Abstract
Background
The first confirmed case of SARS-CoV-2 in North America was identified in Washington state on January 21, 2020. We aimed to quantify the number and temporal trends of out-of-state introductions of SARS-CoV-2 into Washington.
Methods
We conducted a molecular epidemiologic analysis of 11,422 publicly available whole genome SARS-CoV-2 sequences from GISAID sampled between December 2019 and September 2020. We used maximum parsimony ancestral state reconstruction methods on time-calibrated phylogenies to enumerate introductions/exports, their likely geographic source (US, non-US, and between eastern and western Washington), and estimated date of introduction. To incorporate phylogenetic uncertainty into our estimates, we conducted 5,000 replicate analyses by generating 25 random time-stratified samples of non-Washington reference sequences, 20 random polytomy resolutions, and 10 random resolutions of the reconstructed ancestral state.
Findings
We estimated a minimum 287 introductions (range 244-320) into Washington and 204 exported lineages (range 188-227) of SARS-CoV-2 out of Washington. Introductions began in mid-January and peaked on March 29, 2020. Lineages with the Spike D614G variant accounted for the majority (88%) of introductions. Overall, 61% (range 55-65%) of introductions into Washington likely originated from a source elsewhere within the US, while the remaining 39% (range 35-45%) likely originated from outside of the US. Intra-state transmission accounted for 65% and 28% of introductions into eastern and western Washington, respectively.
Interpretation
The SARS-CoV-2 epidemic in Washington was continually seeded by a large number of introductions. Our findings highlight the importance of genomic surveillance to monitor for emerging variants due to high levels of inter- and intra-state transmission of SARS-CoV-2.
Funding Source
None.
Keywords: SARS-CoV-2, Phylogeography, Genomic analysis, Molecular epidemiology
Research in context.
Evidence before this study
We searched PubMed and preprint servers (MedRxiv and BioRxiv) from December 25, 2019 January 31, 2021, for studies examining the introduction and origins of regional severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemics, using keywords including “SARS-CoV-2”, “introduction”, “origins”, and “emergence”. We found numerous studies that examined the zoonotic origins of SARS-CoV-2, and those that describe the origins of SARS-CoV-2 epidemics in various geographies, including Northern California, New York, Boston, Japan, Italy, Greece, Germany, France, Europe, and India. All of these studies employ phylogenetic analysis of SARS-CoV-2 whole genomes. Only two studies have quantified the number of discrete introductions or identified their likely geographic source, namely in the United Kingdom and Switzerland. There also has been significant attention to the origins of SARS-CoV-2 in Washington state (WA), which was the location of the first confirmed SARS-CoV-2 case in North American. When, in late February a second case was identified in WA, preliminary genomic epidemiological analyses suggested that the second case belonged to the same transmission chain as the first and represented substantial cryptic transmission; although subsequent analyses showed that this second case was likely due to a separate introduction. Phylodynamic modelling also suggested that during the first epidemic wave, 3-10% of SARS-CoV-2 cases in WA were attributable to an introduction.
Added value of this study
We present, to our knowledge, the first quantification of the number of distinct introductions and exports of SARS-CoV-2 within WA and their likely geographic source. We conducted phylogenetic analysis of publicly available SARS-CoV-2 genomes (N=11,422). We estimated there was continuous and substantial introduction of SARS-CoV-2 into WA through September 2020, the majority of which originated from elsewhere within the US. We also evaluated potential trends in the size of downstream WA subclades after introduction, and variation in subclade duration and size.
Implications of all the available evidence
Our findings highlight the importance of genomic surveillance to monitor for emerging variants in WA and elsewhere in the US. Monitoring inter- and intra-state transmissions and their origins can be used to determine where public health interventions may be most effective. In addition, because the estimated number of both introductions and exports both decreased soon after WA's “Stay Home, Stay Healthy” order, lockdowns may be effective at immediately reducing inter-state and intra-state transmission of SARS-CoV-2.
Alt-text: Unlabelled box
1. Introduction
The SARS-CoV-2 pandemic likely first emerged in China in late 2019, and by January 2021 there have been over 100 million confirmed cases and over two million deaths due to COVID-19 worldwide [1]. The first confirmed SARS-CoV-2 infection in North America was identified in Washington State (WA), in January 2020. In February a second case was identified in WA [2], and preliminary genomic epidemiological analyses reported that this case belonged to the same transmission chain as the first case and suggested substantial cryptic transmission [3]. While subsequent analyses showed that this second case was likely due to a separate introduction [4], the initial report spurred a rapid public health response that eventually included school closures and a general lockdown (“stay at home” measures) [5].
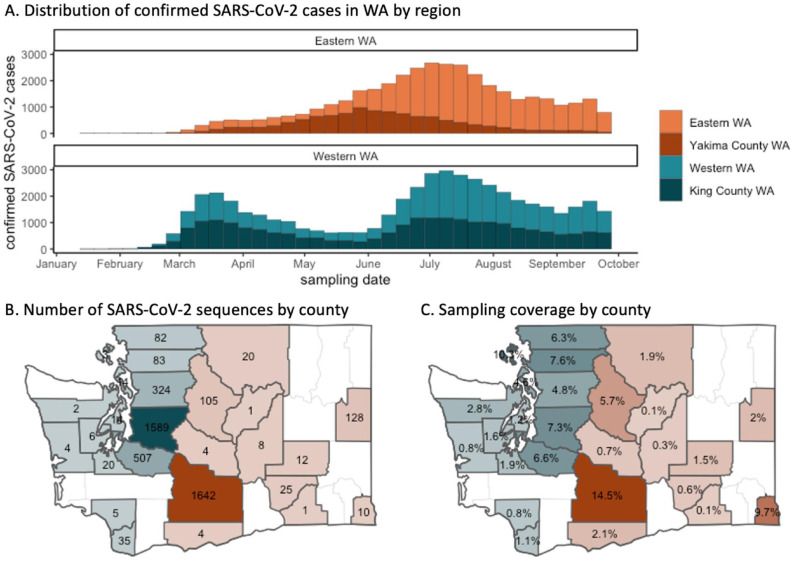
As of January 2021, there have been over 300,000 confirmed cases in WA and over 4,500 deaths due to COVID-19. Within WA the epidemic impact is geographically, demographically, and temporally heterogeneous; there has been substantial variation among counties and ZIP codes in confirmed case counts over time. In particular, eastern and western WA, which are separated by the Cascade mountain range, experienced distinct outbreaks over the spring and summer of 2020 (Figure 1A) and a large proportion of overall confirmed SARS-CoV-2 cases were reported in King and Yakima counties (Figure 1B). As in other locations in the United States (US) and globally, this temporal heterogeneity is likely partially explained by variation in the efficacy of non-pharmaceutical interventions and public health measures such as lockdowns and mask mandates. WA's “Stay-Home Stay-Healthy” order began on March 23rd and required all WA residents to stay home except for essential activities, closed non-essential businesses, and banned gatherings. On May 1, WA announced a plan for reopening in 4 phases, although the majority of WA counties did not progress beyond phase 2 over the summer and fall of 2020 due to high SARS-CoV-2 case counts across the state. A state-wide mask mandate was issued on June 28. Domestic travel into and out of WA state was not restricted prior to November 2020, and international travel was restricted in accordance with federal travel bans imposed on travellers from China, South Korea, Iran, the U.K., and European counties beginning in mid-March.
Figure 1.
Geographic variation in confirmed SARS-CoV-2 cases, genome sequences, and sampling coverage in WA
The SARS-CoV-2 pandemic has resulted in an unprecedented global scientific response, including the rapid sequencing and sharing of SARS-CoV-2 genomes; by October 2020, >120,000 SARS-CoV-2 genomes had been shared online. The analysis of this open data has provided many insights into the zoonotic origin and patterns of global spread of SARS-CoV-2, the development of vaccine candidates, and the surveillance and identification of novel genetic variants of potential public health interest [4,[6], [7], [8], [9], [10]]. The present analysis aimed to combine publicly shared genomic data with limited linked clinical data in order to understand the role of introductions of SARS-CoV-2 during the initial outbreak and through the summer of 2020. Using phylogeographic methods, we aimed to quantify the number, timing, and likely geographic source of out-of-state introductions of SARS-CoV-2 and describe intra-state transmission patterns between eastern and western WA.
2. Methods
Data source
We used full genome SARS-CoV-2 nucleotide sequences obtained from the Global Initiative on Sharing Avian Influenza Data (GISAID, gisaid.org), collected between December 2019 and September 2020. The accession codes for the sequences used in this study are provided in the Digital Supplement. We gratefully acknowledge the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based. Most SARS-CoV-2 sequences available from GISAID are linked to geographic data, including the region, country, and state from which the sequence was sampled. Among sequences sampled in WA over this timeframe and available from GISAID, data on the county of sampling was available for only 53% of sequences. To address this missing data, we obtained additional information on the county from which each sample was obtained from the University of Washington's Virology Lab, which has performed the majority of SARS-CoV-2 full genome sequencing in WA state. Lastly, we used publicly available data on the weekly count of confirmed SARS-CoV-2 cases by county available through the WA Department of Health and estimates of daily SARS-CoV-2 incidence for WA from the Institute for Disease Modeling [11,12]. Confirmed cases of SARS-CoV-2 were defined as the detection of SARS-CoV-2 RNA in a clinical specimen using a molecular amplification detection test by the WA Department of Health.
Sequence analyses
Prior to phylogenetic analyses, we identified and excluded SARS-CoV-2 genome sequences without a complete sampling date, that were incomplete (length <29kbp), or that were low quality (>500 Ns). We aligned sequences with MAFFT and identified SARS-CoV-2 lineages using the PANGOLIN (Phylogenetic Assignment of Named Global Outbreak LINeages, github.com/cov-lineages/pangolin) tool [13,14].
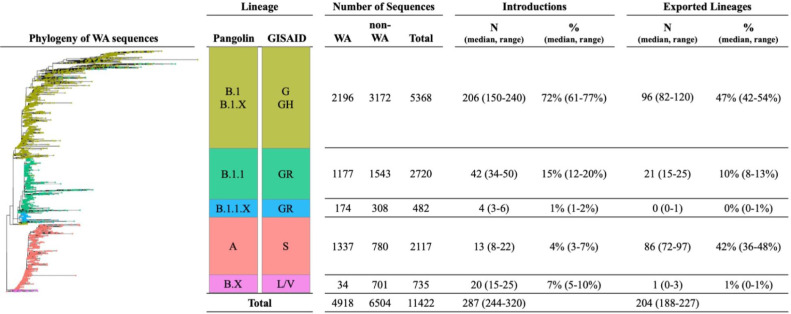
Our analysis pipeline aimed to accommodate issues with SARS-CoV-2 genetic diversity (relative to the timeframes of transmission and accumulation of viral diversity) that lead to challenges with phylogenetic resolution. Notably, SARS-CoV-2 rate of evolution is approximately 10-fold lower than most RNA viruses (approximately 1 × 10−3 substitutions/site/year, or 2 substitutions per month)15 such that the global population of SARS-CoV-2 viruses differed by only 12 nucleotide substitutions by March 2020 [4]. Therefore, we generated a large number of replicate analyses to quantify the uncertainty in our estimates. First, from the full set of global sequences from GISAID (through September 2020) we generated 25 different samples that each included: all WA sequences (N=4918); the closest non-WA sequences for each WA sequence in our sample (based on raw genetic distance; N=5056); a random time-stratified sample of the remaining non-WA sequences (N=1447); and, the Wuhan/Hu-1/2019 reference sequence. Each sample was then stratified by PANGOLIN lineage (A, B.1, B.1.1, B.1.1.X, and B.X, for ). Our choice of five lineages for stratification was based on the phylogeny of WA sequences, and we chose the sequence groups that aligned with monophyletic clades (Figure 2). There are several different nomenclatures for SARS-CoV-2 lineages, therefore, we include both the PANGOLIN and the corresponding GISAID lineages names.
Figure 2.
Estimates number and proportion of SARS-CoV-2 introductions and exports by lineage.
We then reconstructed phylogenies for each of the 25 samples stratified by lineage (N=125 trees) using IQTREE (HKY+G4 substitution model) [16]. Next, for each phylogeny we randomly resolved all polytomies with 20 random resolutions per phylogeny, which resulted in 500 bifurcating trees per lineage. Most of our SARS-CoV-2 phylogenies were characterized by a large number of polytomies; this step allowed us to estimate uncertainty in our estimates that may be due to poor phylogenetic resolution. Next, we time-calibrated each bifurcating tree using the treedater algorithm, assuming a strict molecular clock [17]. We assumed a mean rate of evolution of 0•001 subs/site/year and constrained the rate to be between 0•0009 and 0•0011 substitutions/site/year. This allowed us to estimate to dates of each internal node and the time to most recent common ancestor (MRCA) for each tree.
Lastly, to estimate the number of WA import and export events, we used ancestral state reconstruction to reconstruct the likely state (WA or non-WA) of each node using maximum parsimony methods, implemented with the phangorn package in R. For each phylogeny (i.e. each polytomy-resolved replicate of each PANGOLIN lineage from the 25 sub-samples), we counted the number of introductions to WA and the size of the resulting WA subclades. We identified directional transmission events when the sequential inferred ancestral state (i.e. geographic sampling location) of the internal nodes were not identical when moving from the root of the tree toward the tip. WA subclades were defined as downstream clades that included only WA sequences. We estimated the date of each introduction into WA using the inferred date of the internal node date for the MRCA of each WA subclade. The same approach was used to count lineages exported from WA state, with the opposite direction for each reconstructed node state change.
We also applied ancestral state reconstruction to estimate the likely geographic source of each introduction into WA state (e.g. from outside of the US or from elsewhere within the US), as well as to estimate introductions that occurred within WA state, between eastern and western WA. For the latter of these two analyses, we excluded the 5•5% of WA sequences (N=270) for which the county of sampling was unknown. We included 10 random resolutions of each reconstructed geographic source to account for ambiguous reconstructions.
We provide summary statistics on sequence sampling coverage (overall and by WA county) using both confirmed cases and estimated incident cases as the denominator. Lastly, we estimated the ratio of the number of introductions (representing an individual who acquired SARS-CoV-2 outside of WA) to the total number of WA sequences as a proximal measure of the relative contribution of introductions versus local transmission. This approach allows us to directly compare (via this proxy estimate) our results to those of Müller et al, in which phylodynamic simulation methods estimated that between 1% and 10% of WA cases were imported [18]. For each of our estimates, we report the median and range aggregated across the 5000 replicate analyses. All statistical analyses were conducted using R statistical software version 3.6.2. The use of residual clinical specimens for sequencing was approved by the institutional review board at the University of Washington with a waiver of informed consent.
Role of the funding source
Funders had no role in study design, data collection, analysis, interpretation, or manuscript preparation. The corresponding authors had full access to all study data and final responsibility for the decision to submit for publication.
3. Results
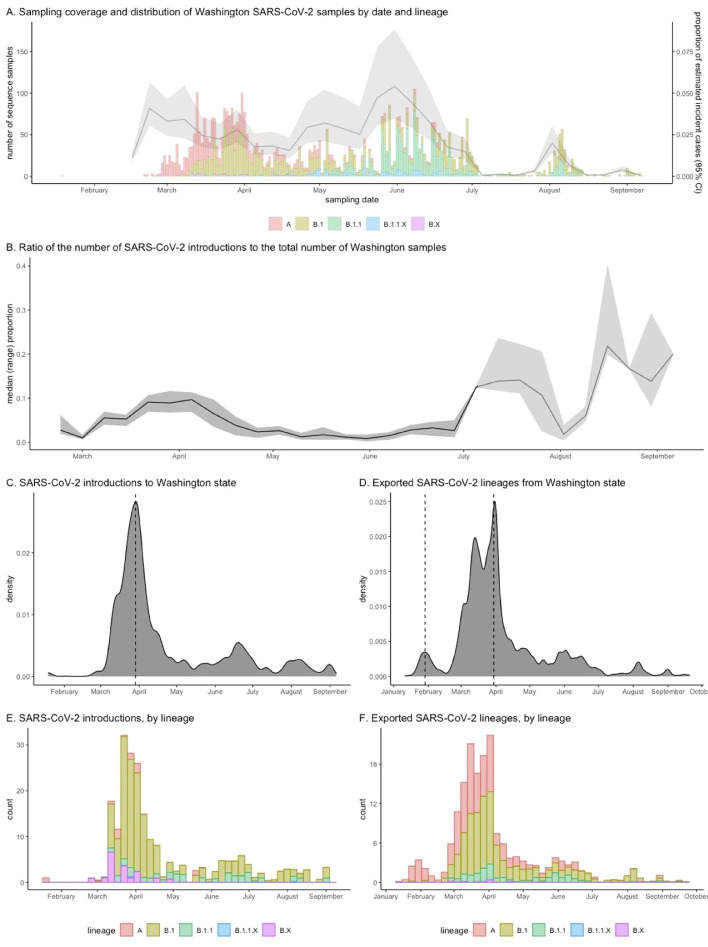
Each analytic sample (replicate) included 11,422 SARS-CoV-2 genomes: 4,918 WA sequences and 6,504 non-WA sequences (Figure 2). Overall, our analysis included sequences for 6% of all confirmed SARS-CoV-2 cases in WA state and 1•8% (95% CI: 1•2-2•8%) of estimated incident SARS-CoV-2 cases [11,12]. The sample coverage for WA state varied modestly between March and June, and decreased significantly in July and late August (Figure 3A).
Figure 3.
Temporal trends in SARS-CoV-2 sampling coverage, introductions and exported lineages for Washington State
We observe temporal trends in prevalent lineage over time (Figure 3A). Early WA samples included mostly sequences of lineages A and B.1. Sequences collected in May and later were comprised largely of B.1.1 lineages. The latter of these (B.1, B.1.1, and B.1.1.X lineages) are all characterized by the Spike 614G variant. There was also substantial county-level variation in sequence sample and coverage (Figure 1B and 1C) and sampling was most representative for King and Yakima counties. Of the 2,688 sequences from western WA, 59% were sampled in King County, and of the 1,960 sequences from eastern WA, 84% were sampled in Yakima County.
We estimate that the were a minimum 287 distinct introductions (range 244-320) of SARS-CoV-2 into WA and 204 exported lineages (range 188-227) through mid-September 2020 (Figure 2). SARS-CoV-2 introductions were primarily B.1 (median 72%, range 61-77%) and B.1.1 (median 15%, range 12-20%) lineages, whereas exported lineages were primarily lineage A (median 42%, range 36-48%) and B.1 (median 47%, range 42-54%).
Most (73%) introductions occurred prior to May 1 and the number of introductions peaked on March 29, 2020, six days following WA's “Stay-Home Stay-Healthy” order (Figure 3). When we stratify by lineage, we observe that lineage A appears to have been introduced at a small number of discrete time points (median 13, range 8-12) while the introduction of lineages B.1 and B.1.1. occur continuously over the spring and summer months. In contrast we observe two waves of exported lineages, the first peaking on January 29, 2020 and the second peaking on March 30, 2020. The first wave of exported lineages was comprised entirely of lineage A while the second wave was predominately lineages A and B.1.
Overall, the ratio of introductions to sampled sequences was 6•1% (range 5•3-6•8%), and is a proxy measure for the relative contribution of introductions to overall incidence. In our analysis, this ratio peaked in late March at 9•7% (range 7•0-11•2%) and fell to around 1-3% in May and June (Figure 3B). After July, our estimate of this ratio is less reliable due to significant under-sampling (Figure 3A).
We estimated that the majority (median 61%, range 55-65%) of introductions into WA state likely originated from a source elsewhere within the US, while the remaining 39% (range 35-45%) of introductions likely originated from outside of the US (Table 1). These proportions did not vary over the study period. We also observed a significant amount of intra-state SARS-CoV-2 transmission. There were a large number of introductions from western WA into eastern WA (median 130, range 115-153), and this comprised the majority (median 65%, range 59-73%) of introductions into the eastern region on the state compared to those from elsewhere in the US (median 20%, range 12-27%) or outside of US (median 15%, range 10-21%). Conversely, there were slightly fewer distinct introductions from eastern WA into western WA (median 94, range 77-115), but these comprised a small proportion of all introductions into the western region on the state (median 28%, range 24-31%) compared to those from elsewhere in the US (median 49%, range 42-57%) or outside of US (median 23%, range 17-29%).
Table 1.
Count and proportion of introductions into Washington state by geographic region
| Likely Geographic Source of Introductions | WA Overall | Western WA | Eastern WA | |||
| Nmedian (range) | %median (range) | Nmedian (range) | %median (range) | Nmedian (range) | %median (range) | |
| US | 177 (126-218) | 61% (51-68%) | 163 (137-191) | 49% (42-57%) | 41 (25-56) | 20% (12-27%) |
| Outside of US | 111 (91-136) | 39% (31-48%) | 77 (57-100) | 23% (17-29%) | 30 (19-41) | 15% (10-21%) |
| Intra-state | ||||||
| Western WA | 130 (115-153) | 65% (59-73%) | ||||
| Eastern WA | 94 (77-115) | 28% (24-31%) | ||||
| Total | 287 (244-320) | 336 (305-366) | 201 (73-187) | |||
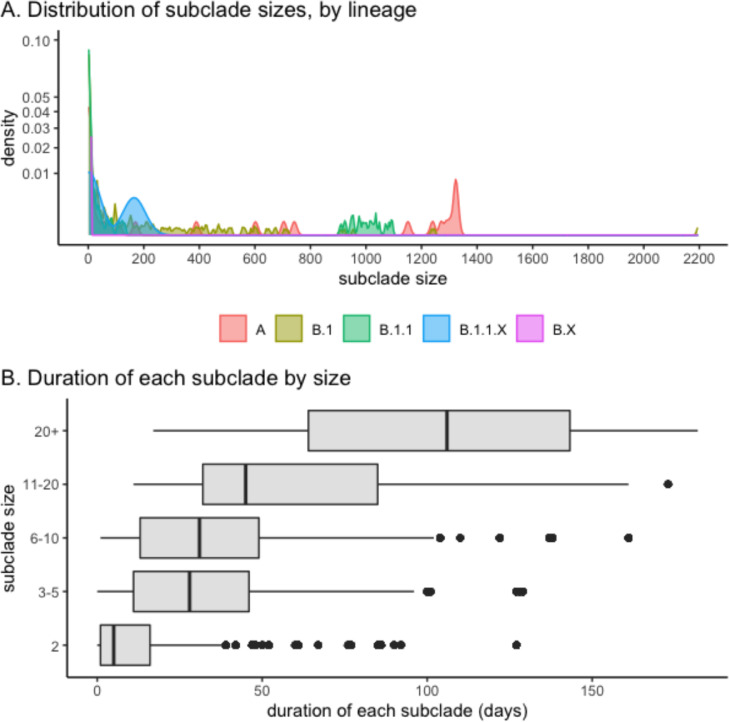
The size of WA subclades resulting from each introduction ranged from 1 to 2,193 sequences (Figure 4A). Most introductions resulted in a single WA sequences (72%) or small subclades of 2 (9%) or 3 to 5 SARS-CoV-2 sequences (8%). The remaining 6% of introductions resulted in moderately sized subclade of 6-20 WA sequences, and 6% resulted in large subclades of 20 or more WA sequences. The largest WA subclades, that included more than 900 WA sequences, were subtypes A and B.1.1. The duration of each subclade – defined as the number of days from first to last sequence collection – was positively correlated with subclade size (Figure 4B). Subclades of just 2 sequences had the shortest duration, with a median of 5 days (IQR 1-16 days), the median duration for small subclades of 3 to 5 sequences was 28 days (IQR 11-46 days). Moderately sized and large subclades had a median duration of 38 days (IQR 20-62 days).
Figure 4.
Size and duration of Washington state subclades resulting from an out-of-state introduction of SARS-CoV-2
Discussion
We found phylogenetic evidence that the SARS-CoV-2 epidemic in WA was seeded by a large number of distinct, ongoing introductions through September 2020. We similarly estimated that a large number of SARS-CoV-2 lineages were exported from WA state during the same period. Introductions appeared to play a significant role early in the epidemic in March and April, but they continued through the summer of 2020. Notably, the peak of introductions on March 29, 2020 coincided exactly with the highest day of test-positivity in WA state [19]. Although many introductions of SARS-CoV-2 result in a single descendent WA sequence, which is suggestive of limited local transmission, the sequence collection time frame for small clades lasted on the order of a week to one and a half months, which suggests these clades represent a modest degree of local transmission. In addition, approximately 12% of introductions resulted in larger subclades, corresponding to long chains of local SARS-CoV-2 transmission.
The majority of introductions of SARS-CoV-2 into WA likely originated from elsewhere within the US. Thus, inter-state travel within the US may play a more important role in sustaining and re-seeding local epidemics, compared to international travel. This is consistent with prior analyses which found that domestic and inter-state travel is a significant source of new SARS-CoV-2 infections, including a Connecticut outbreak that was phylogenetically linked to the initial outbreak in WA state as the likely source [20]. We also observed a significant amount of intra-state transmission between the eastern and western regions of WA. Notably, intra-state transmission account for most transmissions into eastern WA, but the converse was not true. This asymmetrical transmission pattern may be because western WA includes infrastructure for both inter-state and international travel: the Seattle-Tacoma international airport, the seaports, and the Interstate-5 corridor, the main north-south highway that connects several large cities in California, Oregon, and Washington.
We observed distinct temporal trends in introductions and exports by lineage, and lineages with the Spike 614G variant accounted for the majority (88%) of introductions. However, the temporal patterns of introductions and exported SARS-CoV-2 lineages mirrors the temporal shifts of dominant clades that occurred elsewhere in the US and globally [8]. During the study period, none of the variants of concern from the United Kingdom (B.1.1.7 or SARS-CoV-2 VOC 202012/01), South Africa (B.1.351 or 501.V2), or Brazil (P.1), or India (B.1.617.2) were circulating in WA. The first case of B.1.1.7 variant in WA state was sampled from Snohomish County on December 25, 2020; the first case of B.1.351 was sampled in January 2021 in King County; the first case of P.1 was sampled in March 2021, also in King County; and the first case of B.1.617.2 was sampled in April 2021.
Other studies examining the origins of SARS-CoV-2 in Brazil, Northern California, New York, and Boston have also found evidence of multiple introductions [10,[21], [22], [23]]. Our findings are consistent with prior analyses that quantified the relative important of introductions of SARS-CoV-2 [24,25]. Using a similar analytic approach, du Plessis et al. similarly found a large number of introduction of SARS-CoV-2 into the United Kingdom (N=1179, 95% interval 1143-1286) through June 2020; the majority of which resulted in small clades of fewer than 10 sequences [25]. In the United Kingdom, most introductions that occurred prior to May likely originated in Italy, Spain, or France. Another analysis of the relative contribution on introductions to the SARS-CoV-2 epidemic in Switzerland found that most introduced lineages were from neighbouring counties (France, Italy, Germany and Belgium) [24]. Our findings are also consistent with phylodynamic modelling during the first epidemic wave that estimated 3-10% of SARS-CoV-2 cases in WA (excluding Yakima County) were attributable to an introduction; we also observed two distinct waves of introductions, the first which included lineages with the original Spike 614D mutation (i.e. lineage A), followed by the introductions of lineages with the Spike 614G variant (i.e. lineages B.1 and B.1.1) [18].
This study has a number of strengths. To our knowledge, this is the first large-scale phylogenetic analysis to quantify the number of introductions of SARS-CoV-2 in a region of the US or to quantify within-county and within-state transmission patterns using phylogenetic methods. In addition, our findings were robust to the inclusion of different time-stratified random samples of non-WA reference sequences.
Our findings should be interpreted in light of the following limitations. First, we likely are underestimating the true number of introductions due to incomplete sampling. Phylogenetic results need to be interpreted carefully due to incomplete sampling and phylogenetic uncertainty. Because our sampling coverage was only 6% of confirmed SARS-CoV-2 cases, if we were to sequence more genomes, we would find more introductions. Although GISAID and participating laboratories have facilitated the availability of an unprecedented number of publicly available whole genomes of SARS-CoV-2, within specific geographies the sampling coverage is very low and there is significant over-representation of sequences from certain countries (e.g. the United Kingdom, Australia, and the US). Similarly, sampling coverage varied across WA both geographically and temporally; King and Yakima counties account for the majority of SARS-CoV-2 sequences used in our analysis, while other WA counties had significantly lower sampling coverage (Figure 1), In addition, overall WA sampling coverage was very low in July and September 2020 (Figure 3A). Second, due to length-biased sampling, we were unable to assess temporal variation in downstream clade sizes changed over time. Lastly, phylogeographic inference is limited by the low genetic diversity of SARS-CoV-2 as well as the reduction in viral genetic diversity that results from pandemic mitigation measures; therefore, our estimated of the ratio of US to non-US introductions is vulnerable to some degree of bias.
Our findings have several important public health implications. First, our findings highlight the importance of genomic surveillance to monitor for emerging variants in WA and elsewhere in the US due to the high levels of inter- and intra-state transmission of SARS-CoV-2 lineages [26]. Monitoring inter- and intra-state transmissions and their origins can be used to determine where public health interventions may be most effective, such as the relative impact of regional or local versus international travel restrictions. In addition, we observed that number of both introductions and exports fell within the week of the “Stay Home, Stay Health” order, suggesting that lockdowns and travel restrictions may be effective at immediately reducing inter- and intra-state transmission by a reducing overall mobility.
Contributors
DMT and JTH contributed to the study conceptualization, methodology, writing of the original draft, and accessed and verified the data. DMT conducted the formal analysis and visualization. ALG, PR, LS, HX, KRJ, NB, and MLH collected and analysed laboratory data (including genomic sequencing) and contributed to data curation. MF contributed to methodology and interpreted results. All authors reviewed, edited, revised and gave final approval of the Article before submission.
Declaration of interests
ALG reports personal fees from Abbott Molecular, grants from Merck, grants from Gilead, outside the submitted work. DMT, PR, LS, HX, KRJ, NB, MLH, MF and JTH have nothing to disclose.
Acknowledgments
Acknowledgements
We would like to acknowledge Dr. Niket Thakker for providing incidence estimates and reviewing the manuscript, as well as the UW Virology staff who assisted to sequence collection and assembly. We gratefully acknowledge the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based. DMT receives support from the NIH National Institute of Allergy And Infectious Diseases (F31AI152542). JTH and DMT also receive support from the NIH National Institute of Allergy and Infectious Disease (R01AI127232). Funders had no role in study design, collection, analysis, and interpretation of data, writing of the report, or the decision to submit the paper for publication.
Data sharing statement
Full genome SARS-CoV-2 nucleotide sequences and associated metadata are publicly available and can be accessed from the Global Initiative on Sharing Avian Influenza Data (GISAID, gisaid.org). The accession codes for the sequences used in this study are provided in the Digital Supplement. Publicly available data on confirmed SARS-CoV-2 cases are available through the WA Department of Health (https://www.doh.wa.gov/Emergencies/COVID19/DataDashboard). Estimates of daily SARS-CoV-2 incidence for WA from the Institute for Disease Modeling and University of Washington's Virology Lab data on the county each sample was obtained could be made available upon request at the discretion of the authors; requests can be sent to the corresponding author.
Editor note: The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.lana.2021.100018.
Appendix. Supplementary materials
References
- 1.World Health Organization . Feb 12, 2021. Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/(accessed. [Google Scholar]
- 2.McMichael TM, Currie DW, Clark S, et al. Epidemiology of Covid-19 in a Long-Term Care Facility in King County. Washington. N Engl J Med. 2020;382:2005–2011. doi: 10.1056/NEJMoa2005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford T, Greninger AL, Roychoudhury P, et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worobey M, Pekar J, Larsen BB, et al. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370:564–570. doi: 10.1126/science.abc8169. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell SH, Bulger EM, Duber HC, et al. Western Washington State COVID-19 Experience: Keys to Flattening the Curve and Effective Health System Response. J Am Coll Surg. 2020;231:316–324. doi: 10.1016/j.jamcollsurg.2020.06.006. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feb 12, 2021. Global initiative on sharing all influenza data (GISAID) https://www.gisaid.org/(accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacLean OA, Lytras S, Weaver S, et al. bioRxiv; 2020. Natural selection in the evolution of SARS-CoV-2 in bats, not humans, created a highly capable human pathogen. 2020.05.28.122366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dearlove B, Lewitus E, Bai H, et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc Natl Acad Sci U S A. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washington NL, Gangavarapu K, Zeller M, et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv. 2021 doi: 10.1016/j.cell.2021.03.052. 2021.02.06.21251159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Gu W, Federman S, et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thakkar N, Famulare M. One state, many outbreaks: a transmission modeling perspective on current COVID-19 trends in King, Pierce, and Yakima counties What do we already know? Seattle, WA, 2020 https://iazpvnewgrp01.blob.core.windows.net/source/2021-02/reports/pdf/One_state_many_outbreaks.pdf (accessed March 25, 2021).
- 12.Thakkar N, Burstein R, Famulare M. Towards robust, real-time, high-resolution COVID-19 prevalence and incidence estimation What do we already know? Seattle, WA, 2020 https://iazpvnewgrp01.blob.core.windows.net/source/2021-02/reports/pdf/Towards_robust_real_time_high_resolution_COVID_19_prevalence_and_incidence_estimation.pdf (accessed March 25, 2021).
- 13.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambaut A, Holmes EC, O'Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch JW, Capoferri AA, Katusiime MG, Patro SC, Kearney MF. Low genetic diversitymay be an Achilles heel of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24614–24616. doi: 10.1073/pnas.2017726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volz EM, Frost SDW. Scalable relaxed clock phylogenetic dating. Virus Evol. 2017;3 doi: 10.1093/ve/vex025. [DOI] [Google Scholar]
- 18.Müller NF, Wagner C, Frazar CD, et al. medRxiv; 2020. Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State. published online Sept 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randhawa AK, Fisher LH, Greninger AL, et al. Changes in SARS-CoV-2 Positivity Rate in Outpatients in Seattle and Washington State, March 1-April 16, 2020. JAMA - J. Am. Med. Assoc. 2020;323:2334–2336. doi: 10.1001/jama.2020.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fauver JR, Petrone ME, Hodcroft EB, et al. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell. 2020;181:990–996. doi: 10.1016/j.cell.2020.04.021. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Candido DS, Claro IM, de Jesus JG, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 2020;369:297–301. doi: 10.1126/science.abc1917. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemieux JE, Siddle KJ, Shaw BM, et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2020;371:eabe3261. doi: 10.1126/science.abe3261. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadeau S, Beckmann C, Topolsky I, et al. medRxiv; 2020. Quantifying SARS-CoV-2 spread in Switzerland based on genomic sequencing data. 2020.10.14.20212621. [Google Scholar]
- 25.du Plessis L, McCrone JT, Zarebski AE, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;10:eabf2946. doi: 10.1126/science.abf2946. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodcroft EB, Domman DB, Oguntuyo K, et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv. 2021 2021.02.12.21251658. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.