Abstract
This study examines 2 gene-level metrics of variation constraint against protein-truncating variants—probability of loss-of-function intolerance and loss-of-function observed/expected upper bound fraction—and their association with the psychiatric consequences of both recurrent and nonrecurrent gene deletions.
Copy number variants (CNVs) are key etiological contributors to neuropsychiatric disorders. Most psychiatric CNV studies have focused on several dozen loci,1 collectively comprising less than 2% of the genome, where CNVs spontaneously recur sufficiently often to have individually detectable psychiatric associations. We hypothesized that knowledge of gene function could guide the search for nonrecurrent CNVs across the remaining 98%. Specifically, probability of loss-of-function intolerance (pLI) and loss-of-function observed/expected upper bound fraction (LOEUF),2 2 gene-level metrics of variation constraint against protein-truncating variants, have been reported to be uniquely associated with the cognitive consequences of CNVs.3 Here, we show that pLI and LOEUF are similarly associated with the psychiatric consequences of both recurrent and nonrecurrent gene deletions.
Methods
We studied 431 146 self-reported White UK Biobank participants (234 544 females) with International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD-10) codes from linked inpatient, primary care, or death records, excluding participants with neurodevelopmental disorders (ICD-10 codes F70-F89) or who failed CNV quality control (eMethods in the Supplement). The North West Centre for Research Ethics Committee granted ethical approval to UK Biobank, and informed consent was obtained from participants.4 We stratified genes into 8 categories based on Genome Aggregation Database’s pLI scores (low [0-<0.5], medium [0.5-<0.9], high [0.9-<0.99], or extreme [0.99-1]) and recurrence type (recurrent for genes overlapping any of 32 previously defined1 recurrent neuropsychiatric deletion CNV loci [eTable in the Supplement] and nonrecurrent otherwise). For each category, we used Firth logistic regression to test whether carriers of CNVs deleting any gene in the category had higher rates of anxiety disorders (F40/F41), bipolar disorder (F31), major depressive disorder (MDD) (F32/F33), obsessive-compulsive disorder (OCD) (F42), and schizophrenia (F20).
To guard against incorrectly associating the consequences of one CNV category to another, we excluded participants with recurrent deletions when computing associations for nonrecurrent deletions. We also excluded participants with higher-pLI deletions when computing associations for lower-pLI deletions of the same recurrence type. To reduce false-positives, we analyzed CNV calls from 2 different computational pipelines and required them to agree that a gene was fully deleted. As a sensitivity analysis, we replaced pLI with LOEUF, using thresholds capturing similar numbers of genes (low: ≥0.5; medium: 0.4-<0.5; high: 0.3-<0.4; extreme: 0-<0.3).
Results
Nonrecurrent CNVs deleting extreme-pLI genes were found in 787 participants (0.2%). A total of 571 unique extreme-pLI genes were deleted by nonrecurrent CNVs in 1 or more participants (Table), including key neurotransmitter receptors, ion channels, and neurodevelopmental genes.
Table. Nonrecurrent Extreme-pLI Gene Deletion Frequencies.
| Gene | No. (%)a |
|---|---|
| MAPK8 b | 56 (0.013) |
| THOC1 | 20 (0.0046) |
| TJP1 | 19 (0.0044) |
| RANBP2 | 17 (0.0039) |
| INTS6, NXN | 13 (0.0030) |
| BCAR1 | 12 (0.0028) |
| CNOT7, FNDC3A | 11 (0.0026) |
| RB1 b | 10 (0.0023) |
| COL6A1, SMCHD1 | 9 (0.0021) |
| DOPEY1, ELFN1,b TTYH3 | 8 (0.0019) |
| DAAM2, KDM5A, UHRF2, ZNF496 | 7 (0.0016) |
| DLGAP2,b INTS10, PDIA3, PELP1, PRPF39, TRIP13, WNK1 | 6 (0.0014) |
| DLGAP1,b FMN2, LZTS3, NCOR1, SIM1, SMARCA2,c TENM3 | 5 (0.0012) |
| BANP, CACTIN, CSMD1, CSNK1G1, FAM131B, FLT4, FZR1, GSE1, KIAA0947, LMX1A, PTGS2, RAB11FIP3, TERT | 4 (0.0009) |
| AJAP1, ATP13A3, ATP6V1B2, BHLHE40, BMPR1B, CCNA2, CECR2, CENPB, CFLAR, CHRM3,b COL11A1, CPE, DBF4, DCUN1D5, DLC1, EFEMP1, FAM160A2, FNIP1, GLG1, GMPS, GNB2L1, GRIK2,b GSPT1,b HECTD1, IVNS1ABP, KIF26B, KIF3C, LRCH1, MICAL3, NCOA5, PDE10A,b PLXNA1, PLXNB2, PLXND1, PRICKLE1, PSME4, PTBP1, PTPRA,b RERE,c RSPRY1, SH3GLB1, SLC6A3,b SMARCAD1, SMC2, SUGP2, TPR, TTBK2 | 3 (0.0007) |
| ACTR3B, ADAM10,b AFF3, AGFG1, AHCYL2, AHR, ARHGAP42, ASXL2, ATRN, BRINP2,b BRWD1, C17orf85, CADM4, CAND1, CAPZA2, CCNL1, CELF5, CEP170B, CHAMP1,c CHD3,c CNTNAP5, COG3, CPSF6, CUL4A, CUX1,b DKFZP761J1410, DLL4, DMXL1, DNAJC2, DYRK2, EDNRA, EIF5B, ELMSAN1, ETS2, EXOC8, EXT1, EYA3, FAM135A, FBXO38,b FLNC, FRMD5, FRY, FURIN, GGA1, GRID2,b GRM7,b HCN1, HIC2, INO80, INPP4A, ISY1, ISY1-RAB43, ITGB8, ITPR1,c JAG2, JPH3, KCNN2,b KDM6B,c KIF1B,b KLHL2, LRP2, LRRC8B, MAN1A2, MDM2, MKL2, NAV2, NCOA1,c NFAT5, NRIP1, PBX2, PCIF1, PICALM,b PIK3R1, PITPNM3, PRKCD,b PRKG1,b PRPF8, PRR14L, PRRC2B, PSIP1, PSMC2, PSMD11, PSMD13, PTPN12, RBM26, REV1, RFX3,c RFX4, RFX7, RIC8B, ROR1,b SBF1, SEMA6A,b SIPA1L2, SLC12A5,b SLC44A1, SLTM, SMAP1, SRGAP1, STIM2, SYVN1, TERF2, TIPARP, TMEM259, TP63, TRAF6, TRIM2, UBE4B, USP15, USP8, VPS54, XPO4, YAP1, ZBTB44, ZC3H13, ZMYND8,c ZNF654 | 2 (0.0005) |
| ABCA2, ADAMTS6, ADD2, AFTPH, AGO2, AGPAT3, AGPS, AHCTF1, AMBRA1, ANKIB1, ANKRD52, ANKS1B, AP1B1, AP2B1, APPBP2, AQR, ARFGEF2,b ARHGAP29, ARHGAP5, ARHGEF12, ARID2,c ARID3A, ARNT2, ASAP1, ASAP2, ASIC2, ASTN1, ASTN2, ATAD2B, ATP2C1, B4GALT5, BAI3, BAZ2A, BOD1L1, BTBD11, C11orf84, C7orf60, CACNA1I,b CAD, CAMTA2, CASKIN2, CBLL1, CCDC88A, CCNC, CCT2, CDH11, CEP350, CEPT1, CHD1, CHD5, CHD6, CHRM1,b CLASP1,c CLASP2,b CLCN3, CLOCK, CNNM2, COL1A2, COL3A1, CORO1C, CPSF7, CREB1, CSE1L, CSNK1G2, CSNK1G3, CTCF,c CTR9, CUL1, CUL3,c DACH1, DAGLA,b DAPK1, DCAF15, DCC,b DDB1, DDX21, DDX4, DNAJC13, DNAJC6, DNM2, DPF2, DPYSL5,b EED, EEF1G, EIF3D, EIF4ENIF1, ELL2, EP400, EPB41L4B, EPHA7,b ERBB4,b ERP44, ESRP1, EWSR1, EZH2,b FAM160B1, FAM208A, FAT4, FBN2, FBXL17, FBXL5, FBXO33, FBXO41, FBXW2, FBXW7, FGFR2, FNDC3B, FOXJ2, FOXO1, FOXP1,c FRS2, FRYL, GABBR1,b GABRA2,b GALNT13, GANAB, GCLM, GIT2, GNAL, GPS1, GRIN2D,b GRM5,b HGF, HIPK1, HIVEP1, HMBOX1, HMGCR, HMGCS1, HNRNPUL2, HOOK3, HTT, ING3, IPO5, ITPKB, JAK1, JAKMIP2, KCNB1,c KCNH2,b KCNH3,b KDM3A, KDM4B, KDR, KIAA0368, KIAA1429, KIF13A, KIF2A,b KIRREL, KMT2C,c KPNA1, L3MBTL3, LARP4B,c LATS1, LEPR, LPHN1, LPHN2, LRRC4,b MAGI2, MAP1A,c MAP2,b MAP2K4, MAP3K1, MAP4K5, MAPK8IP2,b MAPK8IP3,b MAPKAPK2, MARK2,b MAT2A, MCMBP, MGA, MGAT5, MINK1, MKLN1, MKRN1, MLLT4, MLLT6, MORC2, MPPED2, MRC1L1, MSL1, MTA1, MTF2,c MTMR12, MTMR3, MTPAP, MUC5B, MYCBP2,b MYH10, MYH9, MYO16, NAP1L1, NASP, NCAM1,b NCKAP1,c NELL2, NEURL4, NF2, NFATC3, NFIA, NFKB1, NFKB2, NFKBIA, NISCH, NOL6, NOTCH2,b NRBP1, NRF1, NTNG2,b OLFM1, PACS1,c PACS2,c PAIP1, PAPOLG,c PBRM1, PCGF1, PDS5B, PDZD2, PHC1, PHF19, PHF3,c PIAS2, PIK3CD, PITPNM2, PKN2, PLCG2, PLK2,b PLXNA4, PLXNC1, PNISR, POLR1A, PPARGC1A, PPM1A, PPP1R16B, PPP3CA, PPP3CB, PPP6R3, PPRC1, PRDM16, PREX1, PRKAG2, PRKCG,b PRPF40B, PRPF4B, PSMC6, PSPC1, PTK2,b PTPN4, PTPRM,b PTPRT,b RABL6, RAD21, RALGAPB, RANBP3, RAPGEF6, RB1CC1, RBM14, RBM17, RBM4, REL, RELN,c REST, RFWD2, RFX2, RNF111, RNF2, RNF31, RORB,c RP11-159G9.5, RPL5, RPS6KA5, SART3, SENP6, SERBP1, SF3B2, SF3B3, SFSWAP, SGIP1, SIK2, SKIDA1,c SLC4A4, SLC8A1, SLIT2, SMARCA5, SMARCB1, SMG7, SNAP91, SOX30, SRP54, SSH2, ST18, ST7, STAT1, STAU1, STK39, STXBP5, SUFU, SVEP1, SWT1, TAF5, TBX21, TBX5, TEK,c TENM2,b TEX10, TFAP4, TFCP2L1, TFDP2, THBS1, TLK1, TMEM108, TMEM201, TMEM57, TNIK,b TOP1, TOPBP1, TP53BP1, TP73, TRAF2, TRAPPC10, TRAPPC8, TRIM27, TRIM28, TRIM71, TRIM8, TRIP12,c TRPS1, TSKS, UBN2, UBP1, UCHL1,b ULK1, UNK, USP1, USP42, WDR18, WNK2, WWP1, XPO7, XPOT, XPR1, YLPM1, YTHDC1, YTHDC2, ZBTB16, ZBTB21, ZBTB38, ZC3H12C, ZC3H18, ZC3H7A, ZCCHC6, ZFHX4, ZFR, ZMYND11,c ZNF131, ZNF236, ZNF445, ZNF541, ZNF638, ZNF770, ZNF800, ZNFX1, ZRANB1 | 1 (0.0002) |
Abbreviations: CNV, copy number variant; pLI, probability of loss-of-function intolerance.
Number and percentage of participants for each individual extreme-pLI gene at a nonrecurrent CNV locus. Frequencies are lower than true population prevalences due to the UK Biobank’s healthy participant bias and the exclusion of participants with diagnosed neurodevelopmental disorders.
Manually annotated genes of special interest (eg, ion channel genes, neurotransmitter receptors/transporters, genes involved in neurodevelopment).
Neurodevelopmental disorder genes are listed in the eMethods in the Supplement.
pLI and LOEUF scores were associated with psychopathogenicity (Figure). The 787 participants with nonrecurrent extreme-pLI gene deletions exhibited significantly higher rates of anxiety (odds ratio [OR], 1.45 [95% CI, 1.11-1.88]), bipolar disorder (OR, 2.78 [95% CI, 1.24-6.23]), MDD (OR, 1.44 [95% CI, 1.14-1.81]), OCD (OR, 6.75 [95% CI, 1.25-36.31]), and schizophrenia (OR, 3.79 [95% CI, 1.45-9.94]). Conversely, the 54 413 participants (12.6%) with nonrecurrent low-pLI gene deletions displayed no greater rates of any disorder: anxiety (OR, 0.99 [95% CI, 0.96-1.03]), bipolar disorder (OR, 0.99 [95% CI, 0.87-1.13]), MDD (OR, 1.00 [95% CI, 0.97-1.03]), OCD (OR, 0.95 [95% CI, 0.76-1.19]), or schizophrenia (OR, 0.96 [95% CI, 0.82-1.13]).
Figure. Association of Loss-of-Function–Intolerant Gene Deletions With Risk of 5 Psychiatric Disorders.
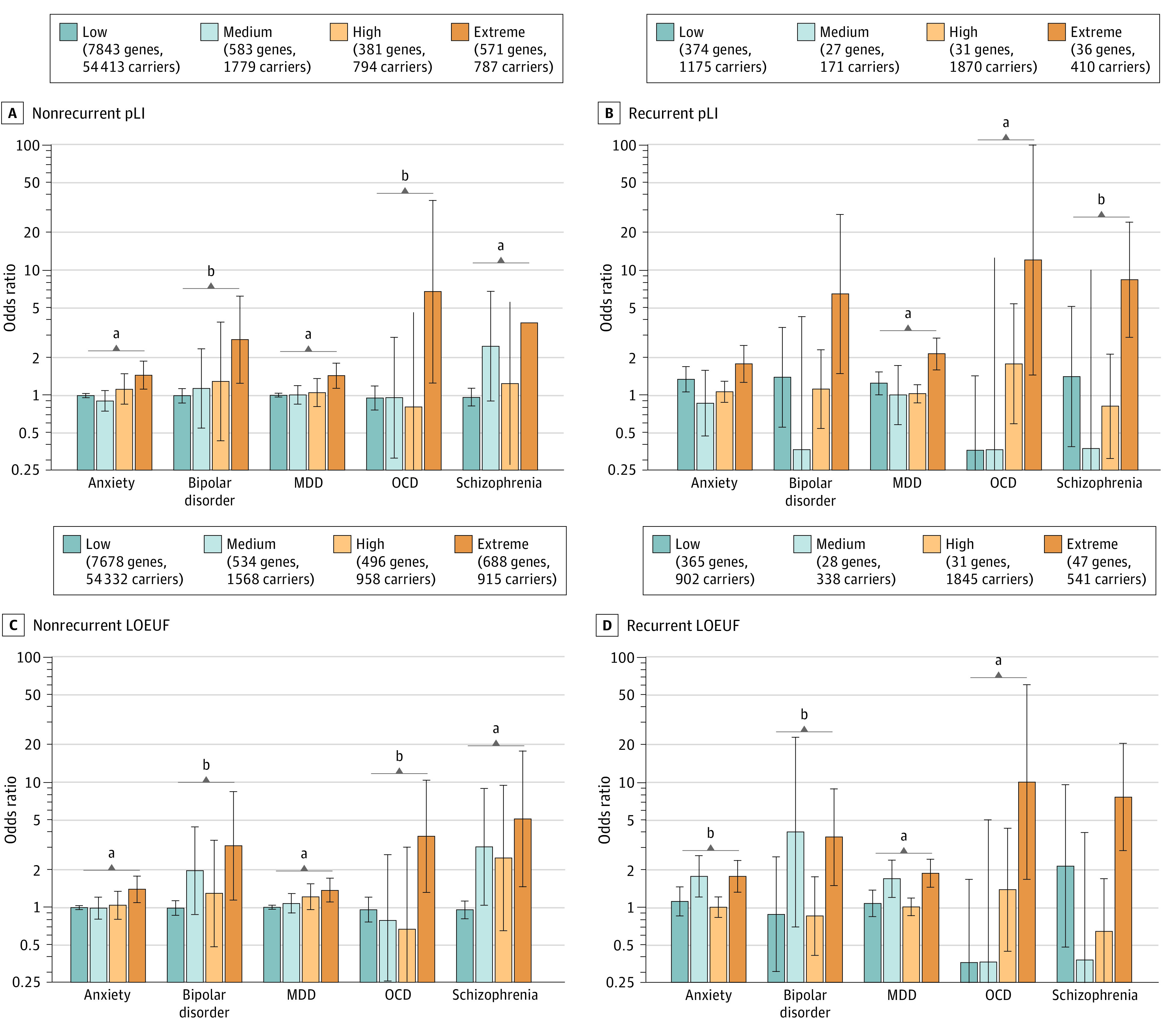
Gene deletions were stratified by probability of loss-of-function intolerance (pLI) (A and B) or loss-of-function observed/expected upper bound fraction (LOEUF) (C and D) and associated with diagnoses of the 5 psychiatric disorders: anxiety (28 820 cases, 340 465 controls), bipolar disorder (1665 cases, 407 363 controls), major depressive disorder (MDD; 35 906 cases, 318 417 controls), obsessive-compulsive disorder (OCD; 703 cases, 429 642 controls), and schizophrenia (827 cases, 407 614 controls). The figure keys indicate how many genes in each category are deleted in at least 1 participant, as well as how many participants carry deletions of at least 1 gene in the category (after excluding participants with recurrent deletions for nonrecurrent categories and excluding participants with higher-pLI deletions for lower-pLI categories of the same recurrence type, as described in the Methods section).
aP < .01 for statistically significant differences between extreme- and low-pLI odds ratios.
bP < .05 for statistically significant differences between extreme- and low-pLI odds ratios.
pLI and LOEUF were also associated with CNV psychopathogenicity. Participants with recurrent CNVs deleting extreme-pLI genes had substantially higher rates of anxiety (OR, 1.78 [95% CI, 1.27-2.51]), bipolar disorder (OR, 6.46 [95% CI, 1.49-28.02]), MDD (OR, 2.15 [95% CI, 1.60-2.88]), OCD (OR, 12.07 [95% CI, 1.45-100.77]), and schizophrenia (OR, 8.39 [95% CI, 2.89-24.37]). Conversely, participants with recurrent deletions of low-pLI genes had only modestly increased risk of anxiety (OR, 1.34 [95% CI, 1.06-1.70]) and MDD (OR, 1.25 [95% CI, 1.01-1.54]) and did not have significantly altered risk of bipolar disorder (OR, 1.39 [95% CI, 0.56-3.50]), OCD (OR, 0.36 [95% CI, 0.09-1.42]), or schizophrenia (OR, 1.41 [95% CI, 0.39-5.13]).
Discussion
While recurrent CNVs are well-known contributors to psychopathology,1 the current study showed that nonrecurrent CNVs are associated with psychiatric disease risk. Gene-level metrics of mutational constraint were associated with psychopathogenic CNVs, both recurrent and nonrecurrent. The 0.2% of participants with nonrecurrent deletions of extreme-pLI genes had significantly higher rates of all 5 psychiatric disorders surveyed, whereas participants with nonrecurrent deletions of only low-pLI genes (the vast majority) had no detectable increase in psychiatric disease risk. Limitations of this study include microarray-based CNV calling, incomplete phenotype ascertainment, and ignoring partial gene deletions, as well as the inability to generalize these findings to other racial and ethnic groups.
These results suggest that interpreting CNVs using mutational constraint metrics, such as pLI, may augment population-based psychiatric genomic screening programs.5 Our approach may ultimately help identify opportunities for early diagnosis and intervention, including personalized therapies targeting specific nonrecurrent CNVs.6
eMethods
eTable. The 32 Recurrent Deletion CNV Regions From Kendall et al. 2019.
eReferences
References
- 1.Kendall KM, Rees E, Bracher-Smith M, et al. Association of rare copy number variants with risk of depression. JAMA Psychiatry. 2019;76(8):818-825. doi: 10.1001/jamapsychiatry.2019.0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285-291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huguet G, Schramm C, Douard E, et al. Genome-wide analysis of gene dosage in 24,092 individuals estimates that 10,000 genes modulate cognitive ability. Mol Psychiatry. 2021;26(6):2663-2676. doi: 10.1038/s41380-020-00985-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203-209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin CL, Wain KE, Oetjens MT, et al. Identification of neuropsychiatric copy number variants in a health care system population. JAMA Psychiatry. 2020;77(12):1276-1285. doi: 10.1001/jamapsychiatry.2020.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodkin JA, Coleman MJ, Godfrey LJ, et al. Targeted treatment of individuals with psychosis carrying a copy number variant containing a genomic triplication of the glycine decarboxylase gene. Biol Psychiatry. 2019;86(7):523-535. doi: 10.1016/j.biopsych.2019.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eTable. The 32 Recurrent Deletion CNV Regions From Kendall et al. 2019.
eReferences


