyme secretion in Aspergillus nidulans. Microbiology. 1998;144:45–53. doi: 10.1099/00221287-144-1-45. [DOI] [PubMed] [Google Scholar]
- 37.Waring R B, May G S, Morris N R. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 38.Weidner G, d'Enfert C, Koch A, Mol P C, Brakhage A A. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr Genet. 1998;33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- 39.Wells G B, Dickson R C, Lester R L. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J Biol Chem. 1998;273:7235–7243. doi: 10.1074/jbc.273.13.7235. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita R, Osherov N, May G S. Localization of wild type and mutant class I myosin proteins in Aspergillus nidulans using GFP-fusion proteins. Cell Motil Cytoskeleton. 2000;45:163–172. doi: 10.1002/(SICI)1097-0169(200002)45:2<163::AID-CM7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 41.Ye X S, Fincher R R, Tang A, O'Donnell K, Osmani S A. Two S-phase checkpoint systems, one involving the functions of both BIME and tyr15 phosphorylation of p34cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 1996;15:3599–3610. [PMC free article] [PubMed] [Google Scholar]
- 42.Ye X S, Fincher R R, Tang A, Osmani A H, Osmani S A. Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA. Mol Biol Cell. 1998;9:3019–3030. doi: 10.1091/mbc.9.11.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye X S, Xu G, Fincher R R, Osmani S A. Characterization of NIMA protein kinase in Aspergillus nidulans. Methods Enzymol. 1997;283:520–532. doi: 10.1016/s0076-6879(97)83041-8. [DOI] [PubMed] [Google Scholar]
- 44.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson B J, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]