SUMMARY
Animals encounter microorganisms in their habitats, adapting physiology and behavior accordingly. The nematode Caenorhabditis elegans is found in microbe-rich environments; however, its responses to fungi are not extensively studied. Here, we describe interactions of C. elegans and Penicillium brevicompactum, an ecologically relevant mold. Transcriptome studies reveal that co-culture upregulates stress response genes, including xenobiotic-metabolizing enzymes (XMEs), in C. elegans intestine and AMsh glial cells. The nuclear hormone receptors (NHRs) NHR-45 and NHR-156 are induction regulators, and mutants that cannot induce XMEs in the intestine when exposed to P. brevicompactum experience mitochondrial stress and exhibit developmental defects. Different C. elegans wild isolates harbor sequence polymorphisms in nhr-156, resulting in phenotypic diversity in AMsh glia responses to microbe exposure. We propose that P. brevicompactum mitochondria-targeting mycotoxins are deactivated by intestinal detoxification, allowing tolerance to moldy environments. Our studies support the idea that C. elegans NHRs may be regulated by environmental cues.
Graphical Abstract
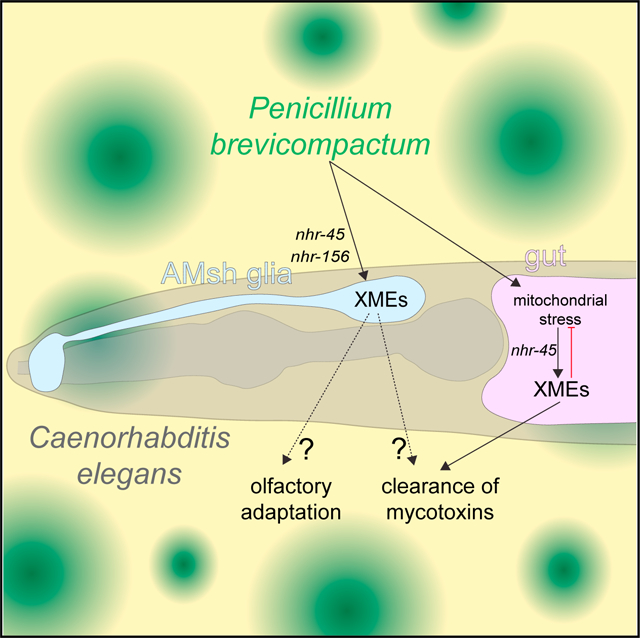
In brief
C. elegans inhabits microbe-rich environments, and there is currently great interest in understanding how they interact with the microbes they encounter in the wild. Wallace et al. describe a transcriptional response that protects C. elegans from mitochondrial toxicity when exposed to the fungal mold Penicillium brevicompactum.
INTRODUCTION
A number of experimental paradigms have been established to study the interactions between C. elegans and model microbial pathogens (Jiang and Wang, 2018; Kim and Ewbank, 2018; Kim and Flavell, 2020; Madende et al., 2020; Radeke and Herman, 2021; Troemel, 2016). Recent work has also explored the interactions between C. elegans and the microbial components of its natural substrates in the wild (Schulenburg and Félix, 2017; Zhang et al., 2017), with most of these studies focusing on bacteria. More than 100 mycotoxins have been identified with nematocidal activities (Li et al., 2007), highlighting the notion that, to survive in its habitat, C. elegans also possess mechanisms to counter fungi. The Penicillium genus includes a large number of abundant fungal species, and the mold Penicillium brevicompactum is found naturally in substrates for wild isolates of C. elegans (Dirksen et al., 2016). Interactions between C. elegans and fungi have not been extensively studied, and effects of Penicillium species, in particular, on C. elegans are virtually unexplored (Huang et al., 2014; Visagie et al., 2014).
Xenobiotic-metabolizing enzymes (XMEs) are key detoxification proteins that include phase I enzymes of the cytochrome P450 (cyp) family, and phase II enzymes of the UDP-glucoronosyltransferase (ugt) and glutathione-S-transferase (gst) families (Hartman et al., 2021). XME functions have been extensively explored in the mammalian liver, where they play a major role in detoxification. XME responses are induced by the nuclear hormone receptors (NHRs) CAR and PXR after drug exposure (Wallace and Redinbo, 2012). The NHR family in C. elegans has undergone massive expansion and sequence diversification (Taubert et al., 2011). NHR genes promote responses to microbe exposure and environmental stress (Jones et al., 2013; Mao et al., 2019; Otarigho and Aballay, 2020; Park et al., 2018; Peterson et al., 2019; Rajan et al., 2019; Shomer et al., 2019; Wani et al., 2021; Ward et al., 2014; Yuen and Ausubel, 2018). Induction of XMEs has also been observed in C. elegans in response to exposure to several model pathogens (Engelmann et al., 2011).
Here, we establish an assay to characterize the interactions between C. elegans and P. brevicompactum. We show that NHRs mediate P. brevicompactum-dependent induction of XMEs in the intestine and in glia, and demonstrate a role for NHRs in protecting C. elegans from the toxic effects of P. brevicompactum exposure.
RESULTS
P. brevicompactum exposure induces expression of stress response genes including XMEs
To establish an assay for investigating interactions between C. elegans and fungal molds, we sequenced the internal transcribed spacer rDNA regions (Schoch et al., 2012) of moldy contaminants occurring spontaneously on NGM (nematode growth medium) plates in our laboratory. Penicillium brevicompactum (ATCC no. 9056) was identified as a common contaminant, and chosen for further study because, in addition to growing readily on NGM plates suitable for C. elegans co-culture, it is found naturally in substrates for wild isolates of C. elegans (Dirksen et al., 2016). To characterize transcriptional changes in C. elegans following P. brevicompactum exposure, whole-animal mRNA was extracted and sequenced from young adult nematodes grown in the presence of the standard food source E. coli OP50, with or without P. brevicompactum co-culture (see Figure S1A for PCA). We identified 100 genes that are significantly upregulated (fold change > 2, p-adj < 0.05, Tables S1 and S3) and 187 genes that are significantly downregulated (fold change < 0.5, p-adj < 0.05, Tables S2 and S3) following P. brevicompactum exposure. Functional analysis using Worm-Cat (Holdorf et al., 2019) showed that more than one-third of the upregulated genes are involved in stress responses (Figure 1A). Statistical enrichment analysis revealed that genes belonging to the stress response sub-categories “detoxification,” “pathogen,” and “heavy metal” are significantly enriched in the dataset of upregulated genes, with genes in the “detoxification” category also showing significant enrichment in the dataset of downregulated genes (Figure 1B). As detoxification genes are the most abundant stress-gene class modulated by P. brevicompactum exposure, we focused on these for further characterization.
Figure 1. Penicillium brevicompactum induces expression of stress response genes including xenobiotic-metabolizing enzymes in C. elegans.
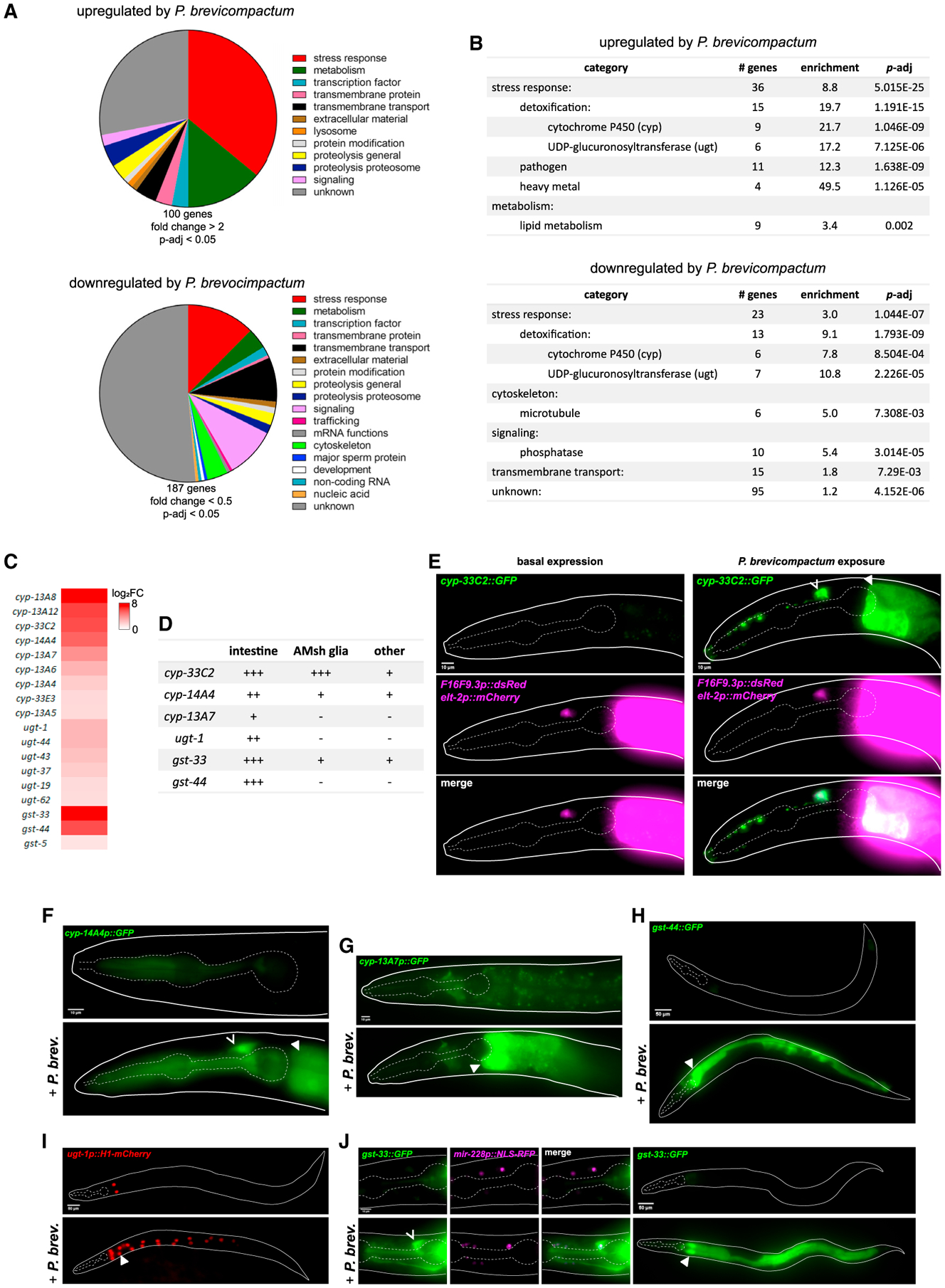
(A) Classification of P. brevicompactum-dependent genes.
(B) Statistically enriched gene categories. (A) and (B) Based on analysis using WormCat (Holdorf et al., 2019).
(C) Heatmap of xenobiotic-metabolizing enzymes (XMEs) induced by P. brevicompactum.
(D) Summary of confirmed P. brevicompactum-induced genes. +++ strong expression; ++ moderate expression; + weak expression. Tissues classified as other are as follows: cyp-33C2, excretory cell, vulva, several undefined cells in the head and tail; cyp-14A4, pharynx; gst-33, excretory cell, pharynx, several undefined cells in the head and tail.
(E) Expression of cyp-33C2∷GFP with F16F9.3p∷dsRed (AMsh glia) and elt-2p∷mCherry (intestine).
(F–J) expression of additional XMEs listed in (D). mir-228p∷NLS-RFP is pan-glial. Open arrowhead, AMsh glia; closed arrowhead, intestine. All data are based on three biological replicates. Scale bars, 10 μm (E, F, G, and J [left]), 50 μm (H, I, and J [right]).
XMEs are major detoxification proteins encoded by cytochrome P450 (cyp), UDP-glucoronosyltransferase (ugt), and glutathione-S-transferase (gst) gene families (Hartman et al., 2021). Eighteen of the P. brevicompactum-induced genes encode XMEs (Figure 1C). To confirm P. brevicompactum-dependent induction, and to identify sites of expression, we examined fluorescent transcriptional reporters for six highly induced XMEs. All showed robust induction, validating our RNA sequencing (RNA-seq) data (Figures 1D–1J). XME induction was consistently observed in the intestine, in line with previous reports of detoxification functions in this tissue (Hartman et al., 2021). XME induction was also observed in other tissues, and most consistently in AMsh glial cells, which are components of the major sensory organ in the head of the animal (Singhvi and Shaham, 2019). In these experiments, animals were grown in the presence of P. brevicompactum from the L1 stage and scored 2 days later as young adults. XME induction is also observed in adults acutely exposed to P. brevicompactum, as demonstrated by cyp-33C2∷GFP induction within 3 h of exposure in both the intestine and AMsh glia (Figure S1B). Induction of XMEs in AMsh glia is particularly interesting given that XMEs are highly expressed in sustentacular cells, the analogous cell type in the mammalian olfactory epithelium (Heydel et al., 2013).
Overall, our sequencing data demonstrate that exposure of C. elegans to P. brevicompactum causes significant changes in gene expression, strongly suggesting a biologically meaningful response to this microbe. Furthermore, the significant enrichment of stress response genes, and specifically of detoxification genes, suggests that P. brevicompactum may constitute a toxic species for C. elegans.
The NHRs nhr-45 and nhr-156 are required for XME induction
To investigate the molecular mechanism through which XMEs are induced by P. brevicompactum, we initially looked at well-characterized innate immune genes of the p38 MAP kinase pathway (Kim et al., 2002), and at cilia genes that are required for chemosensation (Inglis et al., 2007). Mutations in neither gene class affect XME induction (Figure S2A). We also assessed whether AMsh glia are required for the response in the intestine by using a previously published line in which AMsh glia have been genetically ablated (Bacaj et al., 2008), and found that AMsh glia are not required for the intestinal induction of cyp-33C2 (Figure S2A).
In light of the established role of CAR and PXR receptors in the induction of XMEs in the mammalian liver (Wallace and Redinbo, 2012), we next screened available C. elegans nhr RNAi clones (Table S4) for effects on P. brevicompactum-dependent induction of cyp-33C2∷GFP, an XME reporter we developed (Figure 1E). This screen revealed that nhr-45 is required for cyp-33C2 induction in both intestine and AMsh glia, while nhr-156 is partially required for cyp-33C2 induction in AMsh glia only. These results were confirmed using a panel of loss-of-function mutants either previously identified or generated in this study (Figures 2A–2C).
Figure 2. nhr-45 and nhr-156 regulate induction of XMEs.
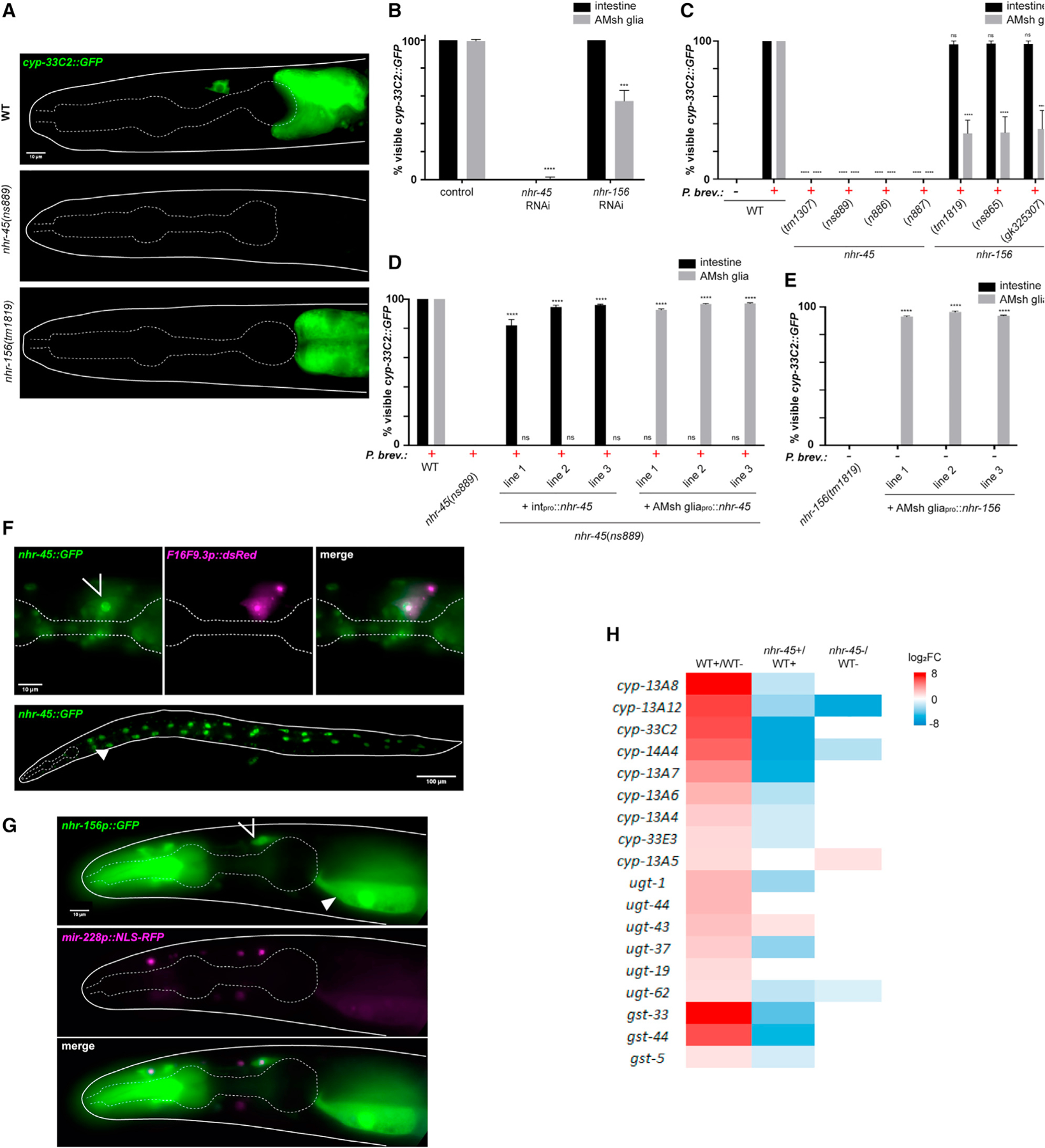
(A) cyp-33C2∷GFP induction following P. brevicompactum exposure.
(B) cyp-33C2∷GFP induction in intestine and AMsh glia in RNAi-treated worms. ****p < 0.0001, ***p < 0.001 (t test). Note that p values cannot be calculated for the intestine because there is no variation between replicates.
(C) cyp-33C2∷GFP induction in intestine and AMsh glia. nhr-45 (tm1307), 1,545 bp deletion with 235 bp insertion; nhr-156 (tm1819), 553 bp deletion; nhr-156 (gk325307), Trp215 > amber. New alleles generated are as follows: nhr-45 (ns886) (14 bp deletion in exon1, generated by CRISPR); nhr-45 (ns887) (11 bp insertion in exon 1, generated by CRISPR); nhr-45 (ns889) (7 bp deletion in exon 1, generated by CRISPR); nhr-156 (ns865) (Gln29 > ochre, generated by EMS mutagenesis). ****p < 0.0001 (ANOVA compared with WT + P. brevicompactum).
(D) cyp-33C2∷GFP induction in intestine and AMsh glia. Intpro is elt-2p for intestinal rescue, AMsh gliapro is F16F9.3p for glial rescue. ****p < 0.0001 (ANOVA compared with nhr-45[ns889]).
(E) Basal expression of cyp-33C2∷GFP in the absence of P. brevicompactum. ****p < 0.0001 (ANOVA).
(F) Expression of nhr-45∷GFP with F16F9.3p∷dsRed (AMsh glia).
(G) Expression of nhr-156p∷GFP with mir-228p∷NLS-RFP (pan-glia). In (F) and (G), open arrow, AMsh glia; closed arrow, intestine.
(H) Heatmap of expression changes of the 18 P. brevicompactum-induced XMEs in nhr-45 partial loss-of-function mutant strain (nhr-45 [ns889]; nsIs910 [elt-2p∷nhr-45]). +, P. brevicompactum exposure;−, control samples not exposed. All data are based on three biological replicates. Error bars, SEM (B, C, D, and E). Scale bars, 10 μm (A, F [top], and G), 100 μm (F [bottom]).
The two NHRs we identified function cell autonomously to induce cyp-33C2 expression. Introduction of an nhr-45 cDNA under the control of intestine- or AMsh glia-specific promoters into nhr-45 mutant animals is sufficient to rescue cyp-33C2∷GFP induction defects in the corresponding tissues (Figure 2D). Interestingly, overexpression of nhr-156 (but not overexpression of nhr-45) is sufficient to induce expression of cyp-33C2 even in the absence of P. brevicompactum (Figure 2E). It is possible that the nhr-156 multicopy DNA array generated in these strains bypasses its physiological regulation, resulting in ligand-independent activity. While this result precludes straight-forward interpretation of our rescue experiments, it provides further evidence that nhr-156 can regulate cyp-33C2∷GFP induction. Furthermore, nhr-156 overexpression in AMsh glia induces cyp-33C2∷GFP expression only in AMsh glia, demonstrating a cell-autonomous function. Consistent with their cell-autonomous functions, expression of nhr-45 and nhr-156 is observed in intestinal cells and AMsh glia, as well as in other cell types (Figures 2F and 2G). In these studies, nhr-45 expression was monitored using a recombineered fosmid encoding a translational fusion to GFP, revealing NHR-45:GFP accumulation in nuclei (Figure 2F). nhr-156 expression was observed using an nhr-156 promoter:GFP transcriptional reporter (Figure 2G). Overexpression of nhr-156 in the intestine, using its endogenous promoter, is sufficient to drive basal expression of cyp-33C2 in the intestine (Figure S2B). This is perhaps surprising, given that nhr-156 is not required for P. brevicompactum-dependent induction of cyp-33C2 in this tissue, and suggests that cell-specific physiological regulation of nhr-156 activity may be compromised when nhr-156 is overexpressed.
nhr-45 is a major regulator of P. brevicompactum-dependent XME induction
To determine if additional XME genes induced by P. brevicompactum are dependent on nhr-45, we aimed to carry out RNA-seq experiments on nhr-45 mutant animals. These studies were complicated by our observation (see below) that P. brevicompactum at high concentrations is toxic to nhr-45 mutant animals and causes developmental defects. To circumvent this issue, we took advantage of an nhr-45 mutant line with a genomically integrated intestine-specific nhr-45-expressing transgene (nsIs910 [elt-2p:nhr-45]) that serendipitously exhibits only minimal rescue of cyp-33C2∷GFP induction in the intestine, but is nonetheless able to rescue the developmental defects. We hypothesized that this strain contains only partial nhr-45 activity in the intestine, such that it can drive partial expression of target genes in a manner that is sufficient to prevent P. brevicompactum toxicity (see below), but still allows nhr-45-dependent transcripts to be identified. We predicted that at least some nhr-45-dependent transcripts could be identified in this strain, although the extent of this dependence may be underestimated.
By comparing nhr-45 mutant animals to wild-type animals after exposure to P. brevicompactum, we observed that 31 of the 100 P. brevicompactum-induced genes we identified require nhr-45 for their full induction (fold-decrease > 2, p-adj < 0.05 when comparing nhr-45 mutant with wild-type animals) (Table S1). Of the 18 inducible XMEs we identified, 14 are nhr-45 dependent (Figure 2H), including cyp-33C2, as predicted. We validated the RNA-seq results by assessing induction of cyp-14A4p∷GFP in the intestine and in AMsh glia, and found that, in both tissues, induction was indeed nhr-45 dependent (Figure S2C and S2D). With a few exceptions, basal expression of XMEs in the absence of P. brevicompactum was not significantly affected in nhr-45 mutants (Figure 2H). These results show that nhr-45 is a major regulator of P. brevicompactum-dependent XME induction in both the intestine and AMsh glia.
P. brevicompactum is toxic for nhr-45 mutant animals that fail to induce XMEs in the intestine
Our transcriptome analysis of P. brevicompactum-inducible genes revealed strong enrichment for stress response genes involved in detoxification. Furthermore, a large number of mycotoxins have been shown to have nematocidal activities (Li et al., 2007). Together, these observations suggest that P. brevicompactum may have toxic effects on C. elegans, and that induction of detoxification enzymes may represent a stress response aimed at clearing mycotoxins. Supporting this idea, we observed that nhr-45 mutants grown on a plate densely seeded with P. brevicompactum develop slowly, and in some cases arrest and die at early larval stages. This is in contrast to wild-type animals, which are able to tolerate P. brevicompactum exposure (Figures 3A and 3B). Importantly, nhr-45 mutant animals show normal developmental profiles in the absence of mold (Figures 3A and 3B), indicating that nhr-45 does not generally affect development. The developmental defect resulting from P. brevicompactum exposure can be rescued by restoring nhr-45 expression in the intestine, but not in AMsh glia (Figure 3C).
Figure 3. P. brevicompactum exposure causes mitochondrial stress and is toxic for nhr-45 mutants with impaired XME induction.
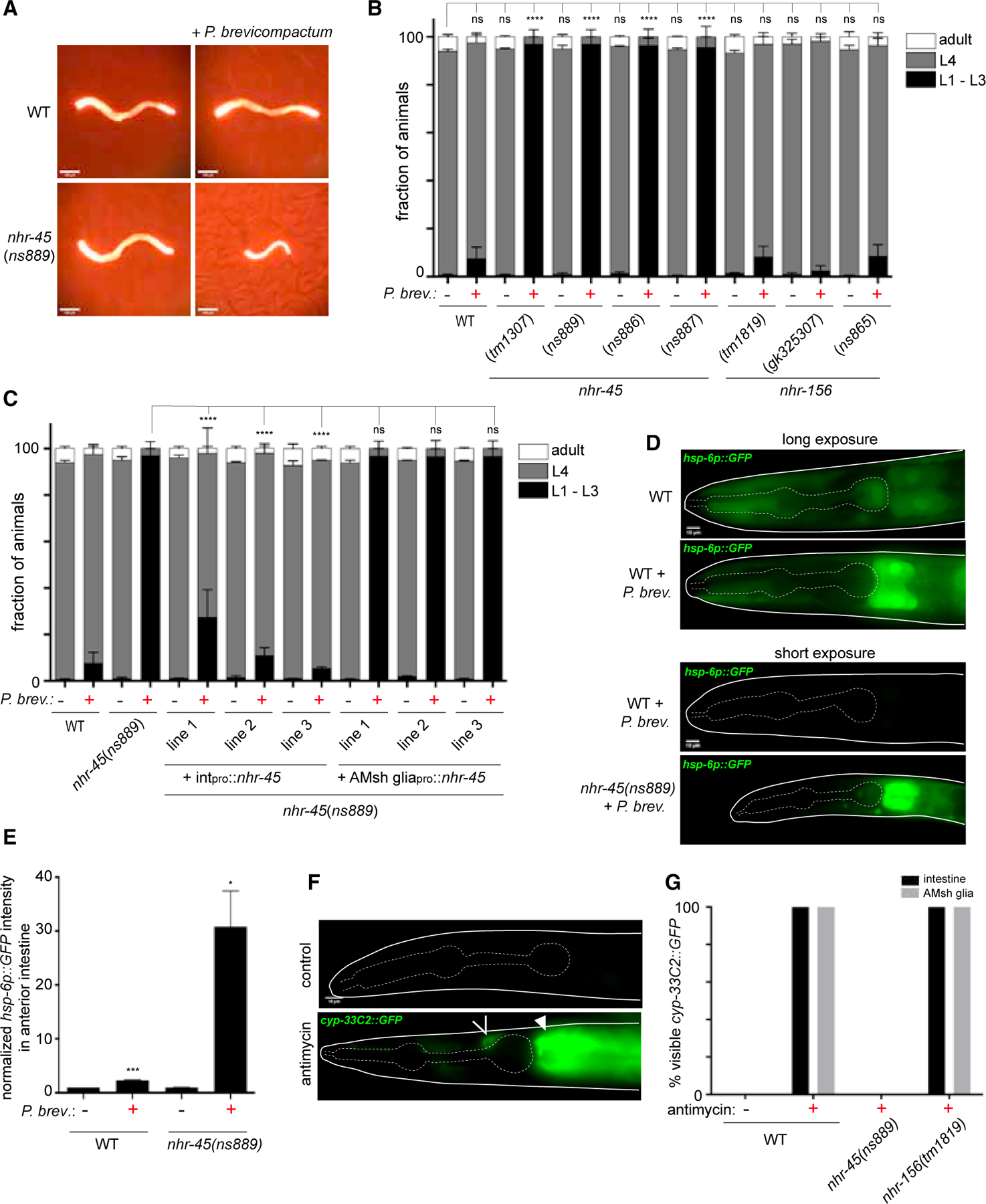
(A) Representative images of indicated strains grown for 2 days from L1 arrest with or without P. brevicompactum exposure. elt-2p∷mCherry is used as a marker to visualize animals.
(B and C) Scoring of developmental stages of indicated strains grown as in (A). (B) ****p < 0.0001 (L4 stage, ANOVA versus WT without P. brevicompactum). (C) ****p < 0.0001 (L4 stage, ANOVA versus nhr-45[ns889] + P. brevicompactum).
(D) hsp-6p∷GFP expression under the indicated conditions.
(E) Quantification of hsp-6p∷GFP fluorescence intensity in anterior intestine, normalized to WT without P. brevicompactum exposure. ***p < 0.001, *p < 0.05 (t test versus corresponding line without P. brevicompactum).
(F) Expression of cyp-33C2∷GFP following antimycin treatment. Open arrowhead, AMsh glia; closed arrowhead, intestine.
(G) Scoring of cyp-33C2∷GFP expression in intestine and AMsh glia following antimycin treatment. All data are based on three biological replicates. Error bars, SEM (B and C); SD (E and G). Scale bars, 100 μm (A), 10 μm (D and F).
nhr-45 was previously shown to regulate XME induction following mitochondrial stress during Pseudomonas aeruginosa infection (Mao et al., 2019). To assess whether mitochondrial stress plays a role in P. brevicompactum toxicity, we measured hsp-6p∷GFP expression (Melber and Haynes, 2018). Wild-type animals exposed to P. brevicompactum show a modest but significant increase in hsp-6 expression in the intestine, particularly in the most anterior cells, compared with animals cultured without P. brevicompactum (Figures 3D and 3E). nhr-45 mutant animals, which fail to induce XME expression and show signs of toxicity on P. brevicompactum, show massive elevation of hsp-6∷GFP in the intestine (Figures 3D and 3E). These results suggest that mycotoxins produced by P. brevicompactum cause mitochondrial stress, with the anterior intestinal cells being particularly sensitive, perhaps because they are more exposed to the environment, and that this stress can be mitigated by inducing expression of nhr-45-dependent detoxifying enzymes. Supporting this idea, we found that applying a known mitochondrial inhibitor, antimycin, phenocopies exposure to P. brevicompactum, resulting in cyp-33C2∷GFP induction in the intestine in an nhr-45-dependent manner (Figures 3F and 3G). Furthermore, we observed that nhr-45 mutants are hypersensitive to the toxic effects of antimycin, consistent with previous observations (Mao et al., 2019). Interestingly, antimycin also induced cyp-33C2∷GFP in AMsh glia, and this also requires nhr-45. However, in contrast to the effect of P. brevicompactum, antimycin-dependent induction of cyp-33C2 in AMsh glia does not require nhr-156, suggesting that additional pathways, other than mitochondrial stress pathways, may underlie the effect of P. brevicompactum on AMsh glia (Figures 3F and 3G).
Together, our results support a model in which the mold P. brevicompactum produces mycotoxins that target mitochondria in the exposed intestinal cells of C. elegans, and that nhr-45-dependent detoxification pathways are required to clear these toxins and allow animals to tolerate moldy environments.
Natural variation in nhr-156 underlies phenotypic diversity in AMsh glial transcriptional responses to microbes
Induction of XMEs has been described after exposure of C. elegans to several model pathogens (Engelmann et al., 2011). We used the cyp-33C2∷GFP reporter to assess whether bacterial pathogens also induce XME expression in AMsh glia. We found that the Serratia marcescens strain Db10 induces expression of cyp-33C2 in AMsh glia (as well as in uncharacterized cells near the anterior bulb of the pharynx), and that this induction also requires nhr-45 and nhr-156 (Figures 4A and 4B).
Figure 4. Natural variations in nhr-156 underlie phenotypic diversity in AMsh glial transcriptional responses to microbes.
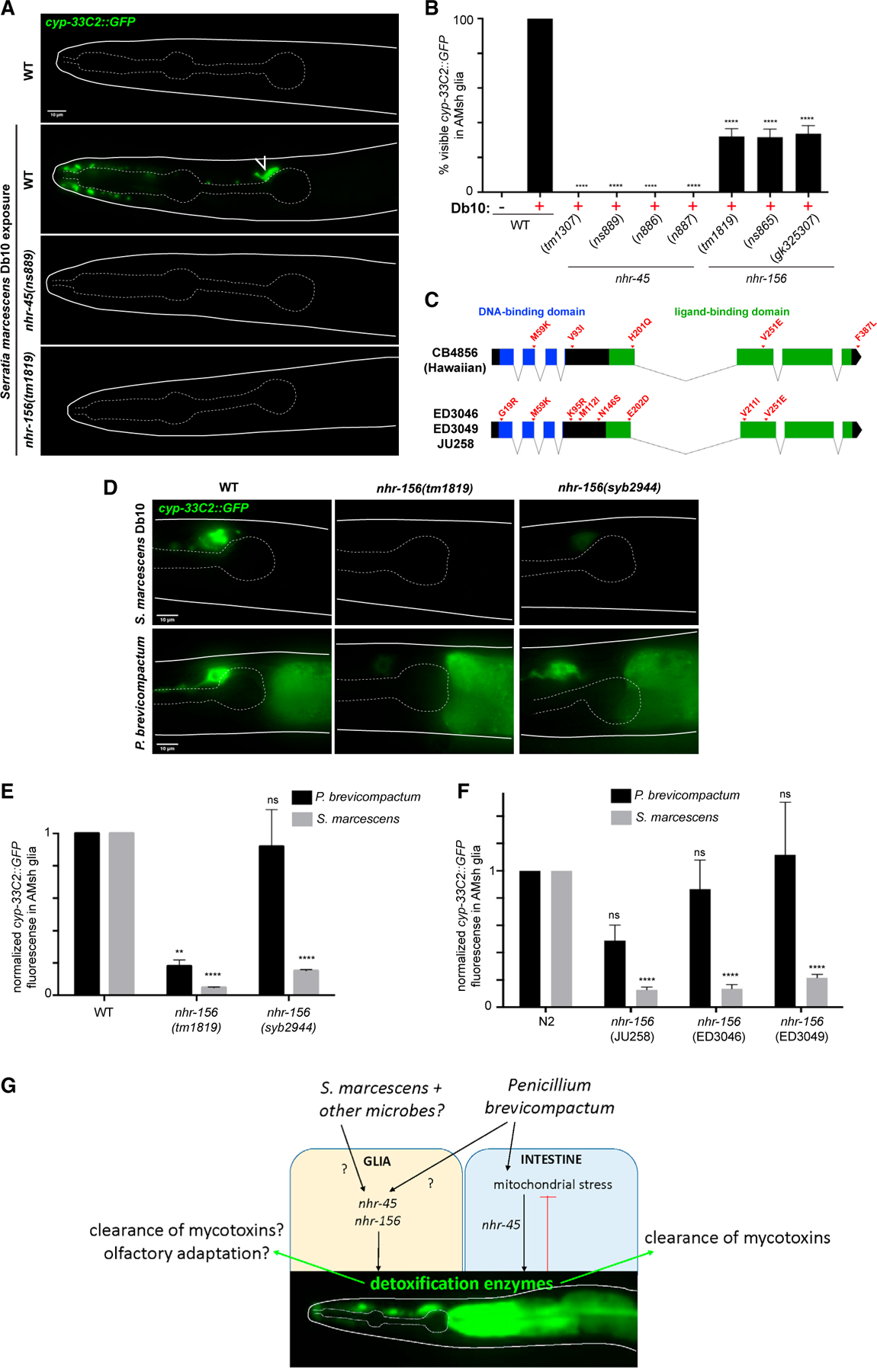
(A) cyp-33C2∷GFP expression following S. marcescens Db10 exposure. Open arrow, AMsh glia.
(B) Scoring of cyp-33C2∷GFP induction in AMsh glia. ****p < 0.0001 (ANOVA versus WT + Db10).
(C) Schematic showing SNPs in the nhr-156 gene that cause amino acid changes.
(D) cyp-33C2∷GFP expression in AMsh glia following exposure to S. marcescens or P. brevicompactum in the indicated strains. syb2944, WT N2 background with the endogenous nhr-156 locus replaced with the Hawaiian version of the gene.
(E and F) Quantification of cyp-33C2∷GFP fluorescence in AMsh glia, normalized to WT for each condition. Genotypes in (F) refer to representative F2 recombinant lines that were generated by crossing cyp-33C2∷GFP from N2 into the indicated isolates. ****p < 0.0001, **p < 0.01 (ANOVA versus corresponding WT). All data are based on three biological replicates.
(G) A model for nhr-45 and nhr-156 contributions to micorbial responses in C. elegans. Error bars, SEM (B); SD (E and F). Scale bars, 10 μm (A and D).
The NHR family in C. elegans has undergone extensive expansion and diversification, and this has led to the idea that divergent NHRs may have evolved to function as receptors for environmental cues (Arda et al., 2010; Robinson-Rechavi et al., 2005; Sladek, 2011; Taubert et al., 2011). We noticed that there are a number of single-nucleotide polymorphisms (SNPs) in nhr-156 sequences of several distinct isolates of C. elegans, including the Hawaiian strain CB4856, compared with the reference N2 Bristol strain. These SNPs result in predicted coding changes in both the DNA-binding and ligand-binding domains (Figure 4C). To assess whether these changes are functionally important, we carried out a series of experiments to determine whether the Hawaiian variant of nhr-156 exhibits alterations in its microbe-dependent activity. We initially crossed the cyp-33C2∷GFP reporter from the N2 background into CB4856, and were surprised to readily isolate recombinant F2 lines in which cyp-33C2∷GFP induction by S. marcescens was lower than observed in N2 (Figure S2A). Partial SNP mapping of the recovered recombinant lines revealed 100% linkage to the Hawaiian variant of nhr-156 (16 of 16 F2 lines). This result suggests that there is a locus near the Hawaiian nhr-156 gene that reduces cyp-33C2∷GFP induction in AMsh glia following S. marcescens exposure. The Hawaiian locus responsible for this phenotype failed to complement nhr-156 loss-of-function alleles, suggesting that nhr-156 itself is the relevant gene.
To confirm these results, we generated the nhr-156 (syb2944) strain in which the N2 nhr-156 locus is replaced with the Hawaiian variant of the gene. Remarkably, this single-gene substitution phenocopies AMsh glia transcriptional defects observed in the Hawaiian recombinant lines (Figures 4D and 4E). Intriguingly, N2 animals bearing the nhr-156 (syb2944) allele still show normal cyp-33C2∷GFP induction in AMsh glia following exposure to P. brevicompactum, demonstrating that the Hawaiian variant of the gene differentially affects the response to different microbes (Figures 4D and 4E).
We also examined three additional C. elegans wild isolates with SNPs in the nhr-156 locus (Figure 4C). Each isolate generated apparently mutant recombinant F2 lines when crossed with the cyp-33C2∷GFP reporter and exposed to S. marcescens (Figure S2A); again with 100% linkage between the phenotype and the variant nhr-156 locus in each case (16 of 16 F2 lines for each isolate). These recombinant lines also show relatively normal cyp-33C2 induction following P. brevicompactum exposure (Figure 4F). As a control, we obtained two wild isolates that do not harbor codon-changing SNPs in the nhr-156 locus (JU1088 and JU1171), and found that these strains generate only fully wild-type recombinant F2 lines when crossed with the cyp-33C2∷GFP marker. Together these results show that divergent sequences in the nhr-156 gene found in different C. elegans strains result in differential transcriptional responses to microbe exposure, supporting the idea that the expansion and diversification of the NHR family may allow NHRs to act as receptors for specific environmental cues.
DISCUSSION
In this study, we characterized the interaction between C. elegans and P. brevicompactum, an ecologically relevant species of mold that has been found in substrates that harbor wild isolates of C. elegans (Dirksen et al., 2016). A large number of pathogens have been described that infect C. elegans and use it as a host. This includes the well-characterized fungal pathogen Drechmeria coniospora, whose spores penetrate the cuticle to infect the epidermis (Kim and Ewbank, 2018), as well as Penicillium marneffei, which colonizes the intestine (Huang et al., 2014). In both cases, fungal infection results in obvious fungal growth in the nematode host, resulting in death. We have not seen any evidence that P. brevicompactum infects C. elegans and uses it as a substrate in this way, and instead favor a model in which it is a toxic species rather than an infectious species for C. elegans. We propose that P. brevicompactum produces mycotoxins that target mitochondria in the intestine and that are normally cleared by XMEs that are induced in a nhr-45-dependent manner (Figure 4G). This model is supported by the previous demonstration that nhr-45 regulates the induction of detoxifying enzymes by mitochondrial stress (Mao et al., 2019), by our observations that P. brevicompactum exposure results in mitochondrial stress in the intestine of nhr-45 mutant animals, and by our findings that the known mitochondrial toxin antimycin phenocopies P. brevicompactum exposure, resulting in XME induction in wild-type animals and toxicity in nhr-45 mutant animals.
Alternative models in which P. brevicompactum spores are ingested and infect the nematode cannot be ruled out. Intriguingly, we observed a significant overlap in the genes induced by P. brevicompactum exposure to those induced by the pathogens P. aeruginosa and S. marcescens, both of which colonize the intestine (Fletcher et al., 2019; Engelmann et al., 2011) (Table S5). The implications of this remains unclear, however, as pathogens that infect the intestine also secrete toxins that contribute to their pathogenicity. Our data support a general model of pathogenesis in C. elegans, in which microbial toxins disrupt core cellular machineries, and this cellular damage triggers host stress responses (Liu et al., 2014; Melo and Ruvkun, 2012).
XMEs in C. elegans have primarily been studied in the intestine (Hartman et al., 2021). In this study we have demonstrated that XMEs are also induced in AMsh glia following P. brevicompactum or S. marcescens exposure. To our knowledge, this is the first demonstration of such induction in glia. While the implications of microbe-dependent XME induction in glia remain under investigation, it is notable that high levels of XME expression are observed in sustentacular glial cells in the mammalian olfactory epithelium (Heydel et al., 2013), as well as in support cells of insect antennae (Leal, 2013), where they have been proposed to regulate olfactory signaling by modifying odorant molecules. Given the essential role that AMsh glial cells play in regulating chemosensory behavior (Bacaj et al., 2008; Wallace et al., 2016), it is fascinating to speculate that XME induction in AMsh glia may regulate the behavioral adaptations that accompany exposure to pathogens (Kim and Flavell, 2020). We note, however, that we did not see defects in aversive conditioning to S. marcescens in nhr-45 or nhr-156 mutants when assessed by lawn-leaving assays (Figure S3B) or defects in survival (Figure S3C). We also checked whether exposure to P. brevicompactum affects subsequent chemosensory behavior, and, while P. brevicompactum exposure results in reduced chemotaxis to the odorant benzaldehyde, this effect does not depend on nhr-45 or nhr-156 (Figure S3D).
The NHR family of C. elegans has undergone expansion as well as sequence diversification, particularly in the ligand-binding domain (Robinson-Rechavi et al., 2005). This has led to the idea that environmental cues rather than endogenous endocrine signals may constitute ligands for many NHRs (Sladek, 2011; Taubert et al., 2011). There are numerous examples of NHRs involved in regulating immune or stress responses following microbe or toxin exposure. This includes nhr-49 and nhr-8, which regulate immune responses to Staphylococcus aureus and P. aeruginosa, respectively (Wani et al., 2021; Otarigho and Aballay, 2020), and nhr-176 and nhr-33/hizr-1, which regulate resistance to thiabendazole and cadmium, respectively (Jones et al., 2013; Shomer et al., 2019). Our observation that natural variations in the nhr-156 gene in wild isolates result in differential transcriptional responses in AMsh glia to microbe exposure supports the idea that NHRs may act as sensors for environmental cues. Future work will investigate the molecular basis for these differences in nhr-156 activity. It is intriguing that the variants share a common amino acid polymorphism in the ligand-binding domain (Figure 4C), raising the possibility that this change underlies differential activation in response to microbial cues from the environment that act as ligands.
Limitations of the study
While our data support a model in which the mold P. brevicompactum produces toxins that cause mitochondrial stress and are toxic for C. elegans, and that these toxins are cleared by the induction of XMEs in the intestine, we have not identified the causative mycotoxins. Further work is needed to identify such toxins using biochemical approaches. Our data on the induction of XMEs in glia highlight that this phenomenon occurs, and our data on variation in the glial transcriptional response suggest that nhr-156 may act as a receptor for environmental cues from microbes. However, the biological significance of the glial XME induction is currently unclear. Further work is needed to elucidate the significance of the glial transcriptional response for detoxification or sensory function.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shai Shaham (shaham@mail.rockfefeller.edu).
Materials availability
Plasmids and strains generated in this study are available by contacting the lead contact.
Data and code availability
Original RNAseq data files have been deposited at GEO with accession number GEO: GSE183331. This paper does not report original code. Any additional data required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Caenorhabditis elegans
Caenorhabditis elegans young adult (approximately 48 h from L1 when grown at 20°C) hermaphrodites were used in this study. Strains were cultured on standard nematode growth media (NGM) plates and fed E. coli OP50. Wild type is Bristol N2. The following mutant alleles were generated in this study (see Key Resources Table for a full list of strains used):
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli OP50 | CGC | OP50 |
| S. marcescens Db10 | CGC | Db10 |
| Chemicals, peptides, and recombinant proteins | ||
| Antimycin A from Streptomyces sp. | Sigma-Aldrich | A8674 |
| Deposited data | ||
| RNAseq raw data | This study | GEO: GSE183331 |
| Experimental models: Organisms/strains | ||
| Penicillium brevicompactum | ATCC | 9056 |
| nsIs775 [pSW81{cyp-33C2∷GFP} + elt-2p∷mCherry] I | This study | OS11850 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nsIs143 (F16F9.3p∷dsRed) X | This study | OS12256 |
| mgIs73 [cyp-14A4p∷GFP∷cyp-14A4 3′ UTR + myo-2p∷mCherry] V | CGC | GR2250 |
| jrsIs1 [cyp-13A7p∷GFP + unc-119(+)] | CGC | SRU1 |
| stIs11634 [ugt-1p∷H1-mCherry + unc-119(+)] | CGC | RW11634 |
| otEx1182 [gst-44∷GFP + rol-6(dom)] | CGC | OH2204 |
| nsEx5944 [pSW74{gst-33∷GFP} + elt-2p∷mCherry] | This study | OS11501 |
| nsEx5944 [pSW74{gst-33∷GFP} + elt-2p∷mCherry]; nsIs698 [mir-228p∷NLS-RFP] | This study | OS11503 |
| nhr-45 (tm1307) X | CGC | VL484 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(tm1307) X | This study | OS12194 |
| nhr-45 (ns886) X | This study | OS12363 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns886) X | This study | OS12386 |
| nhr-45 (ns887) X | This study | OS12364 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns887) X | This study | OS12387 |
| nhr-45 (ns889) X | This study | OS12366 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns889) X | This study | OS12385 |
| nhr-156 (tm1819) V | NBRP | tm1819 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156(tm1819) V | This study | OS12203 |
| nhr-156 (ns865) V | This study | OS12406 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156(ns865) V | This study | OS12751 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156 (gk325307) | This study | OS12199 |
| nhr-156 (ns865) V; nhr-45 (ns889) X | This study | OS12752 |
| nhr-45 (ns889); nsIs910 [elt-2p∷nhr-45cDNA + unc-122p∷GFP] | This study | OS12873 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45 (ns889) X; nsEx6238 [pSW124{elt-2p∷nhr-45 cDNA} + unc-122p∷GFP], gut rescue line 1 of 3 | This study | OS12607 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns889) X; nsEx6239 [pSW124{elt-2p∷nhr-45 cDNA} + unc-122p∷GFP], gut rescue line 2 of 3 | This study | OS12608 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns889) X; nsEx6240 [pSW124{elt-2p∷nhr-45 cDNA} + unc-122p∷GFP], gut rescue line 3 of 3 | This study | OS12609 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns889) X; nsEx6241 [pSW110 {F16F9.3p∷nhr-45 cDNA} + unc-122p∷GFP], glia rescue line 1 of 3 | This study | OS12610 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45(ns889) X; nsEx6242 [pSW110 {F16F9.3p∷nhr-45 cDNA} + unc-122p∷GFP], glia rescue line 2 of 3 | This study | OS12611 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-45 (ns889) X; nsEx6243 [pSW110 {F16F9.3p∷nhr-45 cDNA} + unc-122p∷GFP], glia rescue line 3 of 3 | This study | OS12612 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156(tm1819) V; nsEx6194 [pSW118 {F16F9.3p∷nhr-156 cDNA} + unc-122p∷GFP], glia rescue line 1 of 3 | This study | OS12338 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156(tm1819) V; nsEx6196 [pSW118 {F16F9.3p∷nhr-156 cDNA} + unc-122p∷GFP], glia rescue line 2 of 3 | This study | OS12340 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156(tm1819) V; nsEx6197 [pSW118 {F16F9.3p∷nhr-156 cDNA} + unc-122p∷GFP], glia rescue line 3 of 3 | This study | OS12341 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; che-2 (e1033) X | This study | OS12903 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; dyf-7 (m537) X | This study | OS12094 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; osm-6 (p811) V | This study | OS12095 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; tir-1 (qd4) III | This study | OS11879 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; sek-1 (km4) X | This study | OS12050 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; pmk-1 (km25) IV | This study | OS12051 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nsIs109(F16F9.3p∷DTA(G53E) + unc-122p∷GFP) | This study | OS13492 |
| mgIs73 [cyp-14A4p∷GFP∷cyp-14A4 3′ UTR + myo-2p∷mCherry] V; nhr-45 (ns889) X | This study | OS13109 |
| mgIs73 [cyp-14A4p∷GFP∷cyp-14A4 3′ UTR + myo-2p∷mCherry] nhr-156 (tm1819) V | This study | OS13110 |
| nsIs143 (F16F9.3p∷dsRed) X; nsEx6176 [WRM0636C_B02 (pRedFlp-Hgr) (nhr-45 [15070]∷S0001_pR6K_Amp_2xTY1ce_EGFP_FRT_rpsl_neo_FRT_3xFlagdFRT∷unc-119-Nat) + elt-2p∷mCherry] | This study | OS12342 |
| nsIs698 [mir-228p∷NLS-RFP]; nsEx6179 [pSW121{nhr-156p∷GFP} + elt-2p∷mCherry] | This study | OS12307 |
| zcIs13 [hsp-6p∷GFP + lin-15(+)] V | CGC | SJ4100 |
| zcIs13 [hsp-6p∷GFP + lin-15(+)] V; nhr-45 (ns889) X | This study | OS13183 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156 (syb2944) V | This study | OS13345 |
| CB4856 – C. elegans Hawaiian isolate | CGC | CB4856 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156(CB4856) V | This study | OS13111 |
| JU258 - C. elegans wild isolate | CGC | JU258 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156 (JU258) V | This study | OS13112 |
| ED3046 - C. elegans wild isolate | CGC | ED3046 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156 (ED3046) V | This study | OS13113 |
| ED3049 - C. elegans wild isolate | CGC | ED3049 |
| nsIs775 [cyp-33C2∷GFP + elt-2p∷mCherry] I; nhr-156 (ED3049) V | This study | OS13114 |
| JU1088 - C. elegans wild isolate | CGC | JU1088 |
| JU1171 - C. elegans wild isolate | CGC | JU1171 |
| Recombinant DNA | ||
| pSW81 {cyp-33C2pro (1119bp)∷cyp-33C2∷GFP∷unc-54 3′ UTR} | This study | pSW81 |
| pSW74 {gst-33pro (907bp)∷gst-33∷GFP∷unc-54 3′ UTR} | This study | pSW74 |
| pSW124 {elt-2pro (2908bp)∷nhr-45 cDNA∷unc-54 3′ UTR} | This study | pSW124 |
| pSW110 {F16F9.3pro (2056bp)∷nhr-45 cDNA∷unc-54 3′ UTR} | This study | pSW110 |
| pSW118 {F16F9.3pro (2056bp)∷nhr-156 cDNA∷unc-54 3′ UTR} | This study | pSW118 |
| pSW121 {nhr-156pro (1626bp)∷GFP∷unc-54 3′ UTR} | This study | pSW121 |
| WRM0636C_B02 (pRedFlp-Hgr) (nhr-45 [15070]∷S0001_pR6K_Amp_2xTY1ce_EGFP_FRT_rpsl_neo_FRT_3x FlagdFRT∷unc-119-Nat) | TransgeneOme | nhr-45∷GFP fosmid |
nhr-45 (ns886) (14 bp deletion in exon1, generated by CRISPR); nhr-45 (ns887) (11 bp insertion in exon 1, generated by CRISPR); nhr-45 (ns889) (7 bp deletion in exon 1, generated by CRISPR); nhr-156 (ns865) (Gln29 > ochre, generated by EMS mutagenesis); nhr-156 (syb2944) (endogenous nhr-156 gene removed from N2 background and replaced with the Hawaiian variant of the gene – strain created by SunyBiotech).
Penicillium brevicompactum
Penicillium brevicompactum (ATCC catalog number 9056) was maintained by culturing on nematode growth media (NGM) plates. Cultures were kept at room temperature for 2 weeks or at 4°C for 2 months. Fresh cultures were created by streaking spores from an existing culture. Frozen stocks were created by scraping material from a plate in M9 buffer and freezing at −80°C with glycerol (final concentration 17%). Mold species were genotyped by scraping material from a plate in M9 buffer, briefly centrifuging to pellet material, and lysing in worm lysis buffer (10 mM Tris pH 8.0, 50 mM KCl, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween 20, 0.01% gelatin, 200 μg/mL proteinase K) by incubating at 65°C for 1 h. Genotyping PCRs were performed directly on lysates using ITS, LSU and SSU primers (Schoch et al., 2012).
Serratia marcescens Db10
Serratia marcescens Db10 isolate was cultured in liquid by inoculating LB and growing overnight at 37°C 200 rpm. Liquid culture was seeded on NGM plates and allowed to dry at room temperature for C. elegans exposure.
METHOD DETAILS
Exposure of C. elegans to P. brevicompactum
To expose C. elegans to P. brevicompactum, spores were scraped from a culture plate and added to NGM liquid cultures immediately prior to pouring NGM plates. Plates were left to dry for several days, before being seeded with E. coli OP50. Mold spores were calibrated such that approximately 50% of the surface of the plate was covered by mold when the plates were used for C. elegans assays (this is considered ‘high density’). Unless otherwise stated, assays were performed by plating synchronized L1s on to moldy plates and scoring phenotypes after 2 days of growth at 20°C.
RNA extraction and sequencing
C. elegans were prepared for transcriptome analysis by culturing on plates with or without P. brevicompactum, as described above. To avoid obtaining artifacts due to differences in developmental staging, it is critical that samples are staged correctly. RNA was extracted from samples approximately 48 h from L1 arrest, at which time point the majority of animals had reached the young adult stage. The precise timing was decided empirically for each condition in each experiment, such that some plates were left for up to 52 h if they contained an appreciable number of animals that were still at the L4 stage at the 48-h time point. To overcome the toxicity of P. brevicompactum for nhr-45 mutant animals, we took advantage of the transgenic line nhr-45 (ns889); nsIs910 (elt-2p:nhr-45 cDNA) that contains a partial rescue of nhr-45 function in the intestine. This line showed only minimal rescue of cyp-33C2 induction in the intestine, but showed relatively normal developmental timing on P. brevicompactum. We hypothesized that nhr-45 induces multiple redundantly-acting XME targets, and that the partial restoration of nhr-45 expression was not sufficient to rescue cyp-33C2 induction, but was sufficient to prevent toxicity due to its regulation of other targets. We therefore hypothesized that this line would be a useful tool to identify nhr-45 target genes, albeit with the caveat that we would likely underestimate the extent to which XME expression depends on nhr-45. The sequencing data we obtained confirmed this hypothesis; many of the inducible XMEs required nhr-45 for their full induction, but some still showed partial induction due to the partial nhr-45 rescue. This line was therefore useful for identifying nhr-45 target genes without encountering developmental staging problems.
To extract total RNA, animal pellets (approximate size 100 μL after washing in M9 buffer) were suspended in 4x volume Trizol-LS, vortexed for 5 min and subjected to two rounds of freeze-thawing with additional vortexing for 5 min after each thaw. RNA was then isolated using chloroform extraction following the manufacturer’s guidelines. Total RNA was then subjected to RNeasy column cleanup with DNAse treatment (QIAGEN). cDNA library preparation and sequencing were as performed by The Rockefeller University Genomics Research Center, using Illumina TruSeq stranded mRNA-Seq library preparation kit with polyA selection, and Illumina NextSeq500. Raw sequencing data has been deposited at GEO with accession number GSE183331.
Differential gene expression analysis
Sequencing data was assessed for quality control using FastQC v0.11.12 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) with default parameters. Trimmed fastq files were aligned to WBcel235 using STAR v2.4a aligner with default parameters (Dobin et al., 2013). The alignment results were then evaluated through Qualimap v2.2 (García-Alcalde et al., 2012) to ensure that all the samples had a consistent coverage, alignment rate, and no obvious 5′or 3′ bias. Aligned reads were then summarized through featureCounts on gene (Ensemble gene model Caenorhabditis_elegans.WBcel235.103.gtf). To eliminate the effect of library size, summarized count matrix was normalized through edgeR v 3.28.1 (McCarthy et al., 2012; Robinson et al., 2010). Voom from limma v 3.42.2 was applied to estimate the fold change (Ritchie et al., 2015). A BH-adjusted p value of less than 0.05 (p-adj < 0.05) was used to select genes that have a significant expression change.
Fluorescence microscopy
Imaging was performed using an Axioplan II fluorescence microscope equipped with an AxioCam camera. Images were processed using ImageJ software. Processing involved orienting the samples, cropping to show the region of interest, and setting the upper and lower intensity thresholds for display. All images in a given experiment were processed in an identical manner.
RNA interference
RNAi by feeding was carried out as described previously (Kamath and Ahringer, 2003), with modifications to allow exposure to P. brevicompactum. Overnight cultures of E. coli HT115 carrying pL4440 vectors targeting specific genes (Ahringer and Vidal libraries) were seeded on to P.brevicompactum-NGM plates (described above) supplemented with carbenicillin (25 ug/mL) and IPTG (1 mM). C. elegans were added to plates as L1 larvae and scored 48 h later.
Developmental assay for toxicity
Synchronized L1 larvae were plated on NMG plates seeded with E. coli OP50, with or without P. brevicompactum (see above). Animals were grown at 20°C for 42–44 h, and the percentage of animals at each developmental stage was counted. This time point was chosen because the majority of animals are in the L4 larval stage for wild type animals under normal conditions.
Antimycin treatment
C. elegans were plated on to standard NGM plates with E. coli OP50 as synchronized L1 larvae and allowed to develop at 20°C. The following day, 500 μL of antimycin A from Streptomyces sp. (Sigma-Aldrich) (6.25 μg/mL in M9 buffer) was added to the plate and allowed to diffuse in to the agar. Animals were then assayed after approximately 24 h of exposure.
Serratia marcescens lawn-leaving assays
Assay plates were made by seeding 30 μL of Serratia marcescens Db10 culture on to NGM plates and allowing to dry overnight at room temperature. 20 young adults per replicate were picked on to an assay plate, and the percentage of animals occupying the lawn was calculated at each time point. At least 3 replicates were run for each condition, and data is presented as the mean occupancy of the lawn at each time point.
QUANTIFICATION AND STATISTICAL ANALYSIS
Induction of GFP reporter genes
In some experiments, induction of GFP reporter genes was quantified by scoring the percentage of animals that show visible GFP expression, when assessed by eye using a dissecting microscope. In these experiments, 3 independent replicates were carried out, with at least 30 animals scored blindly per condition per replicate. For rescue experiments, only animals expressing the rescue array, as determined by the presence of the co-injection marker, were scored. Data is presented as the mean of the percentage of animals that show induction across replicates. Significance values were calculated using t-test, for single comparisons, or ANOVA for multiple comparisons. p-values and details of the comparisons performed are indicated in the figure legends.
In other experiments, induction of GFP reporter genes was quantified by measuring fluorescence intensity. 3 independent replicates were carried out, with at least 20 animals per replicate. Animals were imaged at high magnification on an Axioplan II fluorescence microscope (see Method Details above), with identical imaging parameters for every sample in a given experiment. Images were then analyzed blindly using ImageJ software. For each animal, a region of interest was defined and mean pixel intensity was measured. Background subtraction was performed by measuring the mean pixel intensity of a corresponding background region. For each condition in a given experiment, the mean intensity value of all individual animals was calculated. This value was normalized to the corresponding wild-type control in the experiment. Data is presented mean of the normalized fluorescence intensity across the 3 independent replicates. Significance values were calculated using t test, for single comparisons, or ANOVA for multiple comparisons. p values and details of the comparisons performed are indicated in the figure legends.
Developmental assay for toxicity
To assess developmental timing, the percentage of animals at each developmental stage (young adult, larval stage 4, or larval stages 1–3) was scored. 3 independent replicates were carried out, with at least 80 animals scored blindly per condition per replicate. For rescue experiments, only animals expressing the rescue array, as determined by the presence of the co-injection marker, were scored. Data is presented as the mean percentage of animals at each stage across the 3 replicates. Significance values were calculated for the larval stage 4 (L4) category, as this is the stage that the majority of animals develop to under wild-type conditions. Significance values were calculated using ANOVA for multiple comparisons. p-values and details of the comparisons performed are indicated in the figure legends.
Differential gene expression analysis
Full details of the processing of raw sequence files is described above in Method Details section. To calculate significant fold changes, voom from limma v 3.42.2 was applied (Ritchie et al., 2015). A BH-adjusted p-value of less than 0.05 (p-adj < 0.05) was used to select genes that have a significant expression change.
Supplementary Material
Highlights.
The mold P. brevicompactum induces XME expression in C. elegans gut and glia
P. brevicompactum-dependent XME induction is regulated by nhr-45 and nhr-156
Failure to induce XMEs in the intestine results in mitochondrial stress and toxicity
Natural variations in nhr-156 result in phenotypic diversity in the glial response
ACKNOWLEDGMENTS
We thank Shaham lab members for helpful discussions and the Rockefeller University Genomics Resource Center for outstanding technical support. Some strains were provided by the CGC, which is funded by NIH Office of research Infrastructure Programs (P40 OD010440). We thank the National Bio-resource Project (NBRP), Cornelia Bargmann, Jonathan Ewbank, and Howard Hang for strains. S.W.W. was a William N. and Bernice E. Bumpus Foundation postdoctoral fellow. This work was supported in part by NIH grant R35 NS105094 to S.S.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110166.
DECLARATIONS OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Gilst MV, Sequerra R, Doucette-Stamm L, Yamamoto KR, and Walhout AJM (2010). Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol. Syst. Biol 6, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, and Shaham S (2008). Glia are essential for sensory organ function in C. elegans. Science 322, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, et al. (2016). The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, Waterston RH, Hillier LW, and Ewbank JJ (2011). A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. Plos One 6, e19055. 10.1371/journal.pone.0019055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M, Tillman EJ, Butty VL, Levine SS, and Kim DH (2019). Global transcriptional regulation of innate immunity by ATF-7 in C. elegans. Plos Genet. 15, e1007830. 10.1371/journal.pgen.1007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, and Conesa A (2012). Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679. [DOI] [PubMed] [Google Scholar]
- Hartman JH, Widmayer SJ, Bergemann CM, King DE, Morton KS, Romersi RF, Jameson LE, Leung MCK, Andersen EC, Taubert S, et al. (2021). Xenobiotic metabolism and transport in Caenorhabditis elegans. J. Toxicol. Environ. Heal Part B 24, 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydel J, Coelho A, Thiebaud N, Legendre A, Bon AL, Faure P, Neiers F, Artur Y, Golebiowski J, and Briand L (2013). Odorant-binding proteins and xenobiotic metabolizing enzymes: implications in olfactory perireceptor events. Anatomical Rec. 296, 1333–1345. [DOI] [PubMed] [Google Scholar]
- Holdorf AD, Higgins DP, Hart AC, Boag PR, Pazour GJ, Walhout AJM, and Walker AK (2019). WormCat: an online tool for annotation and visualization of Caenorhabditis elegans genome-scale data. Genetics 214, 279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Li D, Xi L, and Mylonakis E (2014). Caenorhabditis elegans: a simple nematode infection model for Penicillium marneffei. PLoS One 9, e108764. 10.1371/journal.pone.0108764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Ou G, Leroux MR, and Scholey JM (2007). The sensory cilia of Caenorhabditis elegans_revised. Wormbook, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, and Wang D (2018). The microbial zoo in the C. elegans intestine: bacteria, fungi and viruses. Viruses 10, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LM, Rayson SJ, Flemming AJ, and Urwin PE (2013). Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS One 8, e69956. 10.1371/journal.pone.0069956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, and Ahringer J (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321. [DOI] [PubMed] [Google Scholar]
- Kim DH, and Ewbank JJ (2018). Signaling in the innate immune response. Wormbook, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, and Flavell SW (2020). Host-microbe interactions and the behavior of Caenorhabditis elegans. J. Neurogenet 34, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, et al. (2002). A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626. [DOI] [PubMed] [Google Scholar]
- Leal WS (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol 58, 373–391. [DOI] [PubMed] [Google Scholar]
- Li G, Zhang K, Xu J, Dong J, and Liu Y (2007). Nematicidal substances from fungi. Recent Patents Biotechnol. 1, 212–233. [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, and Ruvkun G (2014). Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madende M, Albertyn J, Sebolai O, and Pohl CH (2020). Caenorhabditis elegans as a model animal for investigating fungal pathogenesis. Med. Microbiol. Immun 209, 1–13. [DOI] [PubMed] [Google Scholar]
- Mao K, Ji F, Breen P, Sewell A, Han M, Sadreyev R, and Ruvkun G (2019). Mitochondrial dysfunction in C. elegans activates mitochondrial relocalization and nuclear hormone receptor-dependent detoxification genes. Cell Metab 29, 1182–1191.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A, and Haynes CM (2018). UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res 28, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, and Ruvkun G (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otarigho B, and Aballay A (2020). Cholesterol regulates innate immunity via nuclear hormone receptor NHR-8. Iscience 23, 101068. 10.1016/j.isci.2020.101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MR, Ryu S, Maburutse BE, Oh NS, Kim SH, Oh S, Jeong S-Y, Jeong D-Y, Oh S, and Kim Y (2018). Probiotic Lactobacillus fermentum strain JDFM216 stimulates the longevity and immune response of Caenorhabditis elegans through a nuclear hormone receptor. Sci. Rep-Uk 8, 7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ND, Cheesman HK, Liu P, Anderson SM, Foster KJ, Chhaya R, Perrat P, Thekkiniath J, Yang Q, Haynes CM, et al. (2019). The nuclear hormone receptor NHR-86 controls anti-pathogen responses in C. elegans. Plos Genet. 15, e1007935. 10.1371/journal.pgen.1007935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeke LJ, and Herman MA (2021). Take a walk to the wild side of Caenorhabditis elegans-pathogen interactions. Microbiol. Mol. Biol. R 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan M, Anderson CP, Rindler PM, Romney SJ, dos Santos MCF, Gertz J, and Leibold EA (2019). NHR-14 loss of function couples intestinal iron uptake with innate immunity in C. elegans through PQM-1 signaling. eLife 8, e44674. 10.7554/eLife.44674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Maina CV, Gissendanner CR, Laudet V, and Sluder A (2005). Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J. Mol. Evol 60, 577–586. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Consortium FB, List FBCA, Bolchacova E, et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad Sci 109, 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, and Félix MA (2017). The Natural Biotic Environment of Caenorhabditis elegans. Genetics 206, 55–86. 10.1534/genetics.116.195511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomer N, Kadhim AZ, Grants JM, Cheng X, Alhusari D, Bhanshali F, Poon AF-Y, Lee MYY, Muhuri A, Park JI, et al. (2019). Mediator subunit MDT-15/MED15 and nuclear receptor HIZR-1/HNF4 cooperate to regulate toxic metal stress responses in Caenorhabditis elegans. PLoS Genet. 15, e1008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A, and Shaham S (2019). Glia-neuron interactions in Caenorhabditis elegans. Annu. Rev. Neurosci 42, 1–20. [DOI] [PubMed] [Google Scholar]
- Sladek FM (2011). What are nuclear receptor ligands? Mol. Cell Endocrinol 334, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Ward JD, and Yamamoto KR (2011). Nuclear hormone receptors in nematodes: evolution and function. Mol. Cell Endocrinol 334, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER (2016). Host-microsporidia interactions in Caenorhabditis elegans, a model nematode host. Microbiol. Spectr 4. 10.1128/microbiolspec.FUNK-0003-2016. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, Hong S-B, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, and Samson RA (2014). Identification and nomenclature of the genus Penicillium. Stud. Mycol 78, 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, and Redinbo MR (2012). Xenobiotic-sensing nuclear receptors involved in drug metabolism: a structural perspective. Drug Metab. Rev 45, 79–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SW, Singhvi A, Liang Y, Lu Y, and Shaham S (2016). PROS-1/Prospero is a major regulator of the glia-specific secretome controlling sensory-neuron shape and function in C. elegans. Cell Rep. 15, 550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani KA, Goswamy D, Taubert S, Ratnappan R, Ghazi A, and Irazoqui JE (2021). NHR-49/PPAR-a and HLH-30/TFEB cooperate for C. elegans host defense via a flavin-containing monooxygenase. eLife 10, e62775. 10.7554/eLife.62775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JD, Mullaney B, Schiller BJ, He LD, Petnic SE, Couillault C, Pujol N, Bernal TU, Gilst MRV, Ashrafi K, et al. (2014). Defects in the C. elegans acyl-CoA synthase, acs-3, and nuclear hormone receptor, nhr-25, cause sensitivity to distinct, but overlapping stresses. Plos One 9, e92552. 10.1371/journal.pone.0092552.eCollection2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen GJ, and Ausubel FM (2018). Both live and dead Enterococci activate Caenorhabditis elegans host defense via immune and stress pathways. Virulence 9, 683–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Holdorf AD, and Walhout AJ (2017). C. elegans and its bacterial diet as a model for systems-level understanding of host-microbiota interactions. Curr. Opin Biotechnol 46, 74–80. 10.1016/j.copbio.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original RNAseq data files have been deposited at GEO with accession number GEO: GSE183331. This paper does not report original code. Any additional data required to reanalyze the data reported in this paper is available from the lead contact upon request.


