Abstract
To repurpose therapeutics for fibrolamellar carcinoma (FLC) we developed and validated patient-derived xenografts (PDX) from surgical resections. Most agents used clinically, and inhibitors of oncogenes overexpressed in FLC showed little efficacy on PDX. A high-throughput functional drug screen found primary and metastatic FLC were vulnerable to clinically available inhibitors of TOPO1 and HDAC, and to napabucasin. Napabucasin’s efficacy was mediated through reactive oxygen species and inhibition of translation initiation, and specific inhibition of eIF4A was effective. The sensitivity of each PDX line inversely correlated with expression of the anti-apoptotic protein Bcl-xL, and inhibition of Bcl-xL synergized with other drugs. Screening directly on cells dissociated from patient resections validated these results. This demonstrates that a direct functional screen on patient tumors provides therapeutically informative data within a clinically useful time frame. Identifying these novel therapeutic targets and combination therapies is an urgent need, as effective therapeutics for FLC are currently unavailable.
Significance:
Therapeutics informed by genomics have not yielded effective therapies for Fibrolamellar Carcinoma (FLC). A functional screen identified TOPO1, HDAC inhibitors, and napabucasin as efficacious and synergistic with inhibition of Bcl-xL. Validation on cells dissociated directly from patient tumors demonstrates the ability for functional precision medicine in a solid tumor.
Keywords: fibrolamellar carcinoma, drug repurposing, pediatric rare tumors, patient derived xenografts, drug screening, fusion gene
Introduction:
Fibrolamellar carcinoma (FLC) is a fatal liver cancer that affects adolescents and young adults without underlying liver disease. Although rare (1 in 5 million) it is well-defined and consistent across patients, offering a favorable tool for characterizing pathogenesis and for testing the efficacy of therapeutics. Characterization of the FLC transcriptome by RNA-seq revealed a fusion transcript of exon 1 of the heat shock protein DNAJB1, and exons 2-10 of PRKACA, the catalytic subunit of protein kinase A (PKA)(1) in all patients tested. The fusion transcript results from a ~400 kb deletion on chromosome 19 that is present in tumors but not in adjacent non-tumor liver(1-3). The resulting fusion kinase retains the catalytic activity of PKA(1). Use of CRISPR/Cas9 to create the deletion, chimeric fusion gene, and kinase produces tumors that recapitulate the histopathology and the transcriptome of FLC(4,5). Expression of the fusion gene from a transposon also produces tumors, demonstrating that pathogenesis depends on the DNAJB1-PRKACA chimeric protein, not on the loss of genes in the deletion(4). Thus, the chimeric protein is both correlated with and causative of FLC.
FLC is classified as a subset of HCC(6) and treatments for HCC are often used for FLC. However, FLC differs from HCC in its genome, coding and non-coding transcriptome, proteome(7-9), and molecular pathogenesis, suggesting these cancers may require distinct therapies. Currently, patients with FLC are included in the Pediatric Hepatic Malignancy International Therapeutic Trial (PHITT), receiving surgery plus cisplatin and doxorubicin combination therapy. If the tumors are unresectable or metastatic, patients are also given sorafenib, and they are randomized to evaluate the utility of intercalating cycles of gemcitabine and oxaliplatin (NCT03533582). While the use of these drugs in FLC has been reported, their efficacy is not yet known(10). Surgery is the only established curative therapy for FLC, and is usually only palliative for advanced disease(11). For metastatic or anatomically unresectable disease, current chemotherapeutics offer minimal survival advantage(12,13), and the overall 5-year survival ranges between 30 and 45%(14-18). There are no clinically applicable inhibitors of the DNAJB1-PRKACA chimera.
Here, we investigated whether approved or late-phase therapeutics could be repurposed for FLC. We generated a collection of FLC patient-derived xenografts (PDX). Resected tumor tissue was implanted without interim cell culture. The PDX were validated for expression of the fusion transcript and protein, and a histopathology and transcriptome that recapitulated the tumor of origin. We tested a repurposing library of >5000 drugs on cells dissociated from the PDX. The top hits included napabucasin, inhibitors of epigenetic modulators, topoisomerase 1 (TOPO1) and anti-apoptotic proteins. The efficacy of these hits was further validated on PDX implanted in mice. Cells taken directly from freshly resected patient tumors provided additional validation. We show the activity of napabucasin is mediated through inhibition of translation initiation and by activation of reactive oxygen species. We also demonstrate that drug sensitivity inversely correlates with expression of the anti-apoptotic protein Bcl-xL, and that inhibitors of Bcl-xL synergize with other drugs. This is the first large-scale drug screen for FLC using PDX made from fresh tissue without interim cell culture. This screen identifies four novel classes of therapeutics for FLC as well as synergistic drug combinations. Finally, our direct from patient drug screening provides a rapid assay for the personalized profile of therapeutic efficacy against a solid tumor.
Results
PDX of FLC recapitulate the original tumor
Fresh tumor tissue was donated by patients (age range 17-36 years old, 4 female, 2 male) undergoing surgery for FLC. Tumors were cut into pieces and implanted subcutaneously, intra-hepatically or under the kidney capsule. A portion was dissociated into single cells and injected intra-splenically, intra-hepatically or subcutaneously in NSG mice (see Materials and Methods). Tumors were detected by observation and palpation (range 3-12 months). Xenografts were derived from a primary liver tumor (FLC1), a liver recurrence (FLC4), or metastases (FLC2, 3, 5, and 6). The tumors came both from patients who were treatment-naïve and patients who had received prior chemotherapy (Supplementary Table 1).
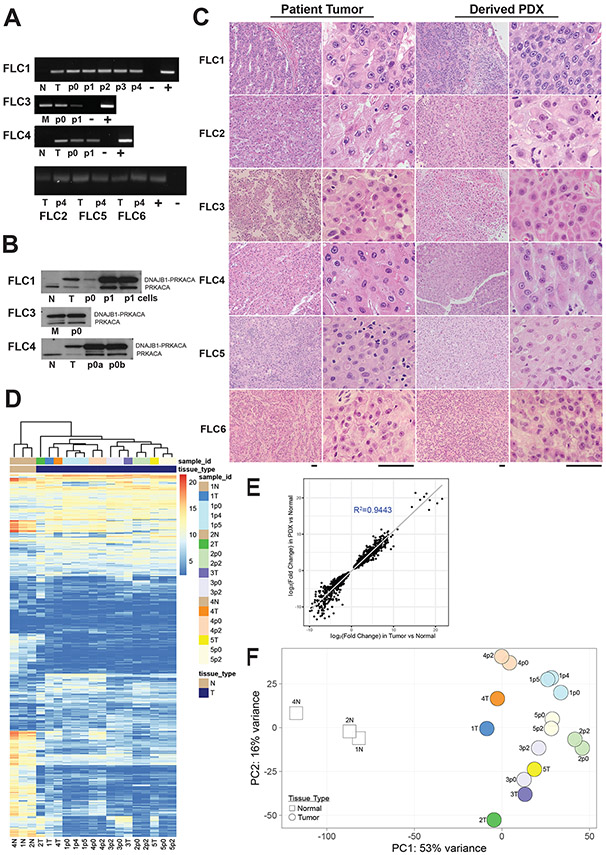
To evaluate whether the PDX model recapitulates the patient tumor, several criteria were assessed. First, the original tumors and the derived PDX were tested by reverse transcriptase PCR (RT-PCR) for expression of the DNAJB1-PRKACA fusion transcript (Figure 1A). In all cases, the fusion transcript was found in the patient tumor and in the derived PDX, but never in the adjacent non-tumor tissue. Through multiple rounds of passaging, xenografts never lost expression of the fusion transcript. The second validation was by Western blotting for the DNAJB1-PRKACA fusion protein (Figure 1B). An antibody that recognizes the shared carboxyl end of PRKACA and DNAJB1-PRKACA demonstrated that while the native protein is found in all samples, the fusion protein was found in every primary tumor and metastasis and PDX, but not in adjacent non-tumor tissue. The third validation was histological analysis. Individual PDXs closely resembled the corresponding tumor of origin, and showed typical features of FLC, including large cells with eosinophilic cytoplasm, prominent nucleoli, “pale bodies” (cytoplasmic inclusion bodies) and areas of fibrosis (Figure 1C). The fourth validation test was based on the transcriptome. We characterized the transcriptome of primary FLC tumors and paired adjacent non-tumor liver (dbGaP Study Accession: phs002435.v1.p1) and identified 509 differentially expressed genes (∣log2∣ fold change≥1, FDR ≤5%) (Supplementary table 2). Unsupervised clustering of tumors, PDX and non-tumor liver showed that tumors clustered with their derived PDX and separated from adjacent non-tumor liver (Figure 1D). The ∣log2∣ fold change of the same differentially expressed genes showed excellent correlation (R2=0.944) between tumors and derived PDX (Figure 1E). Principal component analysis of these differentially expressed genes demonstrated FLC tumors clustered with their derived PDX and away from adjacent non-tumor tissue (Figure 1F). These four criteria support the conclusion that the PDX faithfully replicate important characteristics of their tumors of origin.
Figure 1: Fibrolamellar carcinoma patient derived xenografts recapitulate the original patient tumors.
(A) The fusion transcript for the oncogene DNAJB1-PRKACA was detected by RT-PCR as a 148 bp band in tumor (T), metastases (M) and in all PDX passages in mice (p0, p1, etc) but was not found in the adjacent non-tumor liver (N), (−) negative control, (+) positive control. (B) An antibody that recognizes both PRKACA (41 kDa, lower band) and the oncoprotein, DNAJB1-PRKACA (46 kDa, upper band), revealed both proteins in the patient tumor (T), metastases (M), and each of the passages p0, p1, etc. Only the native PRKACA was found in the adjacent non-tumor liver (N). (C) H&E showed the PDX recapitulate the tumor histopathology (Scale Bar: 50μm). (D) Unsupervised clustering of tumors based on 509 genes differentially expressed between the tumor and adjacent non-tumor liver (∣log2∣ fold change≥1, FDR ≤5%). PDX from liver segregate with liver tumors and PDX from metastases with patient metastases, but away from adjacent non-tumor liver. (E) Gene expression in PDX recapitulates that of the host tumors based on correlation of the ∣log2∣ fold change of the same differentially expressed genes between patient tumors and derived PDX (R2=0.944). (F) Principal component analysis of the same differentially expressed genes for patient tumors (dark color circles), derived PDX (lighter color circles) and adjacent non-tumor liver (white squares).
In vitro screening for drug repurposing
Cells dissociated from the PDX were screened with >5000 compounds (Figure 2A, Supplementary Table 3, the data from the high throughput screens is at: https://osf.io/wzdu6/). We tested compounds eligible for repurposing, including drugs that are approved for clinical use, or are in clinical trials. Pre-clinical and tool compounds were included to help differentiate putative on-target from off-target activities of the clinical drugs.
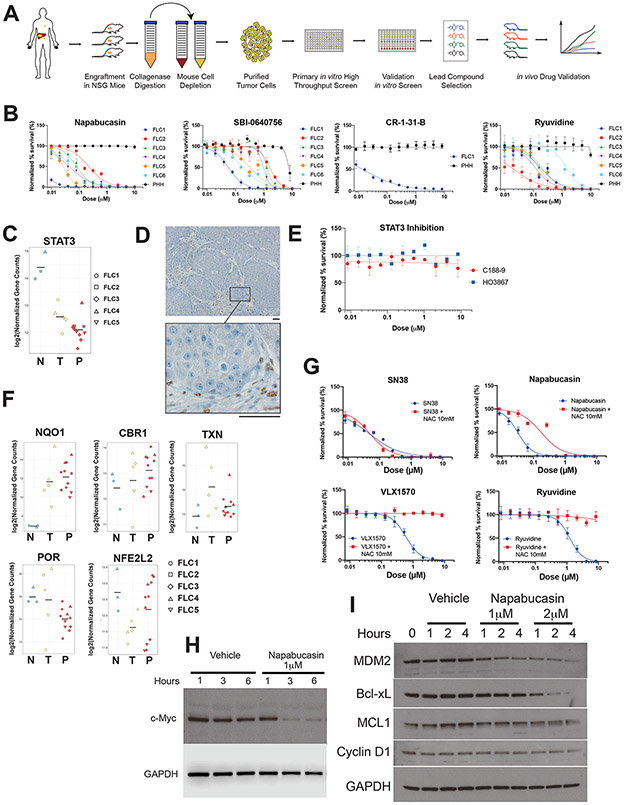
Figure 2: Napabucasin is potent and selective against FLC.
(A) Workflow for generation of PDX and high throughput drug screening. (B) Normalized percent survival of FLC1-6 and PHH treated with napabucasin, additional inhibitors of the EIF4F complex (SBI-0640756 and CR-1-31-B) or Ryuvidine (a SETD8 protein lysine methyltransferase (PKMT) inhibitor). (C) Expression levels of STAT3 by differential expression analysis from RNA-Seq in non-tumor liver (N), tumors (T) and PDX (P). (D) Immunohistochemical staining for pSTAT3 on a section of FLC1, showing positive staining only in the stroma (Scale bar: 50μm). (E) Normalized percent survival of cells from FLC1 treated with the STAT3 inhibitors C188-9 (red) and HO3867 (blue). (F) Differential expression analysis of oxidoreductases (NQO1, CBR1, POR, NFE2L2, TXN in non-tumor liver (N), tumors (T) and PDX (P), from RNA-Seq. (G) Normalized percent survival of cells from FLC1 treated with different drugs in the presence (red) or absence (blue) of N-acetylcysteine (NAC) 10mM. (H,I) Effect of napabucasin (1μM or 2μM) on eIF4F sensitive protein levels of (H) c-myc with GAPDH control and (I) MDM2, Bcl-xL, MCL1, Cyclin D1, C/EBP with GAPDH control, in FLC6 cells.
The screen identified 175 compounds for further evaluation that induced ≥40% killing after treatment with 1μM for 72h. To this list, we then added compounds that are in clinical use against FLC or inhibit targets relevant to the biology of FLC to assess their efficacy (full list is at https://osf.io/wzdu6/). For further validation, these compounds were freshly prepared from powder and then screened using an 11-point dose-response curve (10nM −10μM) on 6 FLC PDXs. Our control cells were primary human hepatocytes (PHH) propagated in immune deficient FNRG mice(19). Drug efficacy was ranked by the average −log10EC50 for FLC1-5, minus the −log10EC50 for the PHH. There was no loss of cell viability during the course of the screen (Supplementary figure 1A).
The drug-sensitivity of the liver tumors (FLC1,4) segregated from the metastases, which were more resistant (FLC2,3,5) (Supplementary figure 1B). The top hit in our screen was napabucasin, the second and third were the epigenetic modulators JIB-04 and ryuvidine; and the fifth was navitoclax, which blocks anti-apopototic proteins.
Validation of agents identified in the primary screen
Napabucasin:
In our efficacy ranking, the top hit was napabucasin, which was potent against all FLC PDX in vitro with an EC50 that ranged from <10 nM to ~300 nM. There was no detectable effect on PHH even at 10 μM (Figure 2B, Supplementary figure 1B). The anti-tumor effect of napabucasin is reported to be mediated by inhibiting p-STAT3 (20,21). However, when we probed for expression levels, STAT3 transcripts were decreased 4-5 fold in FLC tumors relative to the adjacent non-tumor liver (Figure 2C). When protein levels were probed using immunohistochemistry, FLC cells were negative for p-STAT3 staining, and positive cells were only observed in the stroma (Figure 2D). Additionally, specific inhibitors of STAT3, C188-9 and HO3867 had no effect on FLC cells in vitro. (Figure 2E). These results are not consistent with napabucasin affecting FLC through inhibition of STAT3.
The cytotoxicity of napabucasin has alternatively been proposed to be mediated by generation of reactive oxygen species (ROS). The NAD(P)H dehydrogenase [quinone]1 (NQO1), bioactivates napabucasin to form a hydroquinone which, in turn, becomes a semiquinone releasing cytotoxic ROS(22). NQO1 is increased 32-fold in the transcriptome of FLC relative to adjacent normal (7) (Figure 2F), while the levels of other oxidoreductases such as carbonyl reductase 1 (CBR1) or cytochrome P450 oxidoreductase (POR), Nuclear factor erythroid 2-related factor 2 (NFE2L2) and thioredoxin (TXN), which do not bind as effectively to napabucasin, were not similarly elevated (Figure 2F). This mechanism is consistent with the efficacy of Elesclomol, which induces ROS(23) and is sixth in our screen. To test the role of ROS in napabucasin-induced cytotoxicity against FLC, we pre-treated the cells with N-acetyl cysteine (NAC), a precursor of cysteine and glutathione, which scavenges ROS. NAC partially attenuated napabucasin toxicity (Figure 2G). In contrast, NAC had no effect on SN38, the active metabolite of irinotecan, and NAC completely abrogated the toxicity of other agents, including: ryuvidine, VLX1570; and SBI-0640756, NSC697923, and mildly attenuated that of panobinostat, quisinostat, and LY2857785 (Figure 2G, Supplementary Figure 2). These results indicate that ROS generation contributes to some, but not all, of the effects of napabucasin.
Napabucasin has also been reported to inhibit protein synthesis through the eukaryotic initiation factor 4E (eIF4E) and eIF4E-binding protein 1 (4E-BP1); components of the eIF4F complex involved in the initiation of protein synthesis for certain mRNA(21). One of the best-studied oncogenes translationally regulated by the eIF4F complex is c-Myc (24). FLC cells treated with 1 μM napabucasin showed a dramatic decrease in c-Myc levels with no effects on GAPDH (Figure 2H). Treatment with napabucasin lowered the levels of additional proteins whose synthesis is known to be sensitive to the eIF4F complex, including MDM2, Bcl-xL, MCL1, and Cyclin D1 (Figure 2I). SBI-0640756, which also disrupts the complex, was the 19th best compound in the screen (Supplementary figure 1B) and showed a clear therapeutic window against FLC relative to PHH (Figure 2B). Derivatives of silvesterol, such as CR-1-31-B, specifically inhibit the eIF4A RNA helicase and block the translation of mRNA-containing a G-quadruplex(25). Inhibiting eIF4A blocks the synthesis of several key oncogenes, including c-myc and anti-apoptotic proteins such as Bcl-xL and Mcl-1(25). CR-1-31-B was cytotoxic to FLC in the low nanomolar range, with no effect on PHH (Figure 2B). These results indicate that inhibition of eIF4F may substantially contribute to the effects of napabucasin, and eIF4A, and perhaps other members of the initiation complex, may be important therapeutic targets in FLC.
Epigenetic modulators:
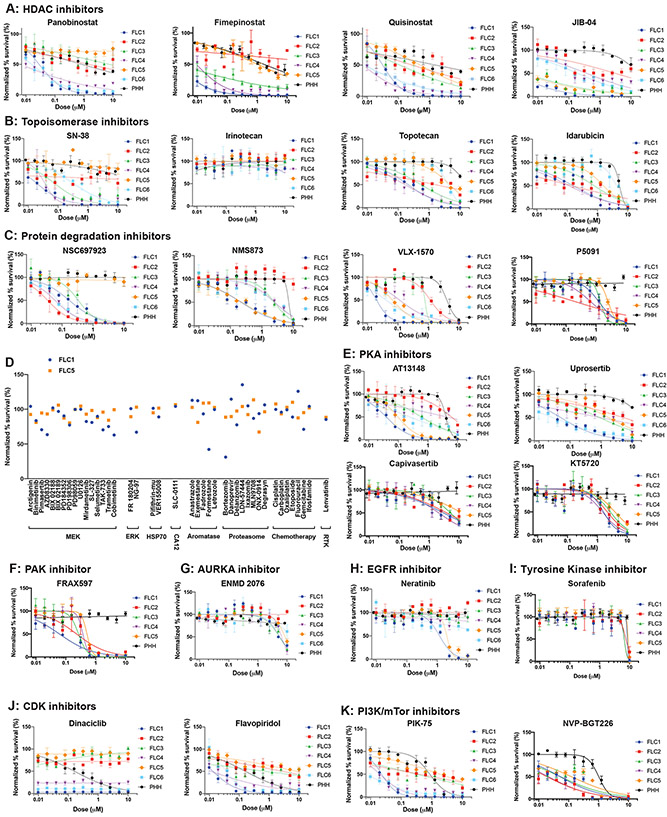
Two of the three highest scoring agents in our screen were epigenetic modulators: JIB-04, a pan-jumonji histone demethylase inhibitor (second in efficacy), and ryuvidine, a SETD8 protein lysine methyltransferase (PKMT) inhibitor (third). Both were effective against all PDX lines (EC50 20-200nM, respectively) and did not affect PHH (EC50>10 μM) ((Supplementary figure 1B, Figure 3A, Figure 2B). Clinical stage epigenetic modulators had a variable effect on different PDX. Quisinostat (46th on the list) and panobinostat (103rd) are pan-histone deacetylase inhibitors (HDACi) that were more effective against the primary liver (FLC1,4) than the metastatic FLC lines (FLC2,3,5) (Figure 3A); while fimepinostat (22nd) another pan-HDACi, affected FLC1,3,4.
Figure 3: Dose response curve validation of drugs from high throughput screen.
(A-J) Dose response curve of drugs tested in triplicate against cells from PDX (FLC1-6) and PHH. Error bars - standard deviation. Drugs were tested at 10μM-10nM with 2-fold serial dilution (apart from specific drugs tested at higher concentrations based on the published EC50). The y-axis shows normalized percent survival calculated as 100–[(positive control–drug response)/(positive control-negative control)*100]. (A) Epigenetic modulators: HDAC inhibitors panobinostat, fimepinostat, quisinostat; and HDM inhibitor JIB-04. (B) Topoisomerase 1 inhbitors SN38, irinotecan, topotecan, and topoisomerase 2 inhibitor idarubicin (more in Supplementary Figure 3A). (C) Inhibitors of the protein degradation pathway: NSC697923, NMS873, VLX-1570 and P5091. (D) Drugs proposed for FLC that were tested in the preliminary screen, but did not meet criteria for validation. Including inhibtors of MEK/ERK, HSP70, CA12, aromatase, proteasome, receptor tyrosine kinases. Y-axis shows normalized percent survival of cells after 1μM treatment for 72h. Blue-FLC1; Orange-FLC5. (E) Protein kinase A inhibitors (more in Supplementary Figure 3B). (F) The PAK1-3 inhibitor FRAX597. (G) The Aurora kinase A inhibitor ENMD-2076 (more in Supplementary Figure C). (H) The ERBB2 inhibitor neratinib (more in Supplementary Figure 3D). (I) The tyrosine kinase inhibitors sorafenib (regorafenib in Supplementary Figure 3E). (J) The CDK inhbitors dinaciclb and flavopiridol (more in Supplementary Figure 3F). (K) PI3K/mTOR inhibitors (more in Supplementary Figure 3G).
Topoisomerase inhibitors:
SN-38 a metabolite of the topoisomerase-1 inhibitor irinotecan, was one of our most efficacious compounds (13th). However, similarly to clinical-stage epigenetic modulators, it also showed a variable response and was effective only against FLC1,3,4. SN-38 was more potent than topotecan or camptothecin, while irinotecan, which requires hepatic metabolism, showed no effect in vitro (Figure 3B, Supplementary Figure 3A). The topoisomerase-2 inhibitor idarubicin showed an effect on some of the FLC PDX while others (aclarubicin, daunorubicin, doxorubicin) did not (Figure 3B, Supplementary Figure 3A).
Other modulators of protein levels:
The top hits in our screen included compounds that, like napabucasin, modulate protein levels independent of transcription. These include NSC697923, the 7th most efficacious compound, an E2 ubiquitin conjugating enzyme (UBE2N) inhibitor, VLX1570, at #31, a deubiquitinase (USP14) inhibitor, P5091, at #38, a deubiquitinase (USP7), and NMS873, an AAA ATPase (VCP/p97) inhibitor at #72. These compounds were effective against different subsets of the FLC PDX (Figure 3C). We did not see any effect of proteasome inhibitors such as bortezomib, ixazomib, delanzomib, ONX-0914, LDN-57444, degrasyn, MLN9708, and danoprevir (Figure 3D).
Inhibitors of PKA:
The oncogenic activity of DNAJB1-PRKACA requires its kinase activity(4). Uprosertib, a PKA inhibitor currently in clinical trials (NCT02093546) was #28 in efficacy and AT13148 was #51. AT13148 and Uprosertib showed efficacy in the nanomolar range against FLC1 and AT13148 showed nanomolar efficacy for FLC1,5. Capivasertib and KT5720 showed uniform efficacy against all FLC PDX lines, but in the micromolar range (Figure 3E). H89 and A-674563 showed no therapeutic window for FLC over PHH (Supplementary Figure 3B). Thus, inhibitors of PKA showed mixed results (Table 1).
Table 1.
PKA Inhibitors tested
| Drug | Ki PKA | Ki against other kinases | EC50 FLC | EC50 PHH |
|---|---|---|---|---|
| H89 | 48nM | PKG 480nM | FLC1 5.82μM FLC2 23.88μM FLC3 18.17μM FLC4 20.94μM FLC5 9.49μM |
41.5 μM |
| KT5720 | 60nM | PKG/PKC >2μM | FLC1 1.61μM FLC2 2.14μM FLC3 2.71μM FLC4 1.48μM FLC5 3.87μM |
>10 μM |
| A-674563 | 16nM | Akt1 11nM CDK2 46nM GSK-3β 110nM ERK2 260nM PKCδ 360nM RSK2 580nM |
FLC1 0.34μM FLC2 0.7μM FLC3 0.88μM FLC4 0.16μM FLC5 3.45μM |
3.45μM |
| Capivasertib (AZD5363) | 7nM | Akt1 3nM Akt2 8nM Akt3 8nM |
FLC1 2.98μM FLC2 3.85μM FLC3 5.15μM FLC4 2.09μM FLC5 12.6μM |
>10 μM |
| AT13148 | 3nM | Akt1 38nM Akt2 402nM Akt3 50nM p70s6k 8nM Rock1 6nM Rock2 4nM |
FLC1 0.039μM FLC2 6.14μM FLC3 0.59μM FLC4 4.29μM FLC5 0.096μM |
4.05 μM |
| Uprosertib (GSK2141795) | 2 nM | Akt1 180nM Akt2 328nM Akt3 38nM |
FLC1 0.049μM FLC2 5.039 μM FLC3 0.87 μM FLC4 0.91 μM FLC5 1.72 μM |
> 10 μM |
Oncogenes in FLC:
Several oncogenes are increased in FLC tumors including aurora kinase A (AURKA), aromatase, p21 activated kinase 3 (PAK3), and members of the epidermal growth factor receptor (EGFR) pathway including ERBB2 (Her2)(7). Consequently, some are targets of clinical trials for FLC. FRAX567 a preclinical group-1 specific PAK inhibitor (IC50=8/13/19nM for PAK 1/2/3), was #12 in our efficacy ranking, showing consistent inhibition against all PDX (Figure 3F). We have previously shown that PAK3 is one of the most highly expressed oncogenes in FLC (7). The effects of inhibiting the other oncogenes that are increased in expression in FLC were not as strong. ENMD-2076, an AURKA inhibitor (IC50=14nM) in a clinical trial for FLC (NCT02234986) was #123 on the list, had a EC50>5μM against the PDX cells (Figure 3G). Aurora Kinase Inhibitor I, a pre-clinical inhibitor (IC50=3.4nM), was #51 with a EC50 of 1-4 μM. However, alisertib, a more potent inhibitor of AURKA (IC50=1.2nM) in phase 3 trials for small cell lung cancer and other malignancies, showed no effect on FLC (Supplementary Figure 3C), suggesting the effect of Aurora Kinase Inhibitor I is likely off-target. Neratinib, an inhibitor both of EGFR (IC50=92nM) and ErbB2 (IC50=59nM) in a clinical trial including FLC (NCT01953926), affected FLC1,5 in the micromolar range (Figure 3H). However, both Erlotinib, used for treating EGFR driven cancers (IC50=2nM), and CP-724714, in trials for tumors expressing ErbB2 (NCT00102895, NCT00055926, IC50=10nM), showed no effect on any FLC lines (Supplementary Figure 3D).
Other clinical compounds for FLC:
Sorafenib and regorafenib, pan-kinase inhibitors used clinically to treat FLC, showed no therapeutic window for FLC over PHH (Figure 3I, Supplementary Figure 3E). Cytotoxic chemotherapies reported to have been used to treat FLC in the clinic, including cisplatin, carboplatin, oxaliplatin, etoposide, fluorouracil, gemcitabine, and ifosfamide(26) [Pediatric Hepatic Malignancy International Therapeutic Trial (PHITT), NCT03533582] showed no to minimal activity against FLC PDX cells (Figure 3D). In a clinical trial of the pan-CDK inhibitor, flavopiridol, two FLC patients had stable disease for 17 months(10). We tested multiple CDK inhibitors, many of which did not pass the primary screen (Supplementary Table 4). Pan-CDK inhibitors including dinaciclib (SCH-727965), and flavopiridol (Figure 3J) and CDK9 inhibitors P276-00, and LY2857785 were potent against FLC1 and FLC4 but not the other PDX, and they were toxic for the PHH (Supplementary Figure 3F).
The mTOR inhibitor, everolimus (IC50=1.6-2.4nM), is being tested in combination with an aromatase inhibitor for FLC (NCT01642186). NVP-BGT226, a dual PI3K/mTOR inhibitor in phase 2 trials for breast cancer, showed potency against all FLC PDX, and PIK-75, a pre-clinical dual PI3K (p110α)/DNA-PK inhibitor (IC50=5.8nM), was potent against FLC1,4 relative to PHH (Figure 3K). In contrast, multiple other mTOR and dual PI3K/mTOR inhibitors (OSI-027, WYE-125132, AZD2014, GDC0980) tested against FLC PDX cells showed no therapeutic window compared to PHH (Supplementary Figure 3G). Likewise, aromatase inhibitors, including letrozole, anastrozole, exemestane, formestane, and fadrozole, did not pass our primary screen (Figure 3D). ERK phosphorylation is increased when the oncogenic driver of FLC, DNAJB1-PRKACA, is expressed in AML12 cells, a mouse cell line overexpressing TGF-alpha(27). Mitogen activated kinase (MEK) inhibitors were reported to be potent against these transduced cells. None of a panel of inhibitors against MEK, including arctigenin, binimetinib, pimasertib, AZD8330, BIX02188, BIX02189, PD184352, PD198306, PD98059, U0126, mirdametinib, SL-327, selumetinib, TAK-733, trametinib and cobimetinib, passed the primary screen (Figure 3D). Similarly, ERK, HSP70 and CA12 inhibitors showed no activity against FLC PDX cells (Figure 3D).
Anti-Apoptotic Proteins:
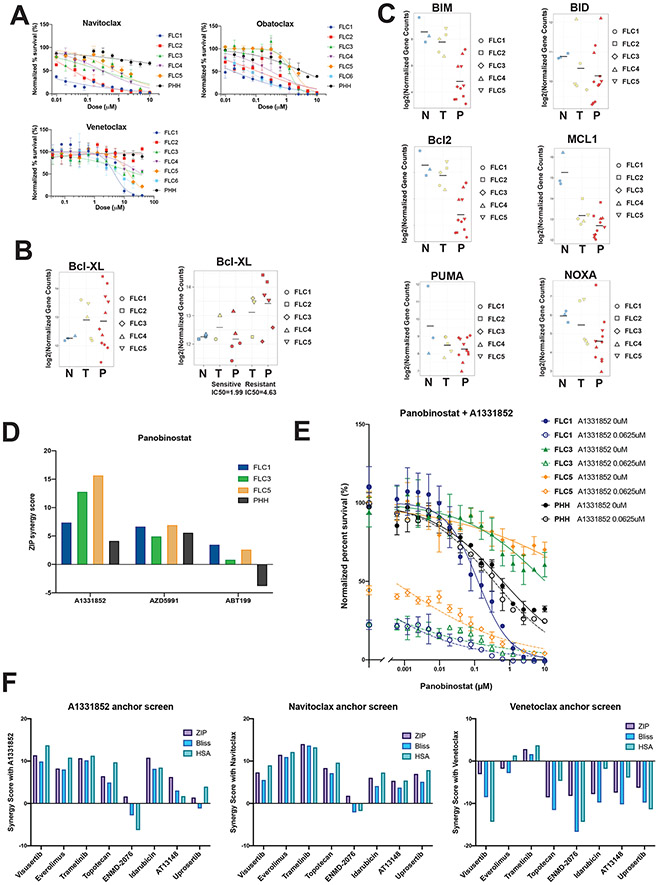
Navitoclax, which inhibits the anti-apoptotic proteins Bcl-2, Bcl-xL and Bcl-w, was the 5th most effective compound (Figure S1, Figure 4A). In contrast, venetoclax, a selective Bcl-2 inhibitor, showed minimal activity against FLC (Figure 4A). Transcripts for Bcl-xL were increased 83% in FLC tumors and derived PDXs compared with adjacent non-tumor liver. In PDX that were more resistant to therapeutics based on average IC50 of compounds tested, Bcl-xL was increased 160% compared to the adjacent non-tumor liver (Figure 4B). Thus, Bcl-xL, but not Bcl-2, may provide a mechanism of apoptosis evasion in FLC. Transcripts for other anti-apoptotic proteins including NOXA, PUMA and BID were not significantly altered, BAX and BAK were increased 50% (7) and MCL1, BIM and Bcl-2 were downregulated (Figure 4C) (7), further strengthening this hypothesis. This led to the suspicion that some of the variability in the response of PDX might be the consequence of differential expression of anti-apoptotic proteins.
Figure 4: Inhibiting Bcl-xL sensitizes FLC but not PHH (next page).
(A) Normalized percent survival of FLC1-6 and PHH with anti apoptotic inhibitors navitoclax (Bcl2/Bcl-xL), obatoclax (Bcl2/Bcl-xL) and venetoclax (Bcl2). (B) Differential expression analysis of Bcl-xL in non-tumor liver (N), tumors (T) and PDX (P), from RNA-Seq (left). Differential expression of Bcl-xL in non-tumor liver, sensitive (FLC1 and 4) versus resistant (FLC 2,3 and 5) tumors and derived PDX (right). (C) Differential expression analysis of additional antiapoptotic (Bcl2, Mcl1) and proapoptotic (BIM, BID, NOXA, PUMA) genes in non-tumor liver, tumors and PDX, from RNA-Seq. (D) The ZIP synergy score of combination therapy with panobinostat and A1331852 (Bcl-xL inhibitor), AZD5991 (Mcl1 inhibitor) and Venetoclax (Bcl2) for FLC1,3,5 and PHH. (E) Dose response curves of panobinostat in the absence (full circles) and presence (empty cicles, dotted lines) of A1331852, showing the normalized percent survival for FLC1,3,5 (blue, green and orange, respectively) and PHH (black). (F) Synergy scores (ZIP, Bliss and HSA) for A1331852, Navitoclax and Venetoclax anchor screens.
Panobinostat, a pan-HDACi (see epigenetic modulators above), was one of the drugs that demonstrated a highly variable response among the sensitive and resistant PDX (Figure S1, 3A). To test whether Bcl-xL was responsible for the variable responses to epigenetic modulators, we used A1331852, a selective Bcl-xL inhibitor. A1331852 synergized with panobinostat, and the extent of synergy correlated to Bcl-xL expression. A more modest synergy was seen with Mcl1 inhibition (AZD5991) and no synergy was observed with Bcl2 inhibition (Venetoclax, ABT199) (Figure 4D). An alternative assay using dose response curves(28) similarly demonstrated synergy between panobinostat and A1331852 in FLC (Figure 4E). This included sensitizing the more resistant lines for which panobinostat was inactive as a single agent. Importantly, treatment with A1331852 did not increase the toxicity of panobinostat in PHH (Figure 4E), and was thus specific for FLC.
Effect of inhibiting Bcl-xL on response to other agents.
To further explore the effect of inhibition of Bcl-xL in FLC we performed an anchor screen on the relatively resistant FLC5. We tested different compounds in combination with either the Bcl-xL inhibitors A1331852 and navitoclax, or the Bcl2 inhibitor venetoclax. These included inhibitors of mTOR (visusertib and everolimus), MEK (trametinib), AURKA (ENMD-2076), PKA (AT13148 and uprosertib), TOPO1 (topotecan) and TOPO2 (Idarubicin). All drugs, apart from ENMD-2076 and uprosertib, showed some synergy with A1331852. Results were similar when drugs were combined with navitoclax, except for uprosertib which now synergized. No drug showed synergy with venetoclax (Figure 4F). Thus, the sensitivity of FLC to a select group of compounds is enhanced by blocking Bcl-xL.
In vivo validation of therapeutics for FLC
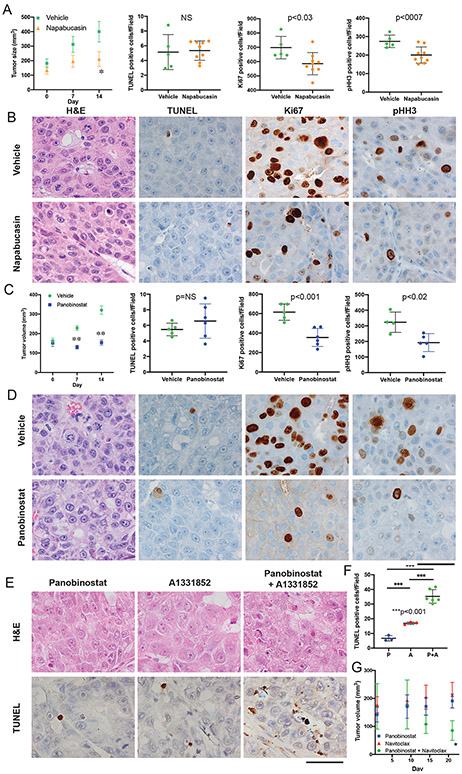
In vitro drug efficacy data may be unrepresentative of outcomes in animal studies. We therefore tested some of our top hits from the in vitro screens in vivo, in mice bearing PDX (see Methods). Napabucasin, the top hit in our screen (Figure S1), reduced growth at 7 and 14 days (44% and 52%) versus vehicle control (70% and 118% p<0.01 Figure 5A). There was a decrease in mitotic figures, Ki67 and pHH3 positive cells in the tumors of the treated mice, and a small increase in TUNEL positive cells (Figure 5A,B). Mice remained active and alert during the course of the napabucasin treatment and experienced an average weight loss of 6.7% compared with 6.2% in the vehicle control treated group. The ratio of tumor volume:mouse weight was stable or decreased for most of the napabucasin treated mice but consistently increased in those treated with vehicle control. To assess the effect of inhibiting eIF4A, CR-1-31B was tested on FLC6, our most aggressive PDX. A statistically significant reduction of growth was observed starting within two weeks versus vehicle control treated mice (Supplementary Figure 4A). It is possible that some of the agents that were negative in the in vitro tests, such as a pan-kinase inhibitor sorafenib, might function by blocking processes only present in intact tissue. However, when sorafenib was tested on FLC6 in vivo, there was no detectable effect on tumor growth (Supplementary Figure 4B).
Figure 5: In vivo validation of drugs against FLC (next page).
(A,B) Effect of napabucasin (10mg/kg) or vehicle control after 14 days on FLC1. (A) Effect of napabucasin on tumor volume in mice (n=6, average±SE, p<0.05) and quantification of the IHC for TUNEL, Ki67 and pHH3. (B) H&E of tumors for mitotic bodies and IHC for TUNEL, Ki67 and pHH3. (C,D) Effect of panobinostat (10mg/kg) or vehicle control after 14 days on (C) Tumor volume (n=5, average±SE, p<0.001) and quantification of the IHC for TUNEL, Ki67 and pHH3 and (D) H&E of tumors for mitotic bodies and IHC for TUNEL, Ki67 and pHH3. (E) H&E of tumors and IHC for TUNEL for mice implanted with FLC1 were treated with vehicle, panobinostat (10mg/kg), A1331852 (25mg/kg), or both for 2 days. Scale bar is 50μM. (F) Quantification of the TUNEL IHC from. (G) Mice implanted with FLC5 were treated with panobinostat (2.5mg/kg), navitoclax (50 mg/kg) or both (n=4, average±SE, *p<0.01).
HDAC inhibitors that scored well in vitro and are available in the clinic were tested in mice. Panobinostat, led to complete inhibition of tumor growth (−7% growth at 7 days; 4% at 14 days) compared with vehicle control (45% at 7 days and 102% at 14 days p<0.0001, Figure 5C). Mice remained active and alert during the treatment. Those treated with panobinostat experienced an average weight loss of 11% compared with 6% in the vehicle control treated group. The tumor volume:mouse weight was stable or decreased in panobinostat treated mice, but increased in mice treated with vehicle control. Panobinostat led to a dramatic decrease in mitotic figures, and in Ki67 and pHH3 positive cells. However, there was no increase in apoptosis, as assayed by TUNEL (Figure 5C,D).
We used the HDAC inhibitor quisinostat to further test the effect of such inhibitors. Treatment of mice with quisinostat led to complete inhibition of tumor growth compared with vehicle control treatment (7% versus 110% growth at 14 days, respectively, p<0.0001, Supplementary Figure 4C). Treatment with quisinostat led to an almost complete disappearance of mitotic figures, Ki67 and pHH3 positive cells, but no significant increase in apoptotic cells as assayed by TUNEL (Supplementary Figure 4C,D). Macroscopic assessment showed necrosis in the drug treated tumors (Supplementary Figure 5).
In treatments of dissociated cells from PDX in vitro, panobinostat synergized with A1331852, an inhibitor of Bcl-XL (Figures 3D,E,F). Administration of these drugs showed synergism in vivo, and after 2 days of treatment, many apoptotic cells were observed with the combined treatment of panobinostat and A1331852 (Figure 5E,F). However, consistent with the previous longer treatments (Figure 5D), no evidence of increased apoptosis was detected in tumors from mice treated with panobinostat alone compared to vehicle (Figure 5F). Furthermore, in FLC5, the most drug-resistant PDX, panobinostat synergized with Navitoclax, resulting in not only in inhibition of tumor growth, but in partial response, with a significant reduction in tumor size (Figure 5G).
Testing in cells directly isolated from patient tumors
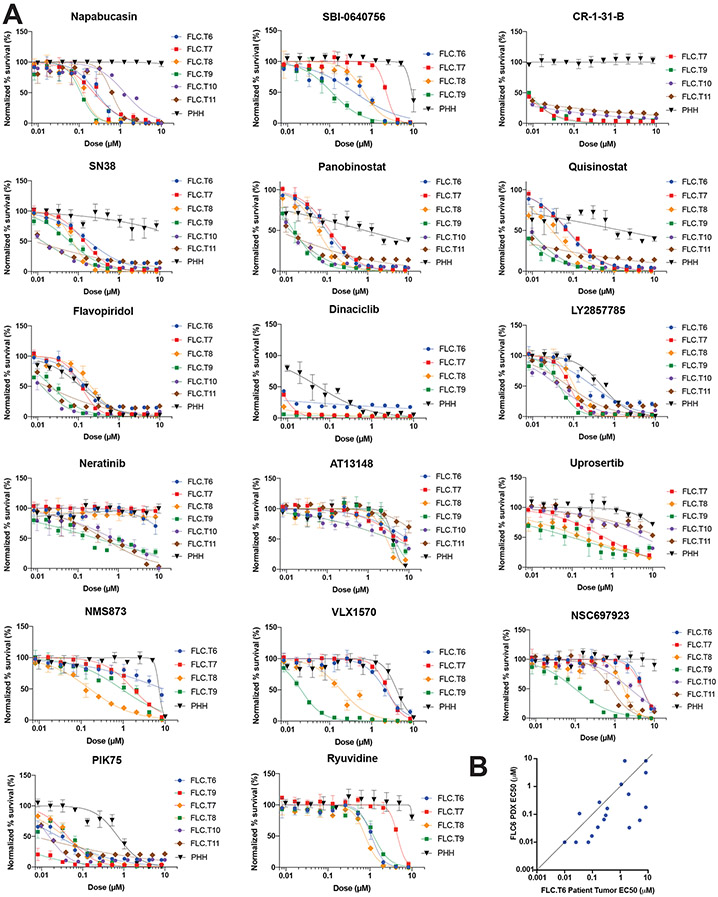
To evaluate whether our screening results in PDX-derived FLC cells were recapitulated in tumor tissue, some of the most efficacious compounds were tested on cells directly isolated from patient tumors. Within a few hours of resection, cells were dissociated and screened against a panel of compounds (Supplementary Figure S6A). All tumor cells were derived from lymph node metastases, with the exception of FLC.T9, which was from ascites fluid (Supplementary Table 5). All cells tested directly from patients were sensitive to the top hits of our in vitro screen including the three inhibitors of the eIF4F complex (napabucasin, CR-1-31-B, and SBI-0640756), the two HDAC inhibitors, (panobinostat and quisinostat), the topo I inhibitor (SN-38), and the dual PI3K (p110α)/DNA-PK inhibitor (PIK-75). There was some sensitivity to uprosertib, dinaciclib, flavopiridol, LY2857785. There were some compounds that did not fare well on PDX, that showed response on a few of the direct-from-patient testing. Most notable is neratinib, currently in a clinical trial for FLC, which had a Ki of 300-700 nM for three of the direct from patient tumor cells, but no effect on three others (Figure 6A).
Figure 6: Direct from patient in vitro screening recapitulates findings from PDX screen.
(A) Dose response curve of drugs tested in triplicate against cells dissociated directly from patient tumors (T6-T11) versus PHH. Error bars – standard deviation. Drugs were tested at 10μM-10nM with 2-fold serial dilution. The y-axis shows normalized percent survival calculated as 100 –[(positive control–drug response)/(positive control-negative control)*100]. Cells were treated for 72h, and normalized percent survival was quantified using Cell Titer Glo. (B) Comparison of the EC50 in cells dissociated from patient tumor FLC.T6 vs cells dissociated from its derived PDX FLC6.
There was a striking similarity among the response profiles of cells isolated from lymph nodes of different FLC patients. The cells from the ascites fluid were more sensitive to some of the drugs in this panel, but their response otherwise resembled that of the lymph nodes. Compounds that inhibit protein degradation pathways (NSC697923 and VLX1570), showed a blunted response in the direct from patient testing (Figure 6A). However, it should be noted that these compounds were not active against cells from all PDX lines either. FLC.T6 was the only tumor for which cells were compared directly from the patient tumor and the derived PDX. Drug sensitivity for FLC.T6 and FLC6 correlated well for most drugs tested and specifically for Napabucasin, Panobinostat, and SN-38; as well as Neratinib. Notable outliers were NSC697923, VLX1570, and AT13148 (Figure 6B and Supplementary Figure 6B).
Discussion:
The development of efficacious therapies for FLC has been stymied by the lack of faithful models. As a result, clinicians have had to rely either on therapies used for other liver cancers, such as HCC, or on therapies that target pathways that are upregulated in FLC. Both approaches are potentially problematic. First, FLC and HCC are distinct diseases. There is little resemblance in the coding and non-coding transcriptome of HCC and FLC(7,8) and the pathogenesis of FLC is rooted in dysregulation of the ecology of PKA signaling(1,29), which has not been implicated in HCC. In addition, there is a lack of published data showing that agents used in treating HCC are efficacious in treating FLC. Finally, pathways that are activated in FLC, or at least increased in expression, may be bystanders irrelevant to pathogenesis or therapy. For example, the increase of PKA activity may be increasing expression of aromatase or AURKA, but these are not necessarily oncogenic in FLC.
To facilitate therapeutic development, we generated several patient-derived xenografts that were validated at the histologic, genomic, transcriptomic and proteomic levels. These were used to test therapeutics that have been used clinically for FLC, target pathways activated in FLC, and, in an agnostic screen of clinical stage drugs, to identify those that could be repurposed for FLC. As a counter-screen we used primary human hepatocytes, similarly propagated and passaged in immune-deficient mice. The top hits were validated in vivo, and then confirmed on tumor cells isolated directly from patient tissue after resection.
Several novel compounds and targets were identified in our screen. Napabucasin was potent against all FLC PDX and did not cause toxicity in the PHH up to the highest dose tested (Figure 2B). Napbucasin inhibits tumor progression and metastasis in osteosarcoma(21) and is being studied in phase 3 clinical trials against several solid tumors. A proposed target of napabucasin is the eIF4F complex. Two other compounds that affect this pathway, SBI-0640756 and CR-1-31-B, also demonstrated specific toxicity in FLC PDX and direct from patient screens (Figures 2B, 6A). SBI-0640756, inhibits eIF4G1 and disrupts the eIF4F complex, blocking the growth of BRAF resistant melanoma(30). CR-1-31B was effective in vitro and in vivo against T-ALL(25) and a similar compound eFT226 is currently in clinical trials for solid tumors (NCT04092673). This raises the possibility of translational regulation as a potent therapeutic target in FLC(25,31,32).
Inhibition of PAK3 was the most efficacious among any of the oncogenes increased in FLC (Figure 3F). PAK3 is increased at the protein and transcript level (log2=7.91, FDR<<10−10) in FLC(7). FRAX567 has activity against malignant meningioma and squamous cell carcinoma in vitro and in vivo(33,34). While there are no clinical PAK1-3 inhibitors currently available, several are under development.
HDAC inhibitors are being developed for several cancers. Panobinostat has activity against class I, IIa and IIb HDACs and is approved as a 3rd line therapy for multiple myeloma in combination with bortezomib and steroids. Quisinostat is in phase 2 clinical trials for cutaneous T cell lymphoma and ovarian cancer (NCT01486277, NCT02948075). Fimepinostat, a dual HDAC (class I, 2b) and PI3K inhibitor, is fast-tracked for diffuse large B-cell lymphoma. The FLC PDX were not uniformly sensitive to these compounds. PDX originating from liver tumors were sensitive and PDX derived from metastases were resistant when assayed in vitro. In vivo treatment with panobinostat or quisinostat led to growth arrest of liver-derived PDX.
Inhibitors of TOPO1 are used for a variety of indications in cancer. Topotecan is approved for cervical, ovarian and small cell lung cancer, and irinotecan is approved for colorectal cancer and its metabolite, SN-38, is in clinical trials for small cell lung cancer (NCT00104754) and colorectal cancer (NCT00311610). Combination therapy with panobinostat and topotecan was shown to have synergistic effects against ovarian cancer in pre-clinical studies(35) and holds potential for FLC.
Activation of anti-apoptotic proteins can lead to evasion of apoptosis in cancer and several inhibitors are currently in clinical trials(36). Navitoclax, which inhibits the anti-apoptotic proteins Bcl2 and Bcl-xL, was one of the top scoring compounds in our assay. In contrast, venetoclax, a selective Bcl2 inhibitor scored towards the bottom of the ranking (Figure 4A, S1). This led us to hypothesize that Bcl-xL contributed to the variations in drug response in FLC. Specific inhibition of Bcl-xL using the tool compound A1331852 sensitized FLC lines that were resistant to panobinostat monotherapy (Figure 4D,E). Synergism was also demonstrated when the TOPO1 inhibitor, topotecan, was combined with A1331852 or navitoclax, but not with venetoclax. This presents the possibility of Bcl-xL inhibition as combination therapy with HDAC or TOPO1 inhibitors. A study combining navitoclax with irinotecan found the safety profile of the combination was consistent with data from monotherapy studies (NCT01009073)(37). Many drugs, which had no detectable activity on their own, synergized with blockers of Bcl-xL. In vivo A1331852 synergized with panobinostat to induce apoptosis (Figure 5E,F), and the combination of navitoclax with panobinostat led to a partial response of the most resistance PDX (Figure 5G). This suggests that strategies for choosing combination therapies should account for the specifics of the mechanisms of drug-sensitivity in FLC.
Various drugs were consistently efficacious against cells dissociate from PDX, preclinical mouse models and cells dissociated from patients. In contrast, most of the drugs currently used in the clinic had minimal activity against FLC. Sorafenib, a multikinase inhibitor which is first line therapy for HCC, as well as other tyrosine kinase inhibitors, did not demonstrate efficacy against FLC in vitro or in vivo (Figure 3I, Supplementary Figure 4B). Chemotherapeutics such as cisplatin, carboplatin, oxaliplatin, etoposide, fluorouracil, gemcitabine, and ifosfamide that are used to treat FLC in the clinic, or are currently in clinical trials showed no utility. Several of the oncogenes found to be upregulated in the FLC transcriptome have been tested in clinical trials including ENMD-2076 for AURKA; everolimus for mTOR, and neratinib for EGFR/ErbB2. Our screen showed no selective toxicity against FLC over human hepatocytes for any of the drugs in clinical trials for these targets (Figure 3D, 3G, 3H). Experimental compounds with even higher specificity for the targets, showed no selective toxicity over primary human hepatocytes. Thus, it will be informative to see the published outcomes of these clinical trials. Recently, inhibition of MEK was proposed for treating FLC(27). We tested a panel of eighteen MAPK/ERK inhibitors, none of which were efficacious against the FLC cells tested (Figure 3D). These results suggest a general caution when using therapeutics to target oncogenes that are increased in expression in a tumor.
PDX in mice are a powerful system for preclinical screening of drugs and have been proposed for precision medicine. However, there are some limitations to the use of the PDX for this purpose. First, the success rate for implantation is 30-35% in our hands. Thus, the PDX would help only a subset of the patients. Second, the PDX took several months to a year to grow. This limits their utility for providing information within a clinically relevant time frame. Third, the PDX may not recapitulate the response of the patient. The physiology of mice is not identical to that of humans, and the human tumor cells are surrounded by mouse stromal cells whose interacting ligands and receptors may not be completely homologous. Mouse strains used for PDX lack a functional adaptive immune system and may not recapitulate the effect of therapeutics on anti-tumor immunity. Fourth, an implanted tumor may not respond to drugs in the same way as a tumor that has developed spontaneously. Fifth, passaging the tumor in a mouse could lead to changes in drug response compared to the original tumor. Finally, it is possible that only a select small subset of the tumor cells implanted in mice can survive and these do not recapitulate the full response of the tumor cells in the patient.
The PDX data have identified promising novel targets and the mechanisms through which they may work. Clinical trials are needed for validation. The screens on tumor cells, isolated from patient tumors shortly after resection, were on the entire set of tumor cells and not subgroup potentially selected by mouse passage. The responses in the direct from patient screens showed similar results across all patients tested, except for drugs inhibiting protein degradation, which were potent only against cells from ascites (n=1) but not from lymph node metastases (n=5). Similarly, both the PDX and the direct from patient cells were sensitive to some of the compounds that seem most promising therapeutically: Blockers of initiation of translation, HDAC or TOPO1. There were some exceptions. Neratinib, showed no effect on PDX, but efficacy in the sub-microlar range on three direct from patient screens and no effect on cells from the other three patients. Is this the consequence of differences in assaying PDX or direct-from-patient cells, or is it patient variability? We compared the results for cells dissociated from PDX FLC6 in figures 2,3,4 with samples derived from the same patient for a direct-from-tumor screen FLC.T6 (Figure 6B, S6B). Fifteen out of eighteen compounds showed similar response in cells dissociated from PDX and direct from patient. The exceptions were two drugs thought to mainly inhibit protein degradation (BLX1570 and NSC697923) and the PKA inhibitor AT13148.
The screening of cells directly from patient tumors has a number of potential advantages. First, the results are available within days of resection or biopsy rather than the months to a year required for PDX. This is critical for informing therapeutic decisions. Second, there is the concern that only a subset cells have grown to form organoids or a PDX. Thus, tests on these model systems may represent the response of a subset of cells. The direct-from-patient test is done directly on a sample, with no intermediate passaging. Third, the direct-from-patient testing gives a personalized profile of a specific patient tumor. This is a step to fulfilling the promise of precision medicine. We have observed that the FLC tumors fall into distinct classes of response. As we accumulate further data from direct from patient screens, individual patient characteristics may become apparent that predict differential responses to individual drugs, much as was revealed by Bcl-xL stratification of the PDX results. Validation of this approach will initially come from clinical case reports of otherwise intractable disease which will test the utility of this method.
Finally, our demonstration that the transcriptome is stable and that the expression of the chimeric oncogene remains stable through consecutive passages of PDX in mice strongly suggests that FLC has become oncogenically addicted to DNAJB1-PRKACA(38,39). Thus, elimination of the oncogene, oncotranscript, or oncoprotein may be an effective therapeutic approach for FLC.
Methods:
Human tissue samples
With Institutional Review Board approval (Rockefeller IRB#SSI-0797, SSI-0855) written informed consent was obtained from patients scheduled for tumor resection. All studies were conducted in accordance with recognized ethical guidelines. The diagnosis of FLC was determined by a pathologist at each institution, and the demonstration of the DNAJB1-PRKACA fusion transcript by RT-PCR and DNAJB1-PRKACA fusion protein by Western blot, as described below. Tumor tissue and adjacent non-tumor liver (if available) not needed for diagnosis and treatment (i.e. “surgical waste”) were collected for the study. After resection, the tissue was placed into cold PBS on ice, cut into 2-3x0.5cm portions and placed into 50mL tubes in Roswell Park Memorial Institute (RPMI 1640 with glutamine) media supplemented with 2% penicillin/streptomycin (p/s). Pieces of tumor were prepared for implantation into mice (see below). The rest of the tumor tissue was cut into 2mm pieces (without mincing) and connective tissue blood vessels, clots, and necrotic tissue, were discarded. Pieces of tumor were also fixed in formaldehyde for histological analysis; and flash frozen or placed into Optimal Cutting Temperature compound (OCT) and frozen for later RNA/protein analysis.
Tumor dissociation
The 2mm pieces of tissue were placed into 50mL Falcon© tubes with RPMI, collagenase 4 (Worthington 1 mg/ml) and DNAse (Roche, 1 μg/ml), and digested while rotating at 37C until digestion was complete (Benchmark scientific Roto-therm). All following steps were done on ice or at 4C. The digested tissue was passed through a 200μm (Pluriselect) strainer using a syringe plunger for remaining pieces, and then through a 100μm strainer (Fisher). The cells were spun down at 300xg for 5 minutes at 4C and the pellet depleted of red blood cells by a 10 sec exposure to 1ml of water followed by the addition of 49mL of PBS. The cells were counted and either implanted into mice or used for in vitro experiments. For in vitro experiments the cells from PDX tumors were subjected to mouse cell depletion according to the manufacturer’s instructions (Miltenyi Biotec).
Mice
For PDX studies NOD-scid-gamma (NSG) mice were purchased from Jackson Laboratories (Bar Harbor, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, strain 005557) and bred at The Rockefeller University animal facility specific-pathogen free (SPF) immune-core. Mice were kept in 12hr light/dark cycle, fed an amoxicillin diet, and had ad libitum access to food and water. Both male and female mice were used for initial implantation of tumors and for passaging PDXs. Female mice 5-8 weeks old were used for in vivo experiments. Mice were inspected at least twice a week for health and tumor growth. Tumors were passaged if they reached 2cm or the mouse displayed signs of illness or weight loss. For passage of PHH, Fah−/− NOD Rag1−/− Il2rgnull (FNRG) mice(40) were transplanted with PHH and subjected to intermittent liver injury(41). After humanization of the liver had plateaued, as determined by human albumin levels in mouse serum, PHH were isolated as described previously(19). All experiments were conducted under animal use protocols approved by Rockefeller University.
Implantation into mice
With IACUC approval (#20027-H) mice were anesthetized using isoflurane and given buprenorphine for analgesia. Pieces of tumor were cut and placed into RPMI on ice. Tumor pieces were implanted subcutaneously, under the kidney capsule or directly into the liver. For subcutaneous implantation a small skin incision was made in the flank area and pieces of tumor were placed between the skin and fascia, before the skin was stapled. For implantation under the kidney capsule, the kidney and retroperitoneum was exposed via abdominal approach and very small incision was made in the kidney capsule. Small pieces of tumor (<0.5mm) were placed under the kidney capsule and gently advanced away from the incision. The kidney was then placed back into the abdominal cavity, before the abdominal wall was sutured and the skin stapled. For implantation directly into the liver, the liver was exposed via abdominal incision and a small area was cauterized. Thin forceps were then used to make a small tunnel in the liver parenchyma and a narrow piece of tissue was placed into the tunnel. The surface edge of the tunnel was then cauterized and the liver was placed back into the abdominal cavity. The abdominal wall was sutured and skin stapled. Cells dissociated from patient or PDX tumors were implanted subcutaneously, into the liver or the spleen. For subcutaneous implantation 5x105-2x106 cells were mixed at a 1:1 ratio with Matrigel (Corning) and injected into the subcutaneous space over the flank. For intrahepatic implantation the 5x105 cells were mixed with Matrigel at a 1:1 ratio and injected directly into the liver. For splenic injection 5x104-5x105 cells in RPMI were injected into the spleen.
Primary screen
Cells dissociated from PDX tumors and depleted of mouse stromal cells were used for high throughput screening. For the primary screen 5093 compounds were dispensed with Janus 384 pintool (Perkin Elmer) into 384 well plates. Cells were plated into the 384 well plates containing screening compounds at 2000 cells/well in Kubota’s medium (PheonixSongs biological) supplemented with 2% p/s and incubated at 37C for 72 hours. Compounds were screened at a final concentration of 1μM. Each plate contained negative control wells treated with DMSO and positive control wells treated with 20 μM chaetocin (Selleckchem #8068). Plates with a Z'-factor <0.5 were disregarded or repeated. The screen was performed in triplicate for FLC1 and FLC5. After 72 hours of incubation Cell titer glo reagent (Promega) was added using Thermo Multidrop Combi (Thermo Scientific) according to the manufacturer’s instructions, and the plates were read for luminescence (BioTek Synergy Neo). The normalized percent cell survival was calculated by
Validation screen
Compounds from the primary screen which produced a survival of <60% at 1μM after 72h were selected for further validation. To these we added inhibitors of protein kinase A inhibitors or pathways up-regulated in FLC, and drugs either suggested or in use for FLC. Compounds were plated in 384 well plates from 10nM-10μM using an 11-point, 2 step serial dilution. Each plate was prepared in triplicate. Cells were plated into the 384 well plates containing the compounds at 2000 cells/well in Kubota’s medium (PheonixSongs biological) supplemented with 2% p/s and incubated for 72 hours. Each plate contained negative control wells treated with DMSO and positive control wells treated with 20μM chaetocin (Selleckchem #8068). Plates with a Z'-factor <0.5 were disregarded or repeated. Compounds were similarly screened against primary human hepatocytes (PHH) grown in humanized mice and isolated as previously described(19). PHH were plated in 384 well plates containing compounds at 5000 cells/well in W10 media (William’s E media supplemented with ITS (BD), peniclllin/streptomycin/gentamycin). PHH were similarly incubated and assayed for luminescence as described for FLC cells above. To measure the role of ROS in the toxicity of compounds, FLC cells were incubated with compounds for 72 hours in the presence or absence of 10mM N-acetyl cysteine. Cells were assayed for luminescence as above. Dose–response curves were generated using GraphPad Prism 8 (GraphPad Software, San Diego, California USA, www.graphpad.com).
Synergy assay
For synergy assays, compounds were plated in a 2-compound combination matrix. Compound A (visusertib, everolimus, trametinib, topotecan, ENMD-2076, Idarubicin, AT13148, or Uprosertib) was plated in a 10-point, 2-fold dilution from 10μM-20nM. Compound B (A-1331852, AZD5991, navitoclax or venetoclax) was plated in an 8-point, 3-fold dilution from 10μM-5nM. Each plate contained negative control wells treated with DMSO, and positive control wells treated with 20 μM chaetocin. Each plate was prepared in triplicate. Cells were plated into the 384 well plates containing the compounds at 2000 cells/well in Kubota’s stemcell growth medium (Phoenix Songs biologicals) supplemented with 2% p/s, incubated for 72 hours, and assayed for luminescence as described above. Dose-response matrix data was analyzed using the web based application SynergyFinder(42). Synergy results are expressed as ZIP, HSA, and Bliss scores(43,44).
Compounds
Compounds for the primary screen were from the High throughput and Spectroscopy Center at The Rockefeller University. These are compound screening libraries that include preclinical drugs, clinically approved drugs, and annotated bioactive compounds with pharmaceutically relevant structures. They were part of the following commercially available libraries: Selleck (Houston, TX), Tocris (Bristol, UK), Pharmakon, Microsource (Gaylorsville,CT), LOPAC, Sigma (Carlsbad, CA), Prestwick Chemical (San Diego, CA), HTSRC Clinical Collection, Rockefeller University (New York, NY); NIH Clinical Collection (provided through the National Institutes of Health Molecular Libraries Roadmap Initiative and distributed by Compound Focus, Inc). Compound stocks were stored in polypropylene 384 well plates at a final concentration of 5 mM in DMSO at −20C. AZD5991 and A1331852 and Navitoclax were purchased from Chemietek. Sorafenib was purchased from MedChemExpress. CR-1-31B was a gift from the Wendel lab. All other compounds were purchased from Selleckchem in powder form and diluted in DMSO to make a stock solution. These were then further diluted in media to reach appropriate final concentrations.
Data from all screening studies are archived and analyzed using the CDD Vault from Collaborative Drug Discovery (Burlingame, CA. http://www.collaborativedrug.com). MarvinSketch (ChemAxon, version 18.28.0) was used for drawing and naming chemical structures. Instant Jchem for Excel (ChemAxon, Budapest Hungary, version 18.28.0) was also used for creating structure spreadsheets and tables. Graphpad PRISM (version 8) was used for curve fitting and data analysis.
Drug formulation for in vivo experiments
Napabucasin, panobinostat, and quisinostat were purchased from Selleckchem and A1331852 and Navitoclax from Chemietek, diluted in DMSO to make stock solutions, aliquoted and stored at −80C. Napabucasin was formulated by heating to 50C for 10 minutes and then sequentially adding 45% PEG300 (Sigma), 5% Tween80 (Sigma) and 45% sterile water, with vortexing after adding each component (Selleckchem). Panobinostat was formulated by sequentially adding 48% PEG300 (Sigma), 2% Tween80 (Sigma) and 48% sterile water, with vortexing after adding each component (Selleckchem). Quisinostat was formulated in 10% hydroxypropyl-b-cyclodextrin (Sigma), 25 mg/ml mannitol (Sigma), in sterile water(45). A1331852 was formulated by sequentially adding 10% Ethanol (Fisher), 60% Phosal 50 PG (Lipoid), and 30% PEG400 (Sigma), and vortexing (MedChemExpress). Navitoclax was formulated in 10% ethanol (Fisher), 30% PEG400 (Sigma), and 60% Phosal 50 PG (Lipoid), with vortexing after adding each component (MedChemExpress). Sorafenib was formulated in 90% corn oil (Selleckchem) with vortexing (MedChemExpress). Working solutions were made fresh prior to administration.
In vivo drug testing
Female NSG mice age 5-8 weeks were used for in vivo studies. Pieces of PDX were implanted subcutaneously in the flank. Mice were followed bi-weekly. Treatment was initiated when tumors reached an average volume of 150mm3 as measured by ultrasound or caliper. Quisinostat 10mg/kg was administered by daily intra-peritoneal (IP) injection for 14 days. Napabucasin or panobinostat were given at 10mg/kg by daily IP injection 5 days on and 2 days off, for a total of 14 days. Navitoclax 50mg/kg or sorafenib 30mg/kg were given by daily oral gavage 5 days on and 2 days off, for a total of 14 days. CR-1-31B was given at 0.1mg/kg by intra-peritoneal injection twice per week for a total of 24 days. A1331852 was given at 25mg/kg by daily intra-peritoneal (IP) injection for 2 days.
Mice received a daily subcutaneous injection of saline and received Nutra-Gel diet bacon flavor (Bio Serv) or DietGel 76A, chocolate flavor (ClearH2O). Mice were monitored daily for health and weight. Tumor size was measured at day 0, 7 and 14 by ultrasound (Vevo 3100, Visual Sonics) or by electronic calipers at day 0 and then twice per week until the end of treatment. Tumor volume was calculated by measuring (length*width*height)/2 for ultrasound or (length*width2/2) for caliper. Mice were sacrificed by ketamine + xylazine injection and cervical dislocation. Tumors were photographed and divided, and pieces were fixed in formalin, frozen as OCT blocks or flash frozen for further analysis. Graphpad PRISM (version 8) was used for data analysis.
Immune Histochemistry Staining
The immunohistochemical analysis by TUNEL,Ki67,PHH3 and pSTAT3 antibodies were performed at Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using Discovery XT processor (Ventana Medical Systems,Roche-AZ).
TUNEL - Slides were manually deparaffinized in xylene, re-hydrated in series of alcohol dilutions (100%, 95% and 70%) and tap water, placed in Discovery XT autostainer, treated with Protease 3 (Ventana Medical Systems,cat#760-2020) for 8 minutes, incubated with Avidin-biotin blocking reagent (Ventana Medical Systems) for 12 minutes and then incubated with TdT (Roche, cat#03333566001, 1000U/ml) and biotin-dUTP (Roche, cat#11093070910, 4.5nmol/ml) labeling mix for 2 hours. Detection was performed using a DAB detection kit (Ventana Medical Systems) according to manufacturer instruction. Slides were counterstained with hematoxylin and coverslipped with Permount (Fisher Scientific).
Ki67hu - Tissue sections were blocked with Background Buster (Innovex,cat#NB306-50) for 30 minutes. Sections were incubated wtih primary mouse monoclonal Ki67 antibody (Dako cat#M7240) at 0.5ug/mL concentration, incubation was done for 6 hours, followed by 60 minutes incubation with a biotinylated mouse secondary antibody (Vector Labs, MOM Kit BMK-2202) at 5.75ug/mL. Detection was performed with secondary antibody blocker, Blocker D, Streptavidin-HRP D (Ventana Medical Systems), according to manufacturer instruction. Slides were counterstained with hematoxylin and coverslipped with Permount (Fisher Scientific).
PHH3 - Tissue sections were blocked with Background Buster (Innovex,cat#NB306-50) for 30 minutes, followed by a 4h incubation with rabbit polyclonal anti-PHH3 antibody (Millipore cat#06-570; 1 ug/ml). Secondary antibody incubation was performed for 32’ with biotinylated goat anti-rabbit IgG (Vector labs, cat#:PK6101; 5.75ug/mL). Blocker D, Streptavidin- HRP and a DAB detection kit (Ventana Medical Systems) were used according to manufacturer instructions.
pStat3 - Tissue sections were blocked with Background Buster (Innovex,cat#NB306-50) for 30’, followed by a 5h incubation with rabbit polyclonal anti-pStat3 antibody (Cell Signaling cat#9135; 0.2 ug/ml). Secondary antibody incubation was performed for 60’ with biotinylated goat anti-rabbit IgG (Vector labs, cat#:PK6101; 5.75ug/mL). Blocker D, Streptavidin- HRP and a DAB detection kit (Ventana Medical Systems) were used according to manufacturer instructions.
Imaging
H&E and IHC slides of original patient tumor tissue and derived FLC PDX tumor tissue were imaged on an Olympus IX83 microscope using a 10x and 60x objective. Images were acquired using an Olympus DP26 camera and cellSense software (Olympus).
RNA isolation, generation of cDNA and PCR
Total RNA from original patient tumors, adjacent non-tumor liver (when available), and derived FLC PDXs was extracted using miRNeasy Mini Kit (Qiagen). RNA concentrations levels were measured using a Nanodrop 2000c (Thermofisher) and purity was assessed by the 260/280 ratio. The miScript II RT Kit (Qiagen) was used to convert total RNA into cDNA according to the manufacturer’s instructions. PCR for the DNAJB1-PRKACA fusion transcript was run for 40 cycles (Forward primer - TTCAAGGAGATCGCTGAGGC; Reverse primer - CTGTGTTCTGAGCGGGACTT, expected amplicon 148kB). Electrophoresis of the PCR product was performed at 100V for 45 minutes using a 2% agarose gel and visualized with SYBR Safe (Life Technologies). Images were acquired with a Gel Doc EZ imager (Bio-Rad).
RNA Sequencing and bioinformatics
RNA concentrations and 260/280 ratios were measured using a Nanodrop 2000c (Thermofisher) and RNA quality was assessed by RIN values (Agilent BioAnalyzer and TapeStation). RNA-seq libraries were prepared using a TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold ribosomal RNA depletion (Illumina). Ribosomal RNA-depleted libraries were sequenced on an Illumina HiSeq X-Ten (BGI Sequencing Services). Quality assessment and trimming were performed using FastQC v0.11.7 and BBDuk (included in BBMap v38.22). Reads were mapped to the human reference genome hg38 supplemented with the EMSEMBL GRCh38.92 gene annotations using STAR v2.6.1(46). Analysis of differential gene expression was conducted in R version 3.5 using DESeq2, excluding rRNA and mt-rRNA genes as well as the immune and stromal signature genes(47). PCA and Heatmaps were generated using the set of genes consistently dysregulated in FLC primary tumors vs adjacent non-tumor liver tissue (FDR 5%) across 3 independent batches of samples (283 up-regulated and 226 down-regulated genes, see Supplementary Material Table 2).
Protein Isolation and Immunoblotting
Total protein from the original patient tumors, adjacent non-tumor liver (when available), and derived FLC PDX was extracted using RIPA buffer (Sigma) supplemented with protease and phosphatase inhibitors (Complete EDTA-free, and PhosSTOP, Roche, Indianapolis, IN). Samples were briefly sonicated on ice and ultracentrifuged for 1 hour at 100,000xg. Supernatants were collected and protein concentrations were measured by a modified Lowry assay (DC protein assay, Bio-Rad). 10ug of protein per sample were diluted with 4x Nupage LDX sample buffer (Life technologies, Carlsbad, CA) containing 10% β-mercaptoethanol. Samples were heated at 100°C for 5 minutes, and then loaded on 4–12% Bis-Tris gels (Nupage, Invitrogen, Carlsbad, CA) and run in MOPS buffer for 50 minutes at 200V. Transfer was performed using the iBlot (Life Technologies, Carlsbad, CA). Membranes were blocked for 1h in 5% milk (Carnation powdered milk) in Tris-buffered saline with Tween (TBST), washed in TBST, and then probed with primary antibodies against PRKACA (Santa Cruz Biotech, PKAα cat Antibody (C-20): sc-903, 1:200) in 5% milk and incubated overnight shaking at 4°C. After washing in TBST, membranes were incubated with horseradish peroxidase-conjugated appropriate secondary antibodies (Sigma, A0545 goat anti rabbit, A9917, goat anti mouse, 1:100,000) in 5% milk in TBST for 1 hour. Membranes were washed in TBST and then incubated with Amersham ECL prime western blotting detection reagent (GE Healthcare), exposed to film in a dark room and then developed.
Supplementary Material
Acknowledgments.
We are grateful for the support from the NIH for grants P50CA210964 (SMS,MST), U54CA243126 (SMS), R01CA205967-04S2 (SMS and AL), the Starr Foundation grant I11-0050 (to SMS and AL), K12CA184746 (MVO), P30CA008748 (MPL, MO, NJC, BAF, MJH, JAS), R01DK085713 (CMR), R01AA027327 (YPJ), F32DK107164 (EM) and fellowships KA4688/1-1 from the Deutsche Forschungsgemeinschaft (MK) and the Belgian American Educational Foundation (KV). Further support was provided by the Robertson Therapeutic Development Fund (SMS, EM and CMR) and by the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust (GL, SMS and EM). This publication was supported in part by the National Center for Advancing Translational Sciences, NIH, through Rockefeller University, Grant #UL1TR001866. We are grateful to the staff and leadership of The Rockefeller University Hospital.
These projects were initiated by critical seed funds from private foundations and we are grateful for the support by the Sohn Foundation (SMS), the Rally Foundation (SMS, JAS, WJH), The Bear Necessities (SMS, JAS, WJH), The Truth365 (SMS, JAS, WJH) and Cannonball Kids’ Cancer (MVO).
Most importantly, we would like to thank all of the fibrolamellar patients, and their caregivers, who have contributed in too many ways to enumerate.
Abbreviations:
- ATR
Ataxia telangiectasia and Rad3-related protein
- AURKA
Aurora kinase A
- AURKB
Aurora kinase B
- Bcl-xL
B-cell lymphoma-extra large encoded by the BCL2-like 1 gene
- Bcl2
B-cell lymphoma 2
- ITS
Insulin, (human) Transferrin, Selenium
- BID
BH3 Interacting Domain Death Agonist
- BIM
Bcl-2-like protein 11
- CA12
Carbonic anhydrase 12
- CDc7
Cell division cycle 7-related protein kinase
- CDK
Cyclin depedent protein kinase
- Cyp19A1
Also know as aromatase or estrogen synthase
- DAB
3,3'-Diaminobenzidine
- DMSO
Dimethyl sulfoxide
- DNA-PK
DNA protein kinase
- EGFR
Epidermal growth factor receptor
- eIF4F
Eukaryotic initiation factor 4F
- ErbB2
erythroblastic oncogene B, also known as Her-2 protooncogene Neu and as epidermal growth factor receptor-2
- FLC
fibrolamellar hepatocellular carcinoma
- H&E
hematoxylin and eosin
- HSA
Highest Single Agent
- HCC
hepatocellular carcinoma
- HDAC
Histone deacetylase
- HSP70
Heat Shock Protein 70
- HTSRC
High Throughput and Spectroscopy Resource Center
- IP
Intraperitoneal
- IHC
Immunohistochemistry
- mTor
mammalian target of rapamycin
- NAC
N-acetyl-cysteine
- NOXA
Phorbol-12-myristate-13-acetate-induced protein 1
- NSG
Nod-Scid-IL2 receptor Gamma null mice
- OCT
Optimal Cutting Temperature compound
- PAK 3
p21-activated kinase 3
- PBS
Phosphate Buffered Saline
- PCR
polymerase chain reaction
- PDX
Patient derived xenografts
- PEG300
Polyethylene glycol molecular weight of 300 Da
- PHH
Primary Human Hepatocytes
- PHH3
Phosphorylated Histone H3
- PHITT
Pediatric Hepatic Malignancy International Therapeutic Trial
- PI3K
Phosphoinositide 3-kinase
- PKA
Protein kinase A
- POR
cytochrome P450 oxidoreductase
- PRKACA
Protein Kinase cAMP-Activated Catalytic Subunit Alpha
- p/s
penicillin and streptomycin
- pSTAT3
phosphorylated STAT3 (Signal Transducer And Activator Of Transcription 3)
- PUMA
p53 upregulated modulator of apoptosis
- RIN
RNA Integrity Number
- RIPA buffer
Radioimmunoprecipitation assay buffer
- ROS
Reactive Oxygen Species
- RPMI
Roswell Park Memorial Institute
- SPF
specific-pathogen free
- SQ
Subcutaneous
- STAT3
Signal Transducer And Activator Of Transcription 3
- TBST
Tris-buffered saline with Tween
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
- ZIP
Zero Interaction Potency
Footnotes
Competing Interests statement
AL discloses consulting and sponsored research agreements with AbbVie, Novartis, and Astra-Zeneca. He is an equity-holding founder of Flash Therapeutics, and is on the SAB of Dialectic Therapeutics and Zentalis Therapeutics.
GL discloses a consulting agreement with Pluristem Therapeutics.
References
- 1.Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014;343(6174):1010–4 doi 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malouf GG, Tahara T, Paradis V, Fabre M, Guettier C, Yamazaki J, et al. Methylome sequencing for fibrolamellar hepatocellular carcinoma depicts distinctive features. Epigenetics 2015;10(9):872–81 doi 10.1080/15592294.2015.1076955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham RP, Jin L, Knutson DL, Kloft-Nelson SM, Greipp PT, Waldburger N, et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2015;28(6):822–9 doi 10.1038/modpathol.2015.4. [DOI] [PubMed] [Google Scholar]
- 4.Kastenhuber ER, Lalazar G, Houlihan SL, Tschaharganeh DF, Baslan T, Chen CC, et al. DNAJB1-PRKACA fusion kinase interacts with beta-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America 2017;114(50):13076–84 doi 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelholm LH, Riaz A, Serra D, Dagnaes-Hansen F, Johansen JV, Santoni-Rugiu E, et al. CRISPR/Cas9 Engineering of Adult Mouse Liver Demonstrates That the Dnajb1-Prkaca Gene Fusion is Sufficient to Induce Tumors Resembling Fibrolamellar Hepatocellular Carcinoma. Gastroenterology 2017;153(6):1662–73 doi 10.1053/j.gastro.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz AG. International classification of diseases for oncology : ICD-O. Geneva: World Health Organization; 2013. viii, 242 pages p. [Google Scholar]
- 7.Simon EP, Freije CA, Farber BA, Lalazar G, Darcy DG, Honeyman JN, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America 2015;112(44):E5916–25 doi 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farber BA, Lalazar G, Simon EP, Hammond WJ, Requena D, Bhanot UK, et al. Non coding RNA analysis in fibrolamellar hepatocellular carcinoma. Oncotarget 2018;9(12):10211–27 doi 10.18632/oncotarget.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darcy DG, Chiaroni-Clarke R, Murphy JM, Honeyman JN, Bhanot U, LaQuaglia MP, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget 2015;6(2):755–70 doi 10.18632/oncotarget.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ang CS, Kelley RK, Choti MA, Cosgrove DP, Chou JF, Klimstra D, et al. Clinicopathologic Characteristics and Survival Outcomes of Patients With Fibrolamellar Carcinoma: Data From the Fibrolamellar Carcinoma Consortium. Gastrointestinal Cancer Research : GCR 2013;6(1):3–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita S, Vauthey JN, Kaseb AO, Aloia TA, Conrad C, Hassan MM, et al. Prognosis of Fibrolamellar Carcinoma Compared to Non-cirrhotic Conventional Hepatocellular Carcinoma. J Gastrointest Surg 2016;20(10):1725–31 doi 10.1007/s11605-016-3216-x. [DOI] [PubMed] [Google Scholar]
- 12.Weeda VB, Murawski M, McCabe AJ, Maibach R, Brugieres L, Roebuck D, et al. Fibrolamellar variant of hepatocellular carcinoma does not have a better survival than conventional hepatocellular carcinoma--results and treatment recommendations from the Childhood Liver Tumour Strategy Group (SIOPEL) experience. Eur J Cancer 2013;49(12):2698–704 doi 10.1016/j.ejca.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Lim II, Farber BA, LaQuaglia MP. Advances in fibrolamellar hepatocellular carcinoma: a review. Eur J Pediatr Surg 2014;24(6):461–6 doi 10.1055/s-0034-1396420. [DOI] [PubMed] [Google Scholar]
- 14.Eggert T, McGlynn KA, Duffy A, Manns MP, Greten TF, Altekruse SF. Fibrolamellar hepatocellular carcinoma in the USA, 2000-2010: A detailed report on frequency, treatment and outcome based on the Surveillance, Epidemiology, and End Results database. United European Gastroenterol J 2013;1(5):351–7 doi 10.1177/2050640613501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology 2004;39(3):798–803 doi 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- 16.Kakar S, Burgart LJ, Batts KP, Garcia J, Jain D, Ferrell LD. Clinicopathologic features and survival in fibrolamellar carcinoma: comparison with conventional hepatocellular carcinoma with and without cirrhosis. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2005;18(11):1417–23 doi 10.1038/modpathol.3800449. [DOI] [PubMed] [Google Scholar]
- 17.Katzenstein HM, Krailo MD, Malogolowkin MH, Ortega JA, Qu W, Douglass EC, et al. Fibrolamellar hepatocellular carcinoma in children and adolescents. Cancer 2003;97(8):2006–12 doi 10.1002/cncr.11292. [DOI] [PubMed] [Google Scholar]
- 18.Njei BK VR; Ditah I Prognosis of Patients with Fibrolamellar Hepatocellular Carcinoma Versus Conventional Hepatocellular Carcinoma: A systematic Review and Meta-analysis. Gastrointest Cancer Res 2014;7(2):49–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Michailidis E, Vercauteren K, Mancio-Silva L, Andrus L, Jahan C, Ricardo-Lax I, et al. Expansion, in vivo-ex vivo cycling, and genetic manipulation of primary human hepatocytes. Proceedings of the National Academy of Sciences of the United States of America 2020;117(3):1678–88 doi 10.1073/pnas.1919035117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Rogoff HA, Keates S, Gao Y, Murikipudi S, Mikule K, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proceedings of the National Academy of Sciences of the United States of America 2015;112(6):1839–44 doi 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo D, Shogren KL, Zang J, Jewison DE, Waletzki BE, Miller AL, 2nd, et al. Inhibition of STAT3 blocks protein synthesis and tumor metastasis in osteosarcoma cells. J Exp Clin Cancer Res 2018;37(1):244 doi 10.1186/s13046-018-0914-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froeling FEM, Swamynathan MM, Deschenes A, Chio IIC, Brosnan E, Yao MA, et al. Bioactivation of Napabucasin Triggers Reactive Oxygen Species-Mediated Cancer Cell Death. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25(23):7162–74 doi 10.1158/1078-0432.CCR-19-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirshner JR, He S, Balasubramanyam V, Kepros J, Yang CY, Zhang M, et al. Elesclomol induces cancer cell apoptosis through oxidative stress. Mol Cancer Ther 2008;7(8):2319–27 doi 10.1158/1535-7163.MCT-08-0298. [DOI] [PubMed] [Google Scholar]
- 24.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004;23(18):3189–99 doi 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 25.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014;513(7516):65–70 doi 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stipa F, Yoon SS, Liau KH, Fong Y, Jarnagin WR, D'Angelica M, et al. Outcome of patients with fibrolamellar hepatocellular carcinoma. Cancer 2006;106(6):1331–8 doi papers2://publication/doi/ 10.1002/cncr.21703. [DOI] [PubMed] [Google Scholar]
- 27.Turnham RE, Smith FD, Kenerson HL, Omar MH, Golkowski M, Garcia I, et al. An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma. eLife 2019;8 doi 10.7554/eLife.44187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-Response Analysis Using R. PloS one 2015;10(12):e0146021 doi 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham RP, Lackner C, Terracciano L, Gonzalez-Cantu Y, Maleszewski JJ, Greipp PT, et al. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1-PRKACA fusion. Hepatology 2018;68(4):1441–7 doi 10.1002/hep.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Pinkerton AB, Hulea L, Zhang T, Davies MA, Grotegut S, et al. SBI-0640756 Attenuates the Growth of Clinically Unresponsive Melanomas by Disrupting the eIF4F Translation Initiation Complex. Cancer research 2015;75(24):5211–8 doi 10.1158/0008-5472.CAN-15-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grzmil M, Hemmings BA. Translation regulation as a therapeutic target in cancer. Cancer research 2012;72(16):3891–900 doi 10.1158/0008-5472.CAN-12-0026. [DOI] [PubMed] [Google Scholar]
- 32.Chio IIC, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, et al. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell 2016;166(4):963–76 doi 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow HY, Jubb AM, Koch JN, Jaffer ZM, Stepanova D, Campbell DA, et al. p21-Activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer research 2012;72(22):5966–75 doi 10.1158/0008-5472.CAN-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow HY, Dong B, Duron SG, Campbell DA, Ong CC, Hoeflich KP, et al. Group I Paks as therapeutic targets in NF2-deficient meningioma. Oncotarget 2015;6(4):1981–94 doi 10.18632/oncotarget.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasim L, Chopra M. Synergistic anticancer effect of panobinostat and topoisomerase inhibitors through ROS generation and intrinsic apoptotic pathway induction in cervical cancer cells. Cell Oncol (Dordr) 2018;41(2):201–12 doi 10.1007/s13402-017-0366-0. [DOI] [PubMed] [Google Scholar]
- 36.Wilson TR, Johnston PG, Longley DB. Anti-apoptotic mechanisms of drug resistance in cancer. Curr Cancer Drug Targets 2009;9(3):307–19 doi 10.2174/156800909788166547. [DOI] [PubMed] [Google Scholar]
- 37.Tolcher AW, LoRusso P, Arzt J, Busman TA, Lian G, Rudersdorf NS, et al. Safety, efficacy, and pharmacokinetics of navitoclax (ABT-263) in combination with irinotecan: results of an open-label, phase 1 study. Cancer Chemother Pharmacol 2015;76(5):1041–9 doi 10.1007/s00280-015-2882-9. [DOI] [PubMed] [Google Scholar]
- 38.Felsher DW. Cancer revoked: oncogenes as therapeutic targets. Nature reviews Cancer 2003;3(5):375–80 doi 10.1038/nrc1070. [DOI] [PubMed] [Google Scholar]
- 39.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science 2002;297(5578):63–4 doi 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 40.de Jong YP, Dorner M, Mommersteeg MC, Xiao JW, Balazs AB, Robbins JB, et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Science translational medicine 2014;6(254):254ra129 doi 10.1126/scitranslmed.3009512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, et al. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nature biotechnology 2007;25(8):903–10 doi 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He L, Kulesskiy E, Saarela J, Turunen L, Wennerberg K, Aittokallio T, et al. Methods for High-throughput Drug Combination Screening and Synergy Scoring. Methods in molecular biology 2018;1711:351–98 doi 10.1007/978-1-4939-7493-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics 2017;33(15):2413–5 doi 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 2020;48(W1):W488–W93 doi 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carol H, Gorlick R, Kolb EA, Morton CL, Manesh DM, Keir ST, et al. Initial testing (stage 1) of the histone deacetylase inhibitor, quisinostat (JNJ-26481585), by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer 2014;61(2):245–52 doi 10.1002/pbc.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi papers2://publication/doi/ 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications 2013;4:2612 doi 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.