Abstract
We report de novo genome assemblies, transcriptomes, annotations, and methylomes for the 26 inbreds that serve as the founders for the maize nested association mapping population. The number of pan-genes in these diverse genomes exceeds 103,000, with approximately a third found across all genotypes. The results demonstrate that the ancient tetraploid character of maize continues to degrade by fractionation to the present day. Excellent contiguity over repeat arrays and complete annotation of centromeres revealed additional variation in major cytological landmarks. We show that combining structural variation with SNPs can improve the power of quantitative mapping studies. Finally, we document variation at the level of DNA methylation, and demonstrate that unmethylated regions are enriched for cis-regulatory elements that contribute to phenotypic variation.
One sentence summary:
A multi-genome analysis of maize reveals previously unknown variation in gene content, genome structure, and methylation.
Maize is the most widely planted crop in the world and an important model system for the study of gene function. The species is known for its extreme genetic diversity, which has allowed for broad adaptation throughout the tropics and intensive use in temperate regions. Nevertheless, most current genomic resources are referenced to a single inbred, B73, which contains only 63-74% of the genes and/or low-copy sequences in the full maize pan-genome (1–4). Moreover, there is extensive structural polymorphism in non-coding and regulatory genomic regions that has been shown to contribute to variation in numerous traits (5). In recent years, additional maize genomes have been assembled, allowing limited characterization of the species pan-genome (2, 6–10). However, comparisons across genome projects are often confounded by differences in assembly and annotation methods.
The maize Nested Association Mapping (NAM) population was developed to study the genetic architecture of quantitative traits (11). Twenty-five founder inbred lines were strategically selected from a larger association panel (12) to represent the breadth of maize diversity, including lines from the non-stiff-stalk temperate heterotic group, lines from tropical and subtropical regions of Africa, Asia, and the Americas, and both sweet corn and popcorn germplasm (13). Each NAM parental inbred was crossed to B73 and selfed to generate 25 populations of 200 recombinant inbred lines that combine the advantages of linkage and association mapping for important agronomic traits (14). Biological infrastructure continues to be developed around these lines (e.g. (15, 16)), but comprehensive genomic resources are needed to fully realize the power of the NAM population.
Consistency and quality of genome assemblies
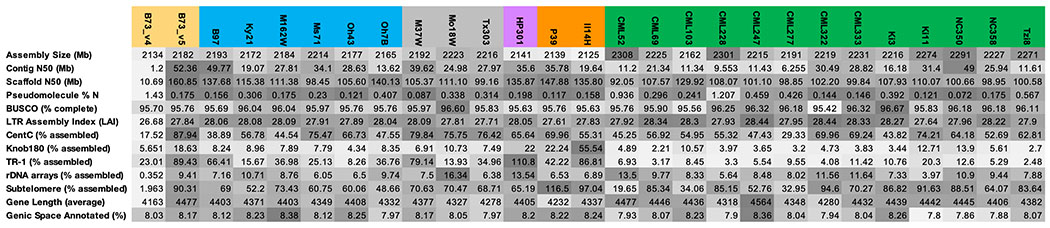
Here we describe assembled and annotated genomes for the 25 NAM founder inbreds and an improved reference assembly of B73 (Table S1). The 26 genomes were sequenced to high depth (63-85X) using PacBio long-read technology, assembled into contigs using a hybrid approach (17), scaffolded using Bionano optical maps, and ordered into pseudomolecules using linkage data from the NAM recombinant inbred lines and maize pan-genome anchor markers (4). Assembly and annotation statistics improve upon nearly all available maize assemblies, including the previous B73 reference genome (18), with the total length of placed scaffolds (2.102-2.162 Gbp) at the estimated genome size of maize, a mean scaffold N50 of 119.2Mb (contig N50 of 25.7 Mbp), complete gene space (mean of 96% complete BUSCOs; (19)), and, based on the LTR Assembly Index (LAI, mean of 28; (20)), full assembly of the transposable-element-laden portions of the genome (Table 1; Table S2). Improvements in contiguity and completeness can be attributed to recent advances in sequence and optical map data, as well as more effective assembly algorithms (21).
Table 1:
Quality metrics for genome assemblies and gene model annotations. Darker shading indicates higher quality. The NAM lines are shaded based on their primary grouping (gold = stiff stalk heterotic group, blue = non-stiff-stalk heterotic group, gray = mixed tropical-temperate ancestry, purple = popcorn, orange = sweet corn, green = tropical).
|
Hp301 and P39 have the lowest amounts of TR-1 and subtelomere repeats, respectively. Our methods can overestimate assembly when repeats are in low abundance (17).
Gene identification and diversity in gene content
We sequenced mRNA from ten tissues for each inbred. These data were used for evidence-based gene annotation of each line, which was then improved using B73 full-length cDNA and expressed sequence tags (ESTs). The evidence set was augmented with ab initio gene models and the gene structures refined for all accessions using phylogeny-based methods. This pipeline revealed an average of 40,621 (standard error (SE) = 117) protein-coding and 4,998 (SE = 100) non-coding gene models per genome. The great majority of genes share orthologs with the grass (Poaceae) family and species in the Andropogoneae tribe of grasses, which includes maize and sorghum (Fig. 1A). The accuracy of the annotations, measured by the congruence between annotations and supporting evidence (Annotation Edit Distance, AED) (22), is higher than previous reference maize annotations (Fig. S1) (2, 6, 10, 18, 23).
Figure 1.
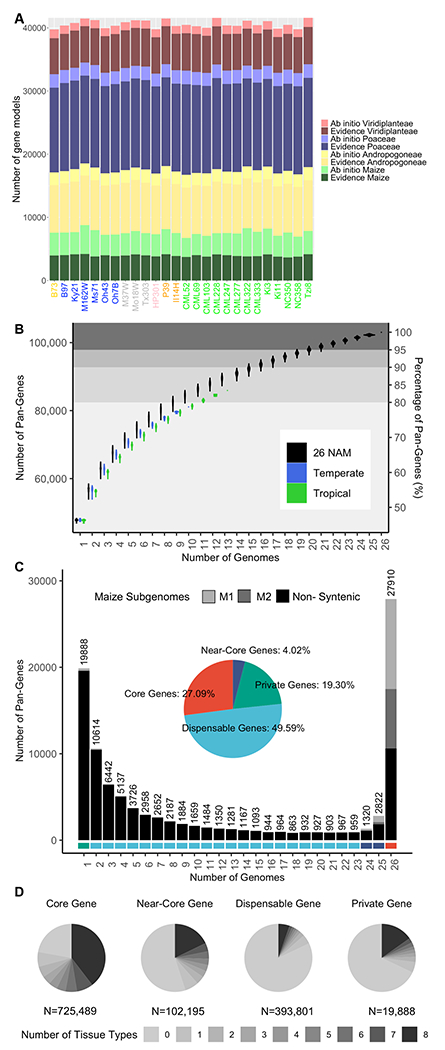
Pan genome analysis of the gene space. A) Pan-genes categorized by annotation method and phylostrata. Genes annotated with evidence have mRNA support whereas ab initio genes are predicted based on DNA sequence alone. Genes within progressing phylostrata - species Zea mays (maize), tribe Andropogoneae, family Poaceae, kingdom Viridiplantae - are more conserved. B) Number of pan-genes added with each additional genome assembly. Order of genomes being added into the pan genome was bootstrapped 1000 times. Tropical lines include (CML52, CML69, CML103, CML228, CML247, CML277, CML322, CML333, Ki3, Ki11, NC350, NC358, Tzi8), temperate lines include (B73, B97, Ky21, M162W, Ms71,Oh43, Oh7B, HP301, P39, and Il14H). C) Proportion of pan-genes in the core, near core, dispensable, and private fractions of the pan-genome. For B and C, tandem duplicates were considered as a single pan-gene and coordinates were filled in when a gene was not annotated, but an alignment with greater than 90% coverage and 90% identity was present within the correct homologous block. D) Number of tissues with expression (RPKM>1) for each gene in each genome based on their pan-genome classification. Tissues in this analysis include (root, shoot, V11 base, V11 middle, V11 tip, anther, tassel, and ear).
We next assessed the gene catalog of the pan-genome. Genes with high sequence similarity, located within blocks of homologous sequence in pairwise comparisons, were grouped together as one pan-gene. In many instances, a gene was not annotated by our computational pipeline, yet at least 90% of the gene was present in the correct homologous location; when this occurred, the pan-gene was considered present (Fig. S2 A; (17)), even though in some cases the absence of annotation may reflect fractionation and/or pseudogenization.
Across the 26 genomes, a total of 103,033 pan-genes were identified. Previous analysis reported ~63,000 pan-genes based on transcriptome assemblies of seedling RNA-seq reads from 500 individuals (1). The superior contiguity of our assemblies and the application of both ab initio and evidence-based annotation using RNA-seq from a diverse set of ten tissues, likely accounts for the increased sensitivity. Over 80% of pan-genes were identified within just ten inbred lines based on a bootstrap resampling of genomes (Fig. 1B). When considered separately, temperate and tropical lines have differentiated sets of pan-genes but show a comparable rate of pan-gene increase as lines are added, suggesting they have similar gene-content diversity (Fig. 1B).
Pan-genes, excluding tandem duplicates (17), were classified as core (present in all 26 lines), near-core (present in 24-25 lines), dispensable (present in 2-23 lines), and private (present in only one line) (Fig. 1C). The portion of genes classified into each of these groups was consistent across genotypes, with an average of 58.41% (SE = 0.07%) belonging to the core genome, 8.23% (SE = 0.05%) to the near-core genome, 31.75% (SE = 0.09%) to the dispensable genome, and 1.60% (SE = 0.08%) private genes (Fig. 1C; Fig. S2 B–C; Table S3). In total, there are 32,052 genes in the core/near-core portion of the pan-genome and 70,981 genes in the dispensable/private portion. The core genes (and gene families enriched for core genes (Table S4)) are generally from higher phylostrata levels (i.e. Viridiplanteae and Poaceae), while those in the near-core and dispensable sets either share orthologs only with closely related species or are maize-specific (Fig. S2 E). Some private genes may be spurious annotations resulting from imperfect masking of repeat sequences, as the majority of core/near-core genes are syntenic to sorghum (57.78%), whereas this is rarely the case for dispensable/private genes (1.83% syntenic). Core genes were expressed in more tissues (Fig. 1D) and had higher transcript abundance (Fig. S2F) when compared to genes present in fewer individuals. However, across the relatively small number of tissues (≥ 8 per line) profiled for this analysis, 18% of dispensable and 32% of private genes were expressed in at least one tissue. A total of 16,751 pan-genes were tandemly duplicated in at least one genome, of which 7,040 were duplicated in a single genome. On a per gene basis in genomes with at least one tandem duplicate the average copy number is 2.20 (SE = 0.01) (Fig. S2 D).
Partial tetraploidy and tempo of fractionation
The maize ancestor underwent a whole-genome duplication (WGD) allopolyploidy event 5-20 MYA ((24, 25), Fig. 2A). Evidence for WGD is found in the existence of two separate genomes that are broken and rearranged, yet still show clear synteny to sorghum (24, 26). Many duplicated genes have since undergone loss, or fractionation, reducing maize to its current diploid state (26, 27). Further, fractionation is biased towards one homoeologous genome (M2, more fractionated) over the other (M1, less fractionated) (26). The M1 and M2 subgenomes are composed almost exclusively of core (87.25%) and near-core (6.19%) pan-genes (Figs. 1C, 2A). The broad architecture of syntenic regions relative to sorghum is consistent across the NAM genomes (Fig. S3).
Figure 2.
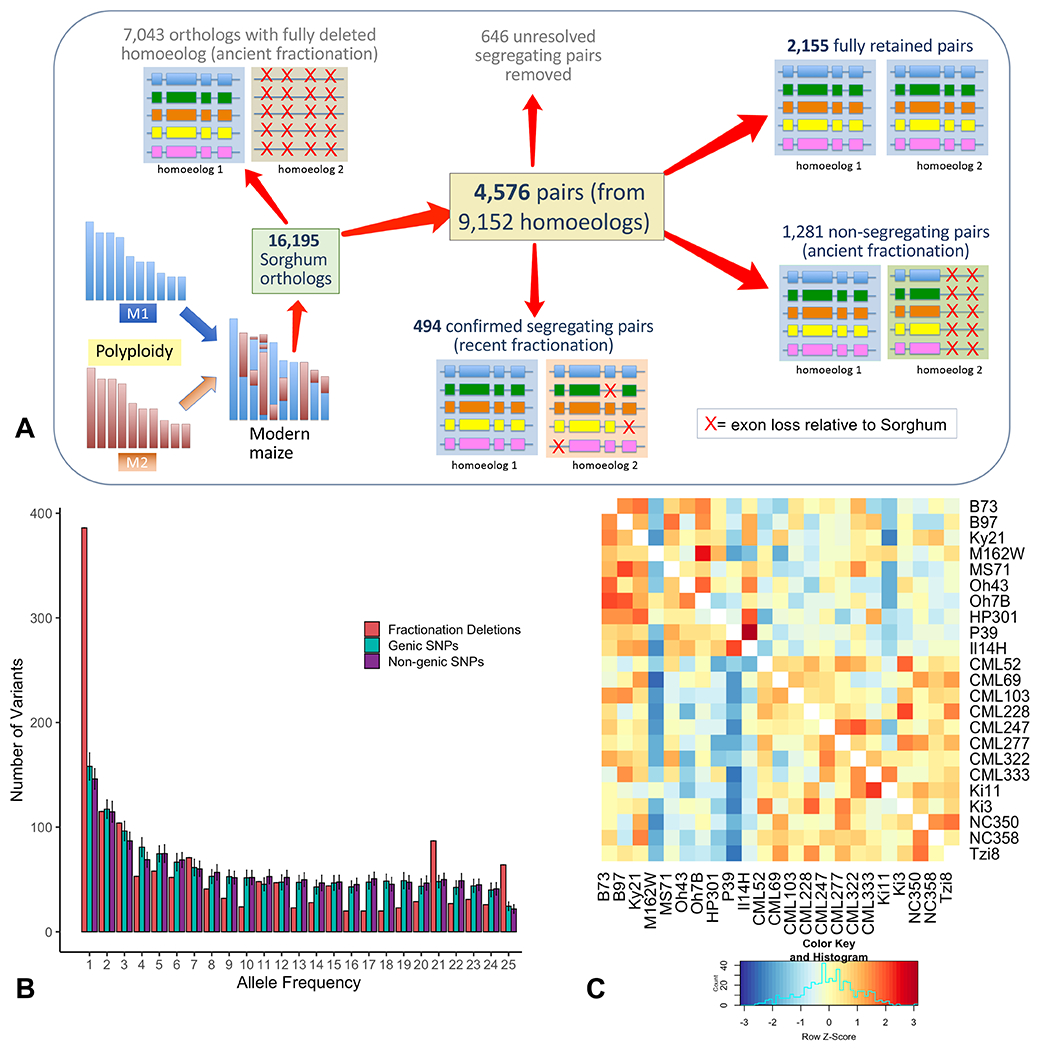
The tempo of fractionation in maize. A) Schematic showing how genes were categorized. 16,195 conservatively chosen orthologs were subdivided into classes representing retained pairs, ancient fractionation, and recent fractionation. B) Unfolded site frequency spectrum (SFS) of segregating exon loss and non-coding SNPs (genic and non-genic) using sorghum to define the ancestral state. C) Heatmap of the number of co-retained exons between any two NAM lines. Lines with mixed ancestry (M37W, Mo18W, Tx303) are excluded. Colors indicate the Z-score (the difference measured in standard deviations between a single pairwise comparison and all others in the row).
Given the ancient timeframe of the WGD in maize and the rapid tempo of fractionation observed in other species (28, 29), little variation in the retention of specific homoeologs is expected at the species level. In fact, prior work in temperate maize suggested that most fractionation occurred before domestication (6, 30). However, our diverse set of genomes allows for a more complete characterization of fractionation within the species. Since fractionation can occur at the level of small deletions (27, 31), we evaluated both partial and complete homoeolog loss beginning with a conservative set of 16,195 maize pan-orthologs. We determined that 7,043 were single-copy orthologs, where the homoeologous gene was likely deleted prior to maize speciation (Fig. 2A). In addition, we identified 4,576 homoeologous pairs (Fig. 2A) of which 2,155 had the same exon structure of the sorghum ortholog in both homoeologs. In 1,281 pairs, at least one copy of the gene differed from its sorghum ortholog, but did not vary among NAM lines, likely representing fractionation that pre-dated Zea mays. Another 1,140 pairs varied across the genomes in their pattern of exon retention, segregating for deletions or structural differences in at least one copy of the gene. This segregating set was manually curated (Dataset S1) to remove loci where exons or flanking sequence could not be confidently identified (Fig. 2A), resulting in a curated set of 494 homoeolog pairs segregating for fractionation, which represents more than 10% of pairs present in the pan-genome. Of these, 281 M2 homoeologs had exon loss compared to 236 M1 homoeologs, a 19% difference (p < 0.05, χ2 test), suggesting ongoing biased fractionation. Analysis of gene ontology terms revealed putative functional differences between fully fractionated and segregating fractionated loci (Fig. S4, Dataset S1).
Population genetic theory predicts mutations segregating within a species, like the segregating fractionation deletions we have identified, arose within the last 4Ne generations, where Ne represents the effective population size of the species. Using the Ne of the maize progenitor teosinte as an upward bound for maize (Ne = 150,000; (32)), we can infer that the majority of segregating fractionation arose within the last 600,000 generations. Therefore, the majority of segregating fractionation substantially post-dates the WGD. Theory also predicts that rare deletions should be younger than those segregating at intermediate frequency. We constructed the unfolded site frequency spectrum (SFS) of segregating fractionation deletions and compared this to the unfolded SFS of non-coding SNPs using sorghum to define the ancestral state (Fig. 2B). The data reveal a similar frequency distribution in deletions and SNPs, with a preponderance of rare variants in both, suggesting that a subset of fractionation may be quite young, with diploidization potentially continuing in modern maize. We also evaluated patterns of co-exon-retention in non-stiff-stalk temperate, tropical, and flint-derived maize, observing population-specific fractionation (Fig. 2C). This variation in homoeolog retention at the population level confirms previous suppositions about the tempo of fractionation (33) and may reflect relaxed constraint on retained homoeologs following domestication and migration of maize to temperate climates.
The repetitive fraction of the pan-genome
Transposable elements (TEs) were annotated in each assembly using structural features and sequence homology (34). Individual TE libraries from each inbred were then combined to form a pan-genome library, which was used to identify TE sequences missed by individual libraries. The annotations reveal that DNA transposons and LTR retrotransposons comprise 8.5% and 74.4% of the genome, respectively (Table S5, Fig. S5). A total of 27,228 TE families were included in the pan-genome TE library, of which 59.7% were present in all 26 NAM founders and 2.5% were unique to one genome (Fig. S6). The average percentage of intact and fragmented TEs were 30.5% and 69.5% (SE = 0.06%), respectively. As reported previously, Gypsy LTR retrotransposon families are more abundant in pericentromeric regions, while Copia LTR retrotransposons are enriched in the gene-dense chromosome arms (Fig. S7) (35). Tropical lines have significantly more Gypsy elements than temperate lines (p = 0.002, t-test), with mean Gypsy content of 1,018 Mbp and 988 Mbp, respectively (Table S5, Fig. S5). This may reflect increasing constraint on Gypsy proliferation in temperate lines that have, on average, smaller genomes (Table 1).
In some maize lines, over 15% of the genome is composed of tandem repeat arrays including the centromere repeat CentC, the two knob repeats knob180 and TR-1, subtelomere, and telomere repeats (36, 37). Repeats of this type remain a major impediment to assembly. A mean of 60% of CentC, 70% of the 4-12-1 subtelomeric sequence (38)), 28.9% of TR-1, 1% of knob180, and 0.09% of rDNA repeat units were incorporated in the final assemblies (Table 1).
A total of 110 (of 260) functional centromeres identified by CENH3 ChIP-seq (39, 40) were fully assembled, and of these 88 are gapless ((Fig. S8A and (40)). Chromosomes with very long CentC arrays (such as chromosomes 1, 6, and 7) often have assembly gaps and the precise location of the centromere could not be determined. However, many centromeres either have fully assembled small CentC arrays or the functional centromeres are located to one side of the CentC tracts in regions dominated by retrotransposons (Fig. 3A). By projecting all centromere locations onto B73, we were able to identify twelve centromere movement events (three on chr5 and chr9, and two on chr3, chr8 and chr10), clarifying and extending prior evidence for centromere shifting (39) (Fig. 3B, Fig. S8B). The variation in CentC abundance and positional polymorphism made it possible to gaplessly assemble at least two variants of all ten centromeres (Fig. S8A).
Figure 3.
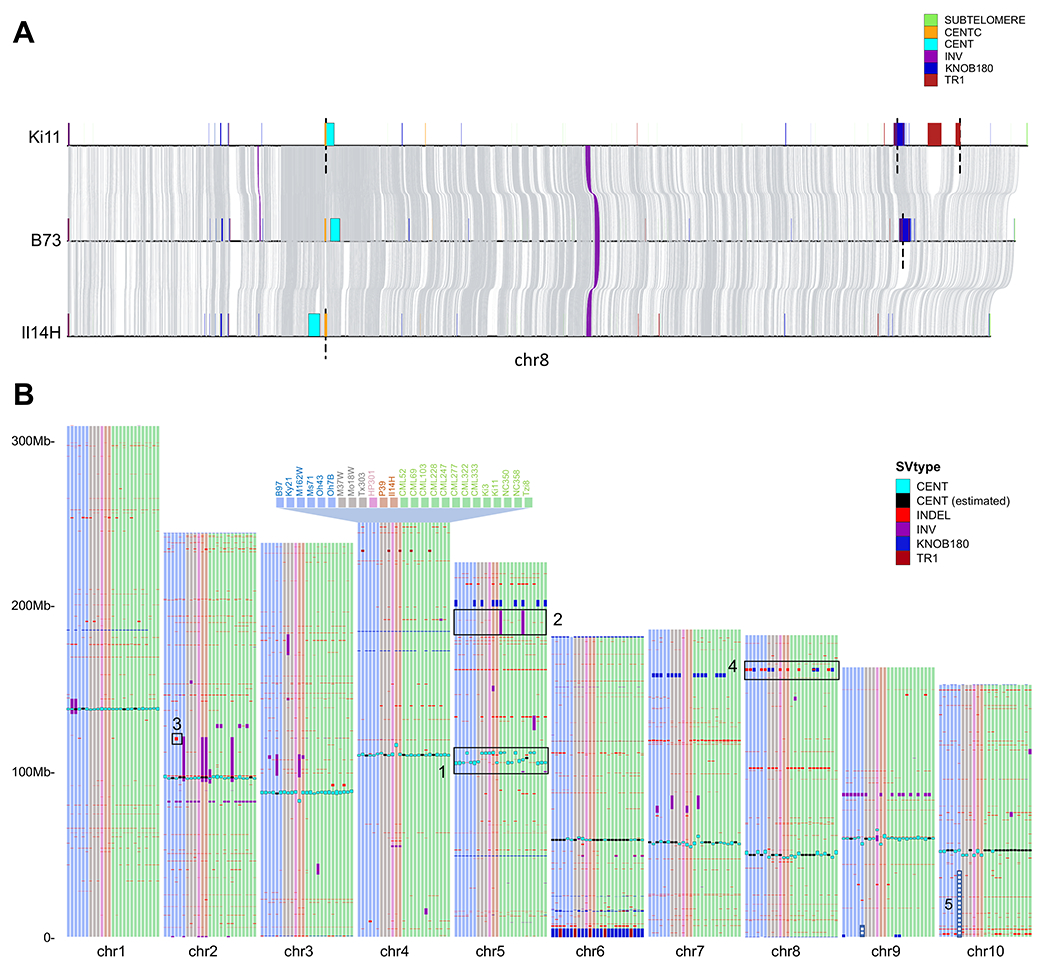
Structural variation in the NAM founders. A) Pairwise alignments between Ki11, B73, Il14H on chromosome 8. Grey links represent syntenic aligned regions; gaps of unknown size (scaffold gaps) are marked by dashed lines. B) Large (>100 kbp) structural variants, centromeres, and knobs across the NAM lines versus the B73 reference. The subset of SVs larger than 1 Mbp were manually curated, and only those containing genes are represented. Features 1-5 highlight major SVs: 1) Multiple centromere movement events; 2) A major inversion previously hypothesized based on suppressed recombination; 3) A large deletion in the Ms71 inbred; 4) Knob polymorphism; 5) Reciprocal translocation between chromosome 9 and 10 in the Oh7B inbred (both segments placed in their standard positions for display).
Both knob180 and TR-1 arrays are subject to meiotic drive and accumulate when a chromosome variant known as Abnormal chromosome 10 (Ab10) is present (37, 41). Although Ab10 is absent from modern inbreds, its legacy remains in the form of many large knobs. The majority of knob180 and TR-1 repeat arrays were identified in mid-arm positions (81.9%) where meiotic drive is most effective. Long knob180 and TR-1 repeat arrays can occur separately, but are more frequently intermingled in fragmented arrays along with transposons (Fig. 3A, Fig. S9) (42). Analysis of classical (cytologically visible) knobs on chromosome 1S, 2S, 2L, 3L, 4L, 5L, 6L, 7L, 8L, and 9S revealed that their locations are syntenic and that several are composed of a series of disjointed smaller knobs (Fig. 3A, Fig. S10). In some lines, knobs are not visible cytologically but can still be detected as smaller arrays at the sequence level; however, many show strict presence-absence variation among the NAM founder inbreds.
Tandem repeat arrays are also commonly found at the ends of chromosome arms (Table S6). Among the 520 chromosome ends, 57.9% contained knob180 repeats and 30.5% contained subtelomere repeats. At least 65.6% of chromosome ends were fully assembled as indicated by the presence of telomere sequences.
Structural variation and impact on phenotype
Comparative analyses among the NAM genotypes to B73 revealed a cumulative total of 791,101 structural variants (SVs) greater than 100 bp in size. Tropical lines, which are the most divergent from B73, include a substantially higher number of SVs than temperate lines (mean = 32,976 versus 29,742; p = 0.00013) (Tables S7, S8). Structural variants are more common on chromosome arms where recombination is highest (Fig. S11), similar to SNPs and other forms of genetic variation (43). Almost half (49.6%) of SVs were <5 kbp in size, with 25.7% being less than 500 bp. Across all size classes SVs are skewed toward rare variants (Fig. S12). Several large SVs were found segregating within the 26 NAM genomes (Fig. 3B), including 35 distinct inversion polymorphisms and 5 insertion-deletion polymorphisms >1 Mbp. For example, a 14.6 Mbp inversion on chromosome 5 in the CML52 and CML322 lines, which was previously hypothesized based on suppressed recombination in the NAM RILs (11), is confirmed here based on assembly. Additionally, there is a 1.9 Mbp deletion with seven genes on chromosome 2 in the MS71 inbred, and a 1.8 Mbp deletion with two genes on chromosome 8 found in eight lines. Our data also capture a very large reciprocal translocation (involving >47 Mbp of DNA) between the short arms of chromosomes 9 and 10 in Oh7B that had been previously detected in cytological studies (38) (Fig. 3B).
The high proportion of rare SVs in maize suggests these may be a particularly deleterious class of variants, as observed in other species (44, 45). Indels and inversions occur in regions that have 49.8% fewer genic base pairs than the genomic background. Furthermore, SVs are 17% less likely to be found in conserved regions than SNPs (odds ratios of 0.27 and 0.58 for SVs and SNPs, respectively, Fisher’s Exact Test, p < 0.001). Approximate Bayesian computation modeling revealed that selection against SVs is at least as strong as that against nonsynonymous substitutions (Fig. S13; See Supplemental Methods). These results suggest that, when they occur, SVs are particularly consequential and relevant to fitness.
To estimate the phenotypic impact of SVs, we assessed the genetic basis of 36 complex traits (14) using 71,196 filtered SVs in 4,027 recombinant inbred lines derived from the NAM founder inbreds (11) (Fig. S14A). The analysis revealed that SVs explain a high percentage of phenotypic variance for disease traits (60.10% ~ 61.75%) and less for agronomic/morphological (20.04% ~ 61.04%) and metabolic traits (4.79% ~ 26.78%). Much of the phenotypic variation was also explained by SNPs, which were much more numerous (288-fold more) relative to our conservative set of SVs (Fig. S14A). When the SNP and SV data were integrated into one linear mixed model, the combined markers only slightly surpassed values from SNPs, consistent with the fact that most SVs are in high linkage disequilibrium with SNPs (Fig. S14A).
We also carried out genome-wide association analyses (GWAS) to identify specific SVs contributing to phenotypic variation for the same suite of traits (Fig. S14B–G). Among the detected GWAS signals, 93.05% overlapped with those identified with SNPs and 6.95% were unique to SVs (no significant SNP detected within 5 Mbp of significant SVs). There was a significant enrichment of SVs associated with phenotypes in genic regions (z = 8.022, p < 1.04e-15; Fig. S15). The most significant association between a SV and a trait not identified using SNP markers was a QTL for northern leaf blight (NLB) on chromosome 10 (Fig. S14F). This SV is within a gene encoding a thylakoid lumenal protein; such proteins could be linked to plant immunity through the regulation of cell death during viral infection (46). We anticipate that the effects of SVs may be even more pronounced in larger association panels where extensive historical recombination may help disentangle their effects from nearby SNPs.
Disease resistance in plants is frequently associated with SV in the form of tandem arrays of resistance genes. Complex arrays of resistance genes are retained, potentially through birth-death dynamics in an evolutionary arms race with pathogens, or through balancing selection for the maintenance of diverse plant defenses (47). Nucleotide-binding, leucine-rich-repeat (NLR) proteins provide a common type of resistance. Our data reveal that there are fewer NLR genes in maize than other Poaceae (Fig. S16) and that most NAM lines have lost the same clades of NLRs as sorghum (Fig. S17). Only one line (CML277) retains the MIC1 NLR clade, which is particularly fast-evolving in Poaceae (48). Nevertheless, there is clear NLR variation among the NAM lines (Fig. S18), and tropical genomes contain a significantly higher number of NLR genes than temperate genomes (t-test, p=0.006), suggesting ongoing co-evolution with pathogens, particularly where disease pressure is high.
The annotated NLR genes were significantly enriched for overlap with SVs (boot-strap permutation test, p<0.001). An extreme example is found at the rp1 (resistance to Puccinia sorghi1) locus on the short arm of chromosome 10, which is known to be highly variable (49). We observed exceptional diversity in the NAM lines with as few as 4 rp1 copies in P39, and as many as 30 in M37W (Table S9). However, due to its repetitive nature, only 18 NAM lines have gapless assemblies of the rp1 locus.
SVs linked to transposons have been shown, through the modulation of gene expression, to underlie flowering-time adaptation in maize during tropical-to-temperate migration (50, 51). Our SV and TE-annotation pipelines identified the adaptive CACTA-like insertion previously reported upstream of the flowering-time locus ZmCCT10 (51). We also surveyed 173 genes linked to flowering-time (52, 53) and discovered three genes (GL15, ZCN10, and Dof21) with TE-derived SVs <5 kbp upstream of their transcription start sites. These SVs distinguish temperate from tropical lines (t < −2.346, p < 0.0358) (Fig. S19) and show significant correlation (F > 8.658, p < 0.001) with expression levels.
Discovery of candidate cis-regulatory elements through DNA methylation
Based on sequence alone, it can be difficult to identify functional sequences in the intergenic spaces. One approach is to score for unmethylated DNA, which provides both a tissue-independent indicator of gene regulatory elements and evidence that annotated genes are active (5, 54, 55). We sequenced enzymatic methyl-seq (EM-seq) libraries from each NAM line and identified methylated bases in three sequence contexts, CG, CHG, and CHH (where H = A, T, or C). Results are consistent across genes and transposons, demonstrating the quality of the libraries (Figs. S20, S21). There is minor variation in total methylation across inbreds, with CML247 being noteworthy for uniformly lower CG methylation in several tissues (Fig. S22). Such natural variation in methylation is also observed in Arabidopsis ecotypes (56).
Each of the three methylation contexts reveal information on the locations of repeats, genes and regulatory elements. mCHH levels are generally low except at heterochromatin borders, whereas mCHG and mCG are abundant in repetitive regions. Both mCHG and mCG are depleted from regulatory elements and mCHG is depleted from exons (57). However mCG is often present in exons (Fig. 4) (58). Thus, to identify unmethylated regions (UMRs) corresponding to regulatory elements and gene bodies, we defined UMRs using a method that takes into account mCHG and mCG but does not exclude high mCG-only regions (the term UMR is used for simplicity; some regions contain CG methylation). Comparison of the 26 methylomes revealed uniformity in number and length of UMRs, averaging about 180 Mbp in total length in each genome (Figs. S23, S24). To confirm the accuracy of the UMR data, we also identified accessible chromatin regions (ACRs) using ATAC-seq for each inbred. We expect chromatin to be accessible mainly in the subset of genes expressed in the tissue sampled (primarily leaves) and to show concordance with UMRs. The data reveal that a mean of 99% of genic and 96% of non-genic (distal) ACRs overlap with UMRs in each genome (Figs. S25, S26).
Figure 4.
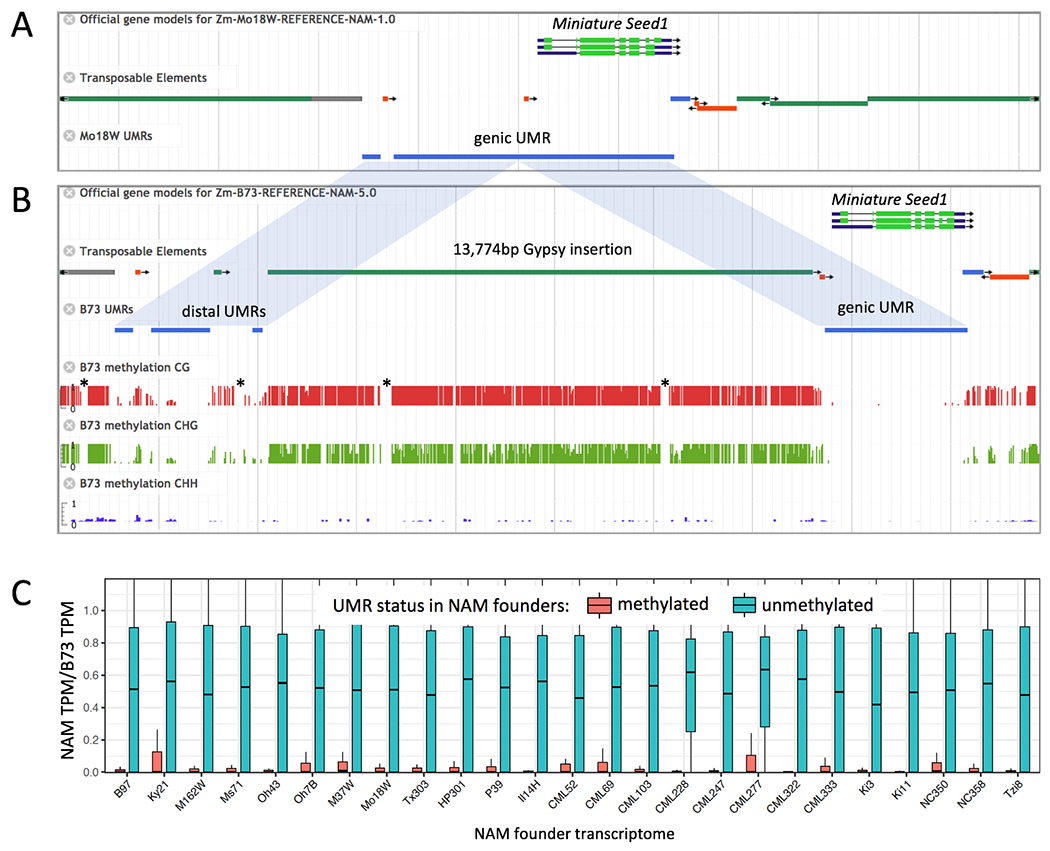
UMR variation across the NAM founders. A) Annotation of the Miniature seed1 gene in the Mo17W inbred. An image from the MaizeGDB browser shows gene, TE, and UMR tracks. TE tracks are color-coded by superfamily: green/grey = LTR, red = TIR, blue = LINE. The grey vertical lines show 2.5 kbp intervals. B) Annotation and underlying methylation data for Miniature seed1 in the B73 inbred. The insertion of a Gypsy element moved part of the proximal UMR to a position 14 kbp upstream from the transcription start site (TSS). Methylation tracks indicate base-pair level methylation values from 0 to 100%. Asterisks indicate gaps in coverage, which are visible in separate tracks (Fig. S28). C) Relationship between methylation and gene expression. UMRs were mapped to B73 to identify UMRs that overlap with TSS. The Y axis indicates the ratio of transcripts per million (TPM, compared to B73) when the region is methylated (red) or unmethylated (teal).
To assess methylation diversity, we mapped UMRs from all inbreds to the B73 genome. Approximately 95% of genic UMRs overlap across genomes in pairwise comparisons (Fig. S27). UMR polymorphism is higher in the intergenic space, particularly among UMRs greater than 5 kbp from genes, where typically ~75% of UMRs overlap (Fig. S27). Even when the UMR sequence is conserved, its position relative to the closest gene may vary dramatically among inbreds. This is exemplified by the Miniature Seed1 gene where a UMR proximal to the promoter in Mo18W is displaced nearly 14 kbp upstream in B73 by a single Huck element (Gypsy LTR superfamily) (Fig. 4). The Huck insertion is present in 23 of 26 genomes, and in two of these (Oh43 and CML322), additional nested TE insertions increased the distance between the gene and the UMR to 27 kbp. Although UMR polymorphism correlates with genetic distance across NAM lines (Fig. S29), UMRs from Tzi8 were not substantially shared with other tropical genomes.
Adaptive variation in DNA methylation has been observed in maize (59), most likely through effects on gene expression. To estimate how well UMRs predict transcription, we identified a conservative subset of UMRs overlapping genes that were unmethylated in B73 but methylated in at least one other methylome. These differentially methylated regions were strongly correlated with differences in gene expression (Fig. 4, Fig. S30). We further evaluated the enrichment of significant GWAS SNPs across 36 traits in UMRs. Based on genome-wide estimates, UMRs show 2.50- to 3.26-fold enrichment across traits for significant associations. Roughly 18% of SNPs identified by GWAS lie outside of genic regions but within UMRs (Table S10), consistent with the view that UMRs can be used to identify functional, non-coding regions (5, 54, 55).
Summary
Our analysis of 26 genomes uncovered variation in both the genic and repetitive fractions of the pan-genome. Tropical, temperate, and flint-derived popcorn/sweet corn germplasm are differentiated in a number of striking ways including their pan-gene complement, homoeolog retention post-polyploidy, abundance of transposable elements, NLR disease-resistance gene copy number, and methylation profiles. The available data will have broad utility for genetic and genomic studies and facilitate rapid associations to phenotyping information. For example, the genic presence-absence variation identified here may be imputed across additional mapping populations to clarify its contribution to heterosis through complementation (60). More generally, these resources should motivate a shift away from the single reference mindset to a multi-reference view where any one of 26 inbreds, each with different experimental and agronomic advantages, can be deployed for the purposes of basic discovery and crop improvement.
Supplementary Material
Acknowledgments:
We appreciate the sequencing services provided by the University of Arizona, Oregon State University, Brigham Young University and the University of Georgia, as well as coordination among sequencing centers provided by Pacific Biosciences. The authors further acknowledge the High Performance Computing facility at Iowa State University (partially funded by NSF 1726447), Minnesota Supercomputing Institute, the Georgia Advanced Computing Resource Center, BlacknBlue high performance computing center at Cold Spring Harbor Laboratory and the participants of the Virtual Maize Annotation Jamboree who evaluated the initial gene predictions for benchmarking and improvements in the final gene annotations.
Funding:
Primary support for this work came from a generous grant from the National Science Foundation (IOS-1744001). Additional support came from NSF IOS-1546727 to CNH, USDA 2018-67013-27571 to CNH, USDA-ARS 8062-21000-041-00D, NSF IOS-1127112 and NIH-OD S10 OD028632 to DW, NSF IOS-1546719 to MBH, NSF IOS-1822330 to JRI and MBH, USDA Hatch project CA-D-PLS-2066-H to JRI, NSF IOS-1856627 to RJS, an NSF Postdoctoral Fellowship in Biology DBI-1905869 to APM, NSF Graduate Research Fellowships 1650042 to AIH and 1744592 to SJS, NSF Research Traineeship (DGE-1545463) to Iowa State University (Trainee SJS), USDA-ARS 58-5030-8-064 to MBH and CMA, USDA-ARS project 5030-21000-068-00D to CMA and MW and NSF IOS-1546657 to JY;
Footnotes
Competing interests: RJS is a co-founder of REquest Genomics, LLC, a company that provides epigenomic services. All other authors declare no competing interests.
Data and materials availability:
Genome assemblies and annotations can be accessed at https://maizegdb.org/NAM_project. Raw data used for the assemblies including PacBio, Illumina, and Bionano data are available through ENA BioProject IDs PRJEB31061 and PRJEB32225. RNA-Seq data is available at ENA ArrayExpress E-MTAB-8633 and E-MTAB-8628. EM-Seq reads are available at ENA ArrayExpress E-MTAB-10088. ATAC-seq reads are available under NCBI GEO accession GSE165787. Other files, tables and supplemental data can be found in CyVerse https://datacommons.cyverse.org/browse/iplant/home/shared/NAM/NAM_genome_and_annotation_Jan2021_release. Links to the NLR trees can be found at https://itol.embl.de/shared/xCJbI9ndshEK. Scripts used to generate and analyze data are available as a Zenodo repository (61).
References:
- 1.Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Peñagaricano F, Lindquist E, Pedraza MA, Barry K, de Leon N, Kaeppler SM, Buell CR, Insights into the maize pan-genome and pan-transcriptome. Plant Cell. 26, 121–135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirsch CN, Hirsch CD, Brohammer AB, Bowman MJ, Soifer I, Barad O, Shem-Tov D, Baruch K, Lu F, Hernandez AG, Fields CJ, Wright CL, Koehler K, Springer NM, Buckler E, Buell CR, de Leon N, Kaeppler SM, Childs KL, Mikel MA, Draft Assembly of Elite Inbred Line PH207 Provides Insights into Genomic and Transcriptome Diversity in Maize. Plant Cell. 28, 2700–2714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin M, Liu H, He C, Fu J, Xiao Y, Wang Y, Xie W, Wang G, Yan J, Maize pan-transcriptome provides novel insights into genome complexity and quantitative trait variation. Sci. Rep 6, 18936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu F, Romay MC, Glaubitz JC, Bradbury PJ, Elshire RJ, Wang T, Li Y, Li Y, Semagn K, Zhang X, Hernandez AG, Mikel MA, Soifer I, Barad O, Buckler ES, High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun 6, 6914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerra MK, Colomé-Tatché M, Johannes F, Rowley MJ, Corces VG, Zhai J, Scanlon MJ, Buckler ES, Gallavotti A, Springer NM, Schmitz RJ, Zhang X, Widespread long-range cis-regulatory elements in the maize genome. Nature Plants 5, 1237–1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun S, Zhou Y, Chen J, Shi J, Zhao H, Zhao H, Song W, Zhang M, Cui Y, Dong X, Liu H, Ma X, Jiao Y, Wang B, Wei X, Stein JC, Glaubitz JC, Lu F, Yu G, Liang C, Fengler K, Li B, Rafalski A, Schnable PS, Ware DH, Buckler ES, Lai J, Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet 50, 1289–1295 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, Spannagl M, Marcon C, Ruban A, Urbany C, Nemri A, Hochholdinger F, Ouzunova M, Houben A, Schön C-C, Mayer KFX, European maize genomes highlight intraspecies variation in repeat and gene content. Nat. Genet 52, 950–957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang N, Liu J, Gao Q, Gui S, Chen L, Yang L, Huang J, Deng T, Luo J, He L, Wang Y, Xu P, Peng Y, Shi Z, Lan L, Ma Z, Yang X, Zhang Q, Bai M, Li S, Li W, Liu L, Jackson D, Yan J, Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet 51, 1052–1059 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Lin G, He C, Zheng J, Koo D-H, Le H, Zheng H, Tamang TM, Lin J, Liu Y, Zhao M, Hao Y, McFraland F, Wang B, Qin Y, Tang H, McCarty DR, Wei H, Cho M-J, Park S, Kaeppler H, Kaeppler SM, Liu Y, Springer N, Schnable PS, Wang G, White FF, Liu S, Chromosome-level Genome Assembly of a Regenerable Maize Inbred Line A188. BioRxiv 10.1101/2020.09.09.289611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer NM, Anderson SN, Andorf CM, Ahern KR, Bai F, Barad O, Barbazuk WB, Bass HW, Baruch K, Ben-Zvi G, Buckler ES, Bukowski R, Campbell MS, Cannon EKS, Chomet P, Dawe RK, Davenport R, Dooner HK, Du LH, Du C, Easterling KA, Gault C, Guan J-C, Hunter CT, Jander G, Jiao Y, Koch KE, Kol G, Köllner TG, Kudo T, Li Q, Lu F, Mayfield-Jones D, Mei W, McCarty DR, Noshay JM, Portwood JL 2nd, Ronen G, Settles AM, Shem-Tov D, Shi J, Soifer I, Stein JC, Stitzer MC, Suzuki M, Vera DL, Vollbrecht E, Vrebalov JT, Ware D, Wei S, Wimalanathan K, Woodhouse MR, Xiong W, Brutnell TP, The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet 50, 1282–1288 (2018). [DOI] [PubMed] [Google Scholar]
- 11.McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, Brown P, Browne C, Eller M, Guill K, Harjes C, Kroon D, Lepak N, Mitchell SE, Peterson B, Pressoir G, Romero S, Oropeza Rosas M, Salvo S, Yates H, Hanson M, Jones E, Smith S, Glaubitz JC, Goodman M, Ware D, Holland JB, Buckler ES, Genetic properties of the maize nested association mapping population. Science. 325, 737–740 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Flint-Garcia SA, Thuillet A-C, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES, Maize association population: a high-resolution platform for quantitative trait locus dissection: High-resolution maize association population. Plant J. 44, 1054–1064 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Yu J, Holland JB, McMullen MD, Buckler ES, Genetic design and statistical power of nested association mapping in maize. Genetics. 178, 539–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace JG, Bradbury PJ, Zhang N, Gibon Y, Stitt M, Buckler ES, Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet. 10, e1004845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, Hermanson PJ, Waters AJ, Starr E, West PT, Tiffin P, Myers CL, Vaughn MW, Springer NM, Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell. 25, 2783–2797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer RJ, Michno J-M, Jeffers J, Hoekenga O, Dilkes B, Baxter I, Myers CL, Integrating Coexpression Networks with GWAS to Prioritize Causal Genes in Maize. The Plant Cell. 30 (2018), pp. 2922–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See supplementary materials.
- 18.Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin C-S, Guill K, Regulski M, Kumari S, Olson A, Gent J, Schneider KL, Wolfgruber TK, May MR, Springer NM, Antoniou E, McCombie WR, Presting GG, McMullen M, Ross-Ibarra J, Dawe RK, Hastie A, Rank DR, Ware D, Improved maize reference genome with single-molecule technologies. Nature. 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Ou S, Chen J, Jiang N, Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Research. 46, e126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou S, Liu J, Chougule KM, Fungtammasan A, Seetharam AS, Stein JC, Llaca V, Manchanda N, Gilbert AM, Wei S, Chin C-S, Hufnagel DE, Pedersen S, Snodgrass SJ, Fengler K, Woodhouse M, Walenz BP, Koren S, Phillippy AM, Hannigan BT, Dawe RK, Hirsch CN, Hufford MB, Ware D, Effect of sequence depth and length in long-read assembly of the maize inbred NC358. Nat. Commun 11, 2288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilbeck K, Moore B, Holt C, Yandell M, Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics. 10, 67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law M, Childs KL, Campbell MS, Stein JC, Olson AJ, Holt C, Panchy N, Lei J, Jiao D, Andorf CM, Lawrence CJ, Ware D, Shiu S-H, Sun Y, Jiang N, Yandell M, Automated update, revision, and quality control of the maize genome annotations using MAKER-P improves the B73 RefGen_v3 gene models and identifies new genes. Plant Physiol. 167, 25–39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swigonová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J, Close split of sorghum and maize genome progenitors. Genome Res. 14, 1916–1923 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Wang J, Jin D, Guo H, Lee T-H, Liu T, Paterson AH, Genome Alignment Spanning Major Poaceae Lineages Reveals Heterogeneous Evolutionary Rates and Alters Inferred Dates for Key Evolutionary Events. Mol. Plant 8, 885–898 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Schnable JC, Springer NM, Freeling M, Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. U. S. A 108, 4069–4074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M, Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8, e1000409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnable JC, Freeling M, Lyons E, Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol 4, 265–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandáková T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA, Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell. 22, 2277–2290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brohammer AB, Kono TJY, Springer NM, McGaugh SE, Hirsch CN, The limited role of differential fractionation in genome content variation and function in maize (Zea mays L.) inbred lines. Plant J. 93, 131–141 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Tang H, Woodhouse MR, Cheng F, Schnable JC, Pedersen BS, Conant G, Wang X, Freeling M, Pires JC, Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics. 190, 1563–1574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beissinger TM, Wang L, Crosby K, Durvasula A, Hufford MB, Ross-Ibarra J. Recent demography drives changes in linked selection across the maize genome. Nature plants 2, 1084 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X, Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 4, 258–268 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Ou S, Su W, Liao Y, Chougule K, Ware D, Peterson T, Jiang N, Hirsch CN, Hufford MB, Benchmarking Transposable Element Annotation Methods for Creation of a Streamlined, Comprehensive Pipeline. Genome Biol 20, 275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon J-M, Westerman RP, Sanmiguel PJ, Bennetzen JL, Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5, e1000732 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilinski P, Albert PS, Berg JJ, Birchler JA, Grote MN, Lorant A, Quezada J, Swarts K, Yang J, Ross-Ibarra J, Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLoS Genet. 14, e1007162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, Ross-Ibarra J, Wallace JG, Kanizay LB, Alabady M, Qiu W, Tseng K-F, Wang N, Gao Z, Birchler JA, Harkess AE, Hodges AL, Hiatt EN, A Kinesin-14 Motor Activates Neocentromeres to Promote Meiotic Drive in Maize. Cell. 173, 839–850.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Albert PS, Gao Z, Danilova TV, Birchler JA, Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet. Genome Res 129, 6–16 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Schneider KL, Xie Z, Wolfgruber TK, Presting GG, Inbreeding drives maize centromere evolution. Proc. Natl. Acad. Sci. U. S. A 113, E987–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang N, Liu J, Ricci WA, Gent JI, Dawe RK, Maize centromeric chromatin scales with changes in genome size. Genetics. 217, iyab020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swentowsky KW, Gent JI, Lowry EG, Schubert V, Ran X, Tseng K-F, Harkess AE, Qiu W, Dawe RK, Distinct kinesin motors drive two types of maize neocentromeres. Genes Dev. 34, 1239–1251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Seetharam AS, Chougule K, Ou S, Swentowsky KW, Gent JI, Llaca V, Woodhouse MR, Manchanda N, Presting GG, Kudrna DA, Alabady M, Hirsch CN, Fengler KA, Ware D, Michael TP, Hufford MB, Dawe RK, Gapless assembly of maize chromosomes using long-read technologies. Genome Biol. 21, 121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, Gore M, Guill KE, Holland J, Hufford MB, Lai J, Li M, Liu X, Lu Y, McCombie R, Nelson R, Poland J, Prasanna BM, Pyhäjärvi T, Rong T, Sekhon RS, Sun Q, Tenaillon MI, Tian F, Wang J, Xu X, Zhang Z, Kaeppler SM, Ross-Ibarra J, McMullen MD, Buckler ES, Zhang G, Xu Y, Ware D, Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet 44, 803–807 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Abel HJ, Larson DE, Regier AA, Chiang C, Das I, Kanchi KL, Layer RM, Neale BM, Salerno WJ, Reeves C, Buyske S, NHGRI Centers for Common Disease Genomics, Matise TC, Muzny DM, Zody MC, Lander ES, Dutcher SK, Stitziel NO, Hall IM, Mapping and characterization of structural variation in 17,795 human genomes. Nature. 583, 83–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leushkin EV, Bazykin GA, Kondrashov AS, Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection. Genome Biol. Evol 5, 514–524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo S, Okamoto M, Iwai T, Iwano M, Fukui K, Isogai A, Nakajima N, Ohashi Y, Reduced Levels of Chloroplast FtsH Protein in Tobacco Mosaic Virus-Infected Tobacco Leaves Accelerate the Hypersensitive Reaction. The Plant Cell. 12 (2000), p. 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizuno H, Katagiri S, Kanamori H, Mukai Y, Sasaki T, Matsumoto T, Wu J, Evolutionary dynamics and impacts of chromosome regions carrying R-gene clusters in rice. Sci. Rep 10, 872 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV, Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hulbert SH, Bennetzen JL, Recombination at the Rp1 locus of maize. Mol. Gen. Genet 226, 377–382 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, Xu G, Tian J, Wang C, Li D, Wu L, Yang X, Jin W, Doebley JF, Tian F, ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. U. S. A 115, E334–E341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, Li K, Yang N, Li Y, Zhong T, Li J, Chen Y, Yan J, Yang X, Xu M, CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. U. S. A 110, 16969–16974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong Z, Danilevskaya O, Abadie T, Messina C, Coles N, Cooper M, A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS One. 7, e43450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y-X, Li C, Bradbury PJ, Liu X, Lu F, Romay CM, Glaubitz JC, Wu X, Peng B, Shi Y, Song Y, Zhang D, Buckler ES, Zhang Z, Li Y, Wang T, Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J. 86, 391–402 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Oka R, Zicola J, Weber B, Anderson SN, Hodgman C, Gent JI, Wesselink J-J, Springer NM, Hoefsloot HCJ, Turck F, Stam M, Genome-wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize. Genome Biol. 18, 137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crisp PA, Marand AP, Noshay JM, Zhou P, Lu Z, Schmitz RJ, Springer NM, Stable unmethylated DNA demarcates expressed genes and their cis-regulatory space in plant genomes. Proc. Natl. Acad. Sci. U. S. A 117, 23991–24000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawakatsu T, Huang S-SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, Chen H, Dubin M, Lee C-R, Wang C, Bemm F, Becker C, O’Neil R, O’Malley RC, Quarless DX, 1001 Genomes Consortium, Schork NJ, Weigel D, Nordborg M, Ecker JR, Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell. 166, 492–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK, CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23, 628–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bewick AJ, Schmitz RJ, Gene body DNA methylation in plants. Curr. Opin. Plant Biol 36, 103–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu G, Lyu J, Li Q, Liu H, Wang D, Zhang M, Springer NM, Ross-Ibarra J, Yang J, Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat. Commun 11, 5539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Springer NM, Ying K, Fu Y, Ji T, Yeh C-T, Jia Y, Wu W, Richmond T, Kitzman J, Rosenbaum H, Iniguez AL, Barbazuk WB, Jeddeloh JA, Nettleton D, Schnable PS, Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5, e1000734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, Ricci WA, Guo T, Olson A, Qiu Y, Della Coletta R, Tittes S, Hudson AI, Marand AP, Wei S, Lu Z, Wang B, Tello-Ruiz MK, Piri RD, Wang N, Kim DW, Zeng Y, O’Connor CH, Li X, Gilbert AM, Baggs E, Krasileva KV, Portwood II JL, Cannon EKS, Andorf CM, Manchanda N, Snodgrass SJ, Hufnagel DE, Jiang Q, Pedersen S, Syring ML, Kudrna DA, Llaca V, Fengler K, Schmitz RJ, Ross-Ibarra J, Yu J, Gent JI, Hirsch CN, Ware D, Kelly Dawe R, HuffordLab/NAM-genomes: publication.prerelease (2021; https://zenodo.org/record/4781590). [Google Scholar]
- 62.Doyle JJ, Doyle JL, A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19, 11–15 (1987). [Google Scholar]
- 63.Luo M, Wing RA, An improved method for plant BAC library construction. Methods Mol. Biol 236, 3–20 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Vasimuddin M, Misra S, Li H, Aluru S, in 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS) (2019), pp. 314–324. [Google Scholar]
- 65.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, Shakir K, Thibault J, Chandran S, Whelan C, Lek M, Gabriel S, Daly MJ, Neale B, MacArthur DG, Banks E, Scaling accurate genetic variant discovery to tens of thousands of samples. Cold Spring Harbor Laboratory (2018), p. 201178. [Google Scholar]
- 66.Lee T-H, Guo H, Wang X, Kim C, Paterson AH, SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 15, 162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, Cramer GR, Delledonne M, Luo C, Ecker JR, Cantu D, Rank DR, Schatz MC, Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM, Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J, ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A, Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol. Biol 48, 453–461 (2002). [DOI] [PubMed] [Google Scholar]
- 72.CyVerse Data Commons, (available at http://datacommons.cyverse.org/browse/iplant/home/shared/commons_repo/curated/Daniel_Laspisa_B73_RefGen_v4CEN_Feb_2019).
- 73.Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, King M, May G, Lin H, A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun 9, 4844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaser R, Sović I, Nagarajan N, Šikić M, Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smit AFA, Hubley R, Green P, RepeatMasker Open-4.0. 2013–2015 (2015). [Google Scholar]
- 76.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh C-T, Emrich SJ, Jia Y, Kalyanaraman A, Hsia A-P, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia J-M, Deragon J-M, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK, The B73 maize genome: complexity, diversity, and dynamics. Science. 326, 1112–1115 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 78.Manchanda N, Portwood JL 2nd, Woodhouse MR, Seetharam AS, Lawrence-Dill CJ, Andorf CM, Hufford MB, GenomeQC: a quality assessment tool for genome assemblies and gene structure annotations. BMC Genomics. 21, 193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ellinghaus D, Kurtz S, Willhoeft U, LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 9, 18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ou S, Jiang N, LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mobile DNA. 10, 48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ou S, Jiang N, LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 176, 1410–1422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seetharam A, Singh U, Li J, Bhandary P, Arendsee Z, Wurtele ES, Maximizing prediction of orphan genes in assembled genomes. BioRxiv 10.1101/2019.12.17.880294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A, Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 29 (2011), pp. 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu R, Dickerson J, Strawberry: Fast and accurate genome-guided transcript reconstruction and quantification from RNA-Seq. PLoS Comput. Biol 13, e1005851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 7 (2012), pp. 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song L, Sabunciyan S, Florea L, CLASS2: accurate and efficient splice variant annotation from RNA-seq reads. Nucleic Acids Res. 44, e98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Venturini L, Caim S, Kaithakottil GG, Mapleson DL, Swarbreck D, Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience. 7 giy093 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25 (2009), pp. 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mapleson D, Venturini L, Kaithakottil G, Swarbreck D, Efficient and accurate detection of splice junctions from RNA-seq with Portcullis. Gigascience. 7, giy131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A, De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat. Protoc 8 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoff KJ, Lomsadze A, Borodovsky M, Stanke M, in Gene Prediction: Methods and Protocols, Kollmar M, Ed. (Springer New York, New York, NY, 2019), pp. 65–95. [Google Scholar]
- 94.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O, Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, Angelova A, Collura K, Wissotski M, Ashley E, Morrow D, Fernandes J, Walbot V, Yu Y, Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 5, e1000740 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang B, Regulski M, Tseng E, Olson A, Goodwin S, McCombie WR, Ware D, A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing. Genome Res. 28, 921–932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu TD, Watanabe CK, GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 21, 1859–1875 (2005). [DOI] [PubMed] [Google Scholar]
- 99.Kent WJ, BLAT—The BLAST-Like Alignment Tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, Lu Z, Olson A, Stein JC, Ware D, Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun 7, 11708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang R-G, Wang Z-X, Ou S, Li G-Y, TEsorter: lineage-level classification of transposable elements using conserved protein domains. BioRxiv 10.1101/800177 (2019). [DOI] [Google Scholar]
- 102.Campbell MS, Holt C, Moore B, Yandell M, Genome Annotation and Curation Using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48, 4.11.1–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tello-Ruiz MK, Naithani S, Gupta P, Olson A, Wei S, Preece J, Jiao Y, Wang B, Chougule K, Garg P, Elser J, Kumari S, Kumar V, Contreras-Moreira B, Naamati G, George N, Cook J, Bolser D, D’Eustachio P, Stein LD, Gupta A, Xu W, Regala J, Papatheodorou I, Kersey PJ, Flicek P, Taylor C, Jaiswal P, Ware D, Gramene 2021: harnessing the power of comparative genomics and pathways for plant research. Nucleic Acids Res. 49, D1452–D1463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Edgar RC, Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 105.Chen M-JM, Lin H, Chiang L-M, Childers CP, Poelchau MF, The GFF3toolkit: QC and Merge Pipeline for Genome Annotation. Methods Mol. Biol 1858, 75–87 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S, InterProScan 5: genome-scale protein function classification. Bioinformatics. 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olson AJ, Ware D, Ranked Choice Voting for Representative Transcripts with TRaCE. Cold Spring Harbor Laboratory; (2020), p. 2020.12.15.422742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E, The Ensembl core software libraries. Genome Res. 14, 929–933 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T, deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thorvaldsdóttir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 112.Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen P-Y, Pellegrini M, BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 14, 774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guo W, Zhu P, Pellegrini M, Zhang MQ, Wang X, Ni Z, CGmapTools improves the precision of heterozygous SNV calls and supports allele-specific methylation detection and visualization in bisulfite-sequencing data. Bioinformatics. 34, 381–387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ricci WA, Unmethylated Regions Encompass the Functional Space Within the Maize Genome. BiorXiv 10.1101/2021.04.21.425900 (2021) [DOI] [Google Scholar]
- 115.Schultz MD, Schmitz RJ, Ecker JR, “Leveling” the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28, 583–585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.gnu.org (available at https://www.gnu.org/software/datamash/).
- 117.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ, Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ, Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol 36, 89–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen S, Zhou Y, Chen Y, Gu J, fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P, Sambamba: fast processing of NGS alignment formats. Bioinformatics. 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gaspar JM, Improved peak-calling with MACS2. Cold Spring Harbor Laboratory; (2018), p. 496521. [Google Scholar]
- 123.Monnahan PJ, Michno J-M, O’Connor C, Brohammer AB, Springer NM, McGaugh SE, Hirsch CN, Using multiple reference genomes to identify and resolve annotation inconsistencies. BMC Genomics. 21, 281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A, MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol 14, e1005944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Team RC, Others R: A language and environment for statistical computing (2013), (available at http://finzi.psych.upenn.edu/R/library/dplR/doc/intro-dplR.pdf).
- 126.Su W, Gu X, Peterson T, TIR-Learner, a New Ensemble Method for TIR Transposable Element Annotation, Provides Evidence for Abundant New Transposable Elements in the Maize Genome. Mol. Plant 12, 447–460 (2019). [DOI] [PubMed] [Google Scholar]
- 127.Kato A, Lamb JC, Birchler JA, Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U. S. A 101, 13554–13559 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Benson G, Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haas BJ, Delcher AL, Wortman JR, Salzberg SL, DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 20, 3643–3646 (2004). [DOI] [PubMed] [Google Scholar]
- 130.Lyons E, Freeling M, How to usefully compare homologous plant genes and chromosomes as DNA sequences: How to usefully compare plant genomes. Plant J. 53, 661–673 (2008). [DOI] [PubMed] [Google Scholar]
- 131.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z, agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC, Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ, Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun 8, 14061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, Integrative genomics viewer. Nat. Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc (1995) (available at 10.1111/j.2517-6161.1995.tb02031.x). [DOI] [Google Scholar]
- 136.Steuernagel B, Witek K, Krattinger SG, Physical and transcriptional organisation of the bread wheat intracellular immune receptor repertoire (2018) (available at https://repository.kaust.edu.sa/handle/10754/628448).
- 137.Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, Stritt C, Roulin AC, Schackwitz W, Tyler L, Martin J, Lipzen A, Dochy N, Phillips J, Barry K, Geuten K, Budak H, Juenger TE, Amasino R, Caicedo AL, Goodstein D, Davidson P, Mur LAJ, Figueroa M, Freeling M, Catalan P, Vogel JP, Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun 8, 2184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sarris PF, Cevik V, Dagdas G, Jones JDG, Krasileva KV, Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de W Van, F. Monteiro, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F, A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana. Cell. 178, 126–1272.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wickham H, ggplot2: Elegant Graphics for Data Analysis (Springer, 2016). [Google Scholar]
- 141.Stamatakis A, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Letunic I, Bork P, Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frith MC, Gentle masking of low-complexity sequences improves homology search. PLoS One. 6, e28819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Frith MC, Kawaguchi R, Split-alignment of genomes finds orthologies more accurately. Genome Biol. 16, 106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Frith MC, Noé L, Improved search heuristics find 20,000 new alignments between human and mouse genomes. Nucleic Acids Res. 42, e59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hamada M, Ono Y, Asai K, Frith MC, Training alignment parameters for arbitrary sequencers with LAST-TRAIN. Bioinformatics (2016), p. btw742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kielbasa SM, Wan R, Sato K, Horton P, Frith MC, Adaptive seeds tame genomic sequence comparison. Genome Research. 21 (2011), pp. 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Song B, Wang H, Wu Y, Rees E, Gates DJ, Burch M, Constrained non-coding sequence provides insights into regulatory elements and loss of gene expression in maize. bioRxiv 10.1101/2020.07.11.192575 (2020). [DOI] [Google Scholar]
- 149.Hubisz M, Pollard K, Siepel A, Package “rphast” (available at https://mran.microsoft.com/snapshot/2017-04-22/web/packages/rphast/rphast.pdf). [Google Scholar]
- 150.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S, Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol 6, e1001025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ogut F, Bian Y, Bradbury PJ, Holland JB, Joint-multiple family linkage analysis predicts within-family variation better than single-family analysis of the maize nested association mapping population. Heredity . 114, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Haller BC, Messer PW, SLiM 3: Forward Genetic Simulations Beyond the Wright–Fisher Model. Molecular Biology and Evolution. 36 (2019), pp. 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia J-M, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, Lai J, Morrell PL, Shannon LM, Song C, Springer NM, Swanson-Wagner RA, Tiffin P, Wang J, Zhang G, Doebley J, McMullen MD, Ware D, Buckler ES, Yang S, Ross-Ibarra J, Comparative population genomics of maize domestication and improvement. Nat. Genet 44, 808–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Clark RM, Tavaré S, Doebley J, Estimating a Nucleotide Substitution Rate for Maize from Polymorphism at a Major Domestication Locus. Mol. Biol. Evol 22, 2304–2312 (2005). [DOI] [PubMed] [Google Scholar]
- 155.Haller BC, SLiM: An Evolutionary Simulation Framework. Note: If you wish to cite SLiM 2 in a publication, please DO NOT cite this manual (unless you are, in fact, specifically referring to this manual--such as citing one of the recipes given here). We expect to have a publication on SLiM 2 out soon; in the meantime, you can cite the paper on the original version of SLiM: Messer, PW (2013). SLiM: Simulating Evolution with Selection and Linkage. Genetics. 194, 1037–1039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ross-Ibarra J, Tenaillon M, Gaut BS, Historical divergence and gene flow in the genus Zea. Genetics. 181, 1399–1413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS, Investigation of the bottleneck leading to the domestication of maize. Proceedings of the National Academy of Sciences. 95 (1998), pp. 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ranere AJ, Piperno DR, Holst I, Dickau R, Iriarte J, The cultural and chronological context of early Holocene maize and squash domestication in the Central Balsas River Valley, Mexico. Proceedings of the National Academy of Sciences. 106 (2009), pp. 5014–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Csilléry K, François O, Blum MGB, abc: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol 3, 475–479 (2012). [Google Scholar]
- 160.Koster J, Rahmann S, Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 28 (2012), pp. 2520–2522. [DOI] [PubMed] [Google Scholar]
- 161.Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S, Lepak NK, Li H, Mitchell SE, Pressoir G, Peiffer JA, Rosas MO, Rocheford TR, Romay MC, Romero S, Salvo S, Sanchez Villeda H, da Silva HS, Sun Q, Tian F, Upadyayula N, Ware D, Yates H, Yu J, Zhang Z, Kresovich S, McMullen MD, The genetic architecture of maize flowering time. Science. 325, 714–718 (2009). [DOI] [PubMed] [Google Scholar]
- 162.Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES, Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet 43, 159–162 (2011). [DOI] [PubMed] [Google Scholar]
- 163.Poland JA, Bradbury PJ, Buckler ES, Nelson RJ, Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. U. S. A 108, 6893–6898 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hung H-Y, Browne C, Guill K, Coles N, Eller M, Garcia A, Lepak N, Melia-Hancock S, Oropeza-Rosas M, Salvo S, Upadyayula N, Buckler ES, Flint-Garcia S, McMullen MD, Rocheford TR, Holland JB, The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity . 108, 490–499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, Holland JB, Flint-Garcia S, McMullen MD, Buckler ES, Rocheford TR, Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 7, e1002383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hung H-Y, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, McMullen MD, Ware D, Buckler ES, Doebley JF, Holland JB, ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. U. S. A 109, E1913–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, Balint-Kurti PJ, Holland JB, Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet 43, 163–168 (2011). [DOI] [PubMed] [Google Scholar]
- 168.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group, The variant call format and VCFtools. Bioinformatics. 27 (2011), pp. 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA, From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinformatics 43 (2013), doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA, Stacks: an analysis tool set for population genomics. Mol. Ecol 22, 3124–3140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B, High-throughput genotyping by whole-genome resequencing. Genome Res. 19, 1068–1076 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES, TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 23, 2633–2635 (2007). [DOI] [PubMed] [Google Scholar]
- 175.Yang J, Hong Lee S, Goddard ME, Visscher PM, GCTA: A Tool for Genome-wide Complex Trait Analysis. The American Journal of Human Genetics. 88 (2011), pp. 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu S, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B, Mattison A, Morishige DT, Grimwood J, Schmutz J, Mullet JE, The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies and annotations can be accessed at https://maizegdb.org/NAM_project. Raw data used for the assemblies including PacBio, Illumina, and Bionano data are available through ENA BioProject IDs PRJEB31061 and PRJEB32225. RNA-Seq data is available at ENA ArrayExpress E-MTAB-8633 and E-MTAB-8628. EM-Seq reads are available at ENA ArrayExpress E-MTAB-10088. ATAC-seq reads are available under NCBI GEO accession GSE165787. Other files, tables and supplemental data can be found in CyVerse https://datacommons.cyverse.org/browse/iplant/home/shared/NAM/NAM_genome_and_annotation_Jan2021_release. Links to the NLR trees can be found at https://itol.embl.de/shared/xCJbI9ndshEK. Scripts used to generate and analyze data are available as a Zenodo repository (61).


