Abstract
Most mitochondrial proteins are synthesized as precursors that carry N-terminal presequences. After they are imported into mitochondria, these targeting signals are cleaved off by the mitochondrial processing peptidase (MPP). Using the mitochondrial tandem protein Arg5,6 as a model substrate, we demonstrate that MPP has an additional role in preprotein maturation, beyond the removal of presequences. Arg5,6 is synthesized as a polyprotein precursor that is imported into mitochondria and subsequently separated into two distinct enzymes. This internal processing is performed by MPP, which cleaves the Arg5,6 precursor at its N-terminus and at an internal site. The peculiar organization of Arg5,6 is conserved across fungi and reflects the polycistronic arginine operon in prokaryotes. MPP cleavage sites are also present in other mitochondrial fusion proteins from fungi, plants, and animals. Hence, besides its role as a “ticket canceller” for removal of presequences, MPP exhibits a second conserved activity as an internal processing peptidase for complex mitochondrial precursor proteins.
INTRODUCTION
All cellular processes are carried out by proteins, linear chains of amino acids that fold into three-dimensional structures. While the amino acid sequence of a protein is primarily determined by its DNA sequence, many proteins are additionally modified by proteolytic cleavage after their synthesis. Processing of polypeptides at their N-termini is pervasive in both prokaryotic and eukaryotic proteomes. For instance, amino-terminal methionine is removed from many polypeptides when they emerge from ribosomes and the new N-terminus is a crucial determinant of protein stability (Bradshaw et al., 1998; Frottin et al., 2006; Varshavsky, 2011). The majority of intracellular protein-targeting signals are located at the N-terminus and cleaved off upon arrival at the correct cellular destination. For example, around two-thirds of nuclear-encoded mitochondrial proteins are synthesized as precursors that carry an N-terminal mitochondrial targeting sequence (MTS) or presequence that directs them to mitochondrial surface receptors (von Heijne, 1986; Becker et al., 2019; Bykov et al., 2020). These preproteins are imported into mitochondria via the translocases of the outer (TOM complex) and inner mitochondrial membrane (TIM23 complex; Chacinska et al., 2009; Pfanner et al., 2019). Their MTS is cleaved by the mitochondrial processing peptidase (MPP). In some cases, the new N-terminus of the polypeptide generated thereby is further shortened by cleavage of single amino acids or short peptides by the proteases Icp55 or Oct1 before the matured protein folds into its native structure (Naamati et al., 2009; Vögtle et al., 2009, 2011; Calvo et al., 2017; Poveda-Huertes et al., 2017). Correct processing of precursor proteins in the matrix is crucial to maintain mitochondrial function and proteostasis. Dysfunctional preprotein maturation in mitochondria results in proteome instability, aggregation of incorrectly processed precursors, and proteotoxic stress (Poveda-Huertes et al., 2020), and was observed in models of Alzheimer’s disease (Mossmann, Vögtle, et al., 2014).
Internal cleavage of polypeptides is less frequent than proteolytic removal of N-terminal prepeptides, but equally relevant to cellular physiology and organismal health. Examples of medically relevant polypeptides that undergo internal proteolytic processing during their biosynthesis include insulin and the amyloid precursor protein (APP; Steiner and Oyer, 1967; Müller et al., 2017). In most cases, peptides are removed to yield the mature form of a single protein. However, some genes encode fusion proteins that are synthesized as single precursors and are then separated into distinct, functional proteins by proteolytic cleavage. A prominent example is ubiquitin, which is encoded as a fusion with subunits of the ribosome or as head-to-tail repeats of several ubiquitin monomers that are rapidly separated by deubiquitinating proteases (Ozkaynak et al., 1984; Finley et al., 1989; Gemayel et al., 2017). Polyproteins also frequently occur in viral genomes, including HIV and SARS-CoV-2, where the cleavage products often form protein complexes (Yost and Marcotrigiano, 2013; Krichel et al., 2020; Zhang et al., 2020). In eukaryotic genomes, such organization is rare, despite its obvious advantage of stoichiometric coexpression of functionally related proteins.
Here we report a notable exception: The ARG5,6 gene of Saccharomyces cerevisiae encodes both acetyl glutamate kinase (Arg6) and the acetyl glutamyl-phosphate reductase (Arg5), two enzymes that catalyze the second and third steps of arginine biosynthesis in the mitochondrial matrix (Minet et al., 1979). These two enzymes are synthesized as a single precursor protein that is posttranslationally cleaved into separate polypeptides. Arg6 and Arg5 then form a complex with the acetyl glutamate synthase Arg2 (Abadjieva et al., 2001; Pauwels et al., 2003). Here, we investigate the biogenesis of Arg5,6 in more detail and identify MPP as the protease that is responsible for processing of the precursor into two functional enzymes. We demonstrate that Arg6 and Arg5 can be imported into mitochondria separately, where they still form a functional enzyme. However, its organization as a composite precursor that is matured by MPP is highly conserved across fungi. These findings broaden our view of MPP as a processing peptidase with a more general role in mitochondrial preprotein maturation, reaching beyond its canonical function of removing mitochondrial targeting signals.
RESULTS
Arg5,6 is processed by mitochondrial processing peptidase in the mitochondrial matrix
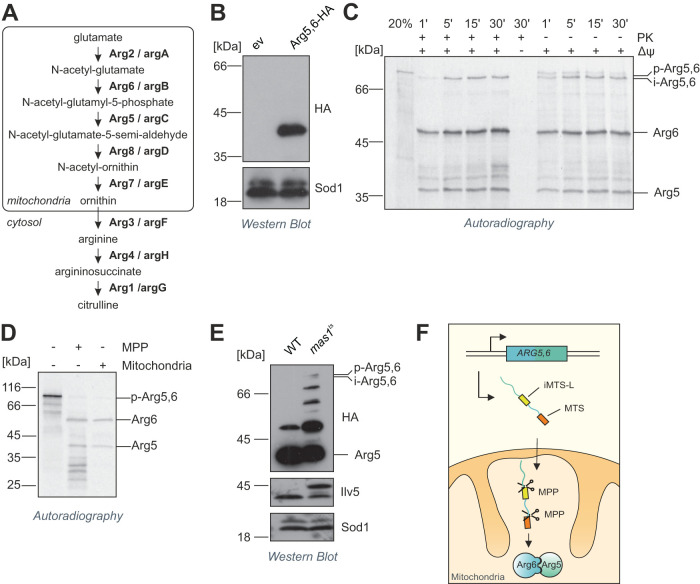
Acetylglutamate kinase (Arg6) and acetyl glutamyl phosphate reductase (Arg5) are located in the mitochondrial matrix, where they catalyze the second and third steps of the biosynthesis of arginine from glutamate (Figure 1A; Minet et al., 1979; Morgenstern, Stiller, Lübbert, Peikert, et al., 2017). The composite precursor protein that is synthesized from the ARG5,6 gene contains an MTS (Vögtle et al., 2009), which has been suggested to direct the single precursor into mitochondria, where it is subsequently cleaved into two separate polypeptides (Boonchird et al., 1991b). Indeed, when we expressed Arg5,6 with a C-terminal hemagglutinin (HA) tag, immunoblotting of cell lysates against the HA epitope revealed only a single band at around 40 kDa, much less than the expected mass of 90 kDa of the composite precursor protein (Figure 1B). Obviously, the precursor is rapidly and efficiently cleaved in vivo, which yields a C-terminal Arg5 fragment.
FIGURE 1:
Arg5,6 is a composite mitochondrial precursor that is processed twice by MPP in the mitochondrial matrix. (A) Schematic representation of arginine biosynthesis in S. cerevisiae. Shown in bold are the enzymes that catalyze the respective steps as well as their E. coli orthologues. (B) When Arg5,6 was C-terminally HA-tagged, immunoblotting revealed a single band at 40 kDa, indicating proteolytic cleavage of the 90-kDa precursor protein. (C) Radiolabeled Arg5,6 precursor was incubated with isolated mitochondria for the indicated times and analyzed by SDS–PAGE and autoradiography. Nonimported material was digested with proteinase K (left half). Twenty percent of the total lysate used per import lane was loaded for control. The membrane potential (Δψ) was dissipated with VAO (valinomycin, antimycin, oligomycin). p, precursor, i, intermediate. (D) His-tagged MPP was expressed and purified from E. coli. Radiolabeled Arg5,6 precursor was incubated with isolated mitochondria for 15 min or purified MPP for 90 min. The processing of Arg5,6 was analyzed by SDS–PAGE, Western blotting, and autoradiography. (E) C-terminally HA-tagged Arg5,6 was expressed in wild-type and mas1ts cells. Cultures were grown at 37°C overnight to impair MPP activity. The processing of Arg5,6 was analyzed by SDS–PAGE and immunoblotting. Ilv5 is a presequence-containing matrix protein. Sod1 is localized to the cytosol and the intermembrane space and carries no presequence. (F) Arg5,6 is imported into the mitochondrial matrix and cleaved twice by MPP: once at the N-terminus to remove the presequence, and once internally at an iMTS-L to separate Arg6 and Arg5.
To analyze the mechanistic basis of this unusual biogenesis, we sought to reconstitute the biogenesis and processing of Arg5,6 in an in vitro system. To this end, we synthesized a radiolabeled Arg5,6 precursor in reticulocyte lysate and incubated it with isolated yeast mitochondria. We observed that the precursor of around 90 kDa was efficiently processed to a slightly smaller intermediate form—indicating the removal of the N-terminal MTS—and further into several smaller fragments, most prominently two polypeptides of around 40 and 50 kDa, which correspond to the C-terminal Arg5 and the N-terminal Arg6, respectively (Figure 1C). The intermediate and the mature polypeptides, but not the Arg5,6 precursor, were protected from digestion with externally added proteinase K. This shows that these polypeptides were translocated across the mitochondrial outer membrane. Both import and processing were dependent on the mitochondrial inner membrane potential (Δψ), as expected for an MTS-containing matrix protein (Garg and Gould, 2016; Schendzielorz et al., 2017; Sato et al., 2019). We conclude that Arg5,6 is imported into the mitochondrial matrix via the presequence pathway and cleaved into separate polypeptides inside mitochondria.
How is the Arg5,6 precursor processed to give rise to the Arg6 and Arg5 enzymes? A number of proteases in the mitochondrial matrix are described (Quiros et al., 2015; Veling et al., 2017). However, most of them are either implicated in degradation and turnover of proteins (such as the Lon protease Pim1) or known to remove short peptides or single amino acids from the N-termini of mitochondrial precursor proteins (such as Oct1 or Icp55), but not for internal cleavage of proteins into two mature parts (Suzuki et al., 1994; Wagner et al., 1994; Vögtle et al., 2009, 2011; Poveda-Huertes et al., 2017). For MPP, the peptidase responsible for the cleavage of the N-terminal MTS, a notable exception was recently reported: the composite precursor protein Atp25 contains internal MPP cleavage sites at which the protein is split into two functionally unrelated polypeptides (Woellhaf et al., 2016). To test whether MPP can cleave the Arg5,6 precursor, we purified MPP from Escherichia coli expressing His-tagged Mas1 and Mas2 (the two subunits of MPP). Incubation of radiolabeled Arg5,6 precursor protein with MPP resulted in the formation of smaller fragments whose size perfectly matched that of those that were generated after import into isolated mitochondria (Figure 1D). Proper processing was blocked when EDTA was added to the reaction (Supplemental Figure 1A), which inhibited the metalloprotease MPP by chelating divalent cations (Luciano et al., 1998).
To further confirm that MPP separates Arg6 and Arg5, we used a temperature-sensitive mas1ts yeast strain in which the catalytic subunit of MPP, Mas1, can be specifically inactivated by increasing the temperature (Yaffe et al., 1985; Witte et al., 1988; Burkhart et al., 2015; Poveda-Huertes et al., 2020). We expressed HA-tagged Arg5,6 in this strain and shifted the culture to 37°C for 12 h. In the mas1ts mutant, but not in the wild type, we observed the accumulation of both the unprocessed Arg5,6 precursor and of the intermediate form in which the presequence was removed, but no internal cleavage was detected. The levels of mature Arg5 were reduced accordingly in the mas1ts strain (Figure 1E). Several bands running between the sizes of the precursor and the Arg5 fragment indicated either processing intermediates or, perhaps more likely, degradation products of the unprocessed Arg5,6 precursor. Note that the precursor form of the presequence-containing matrix protein Ilv5 accumulated in the mas1ts mutant, indicating that MPP activity was indeed impaired under these conditions.
To exclude secondary effects of the long-lasting MPP inactivation, we isolated mitochondria from mas1ts cells grown under permissive conditions and incubated the mitochondria at 37°C for 20 min before we added radiolabeled precursor proteins. This pretreatment partially inactivated MPP and resulted in increased accumulation of unprocessed Arg5,6 precursor and intermediate forms after import into mas1ts mitochondria, thus mirroring the results obtained in vivo (Supplemental Figure 1B).
In summary, we conclude that Arg5,6 is imported into the mitochondrial matrix and processed twice by MPP. A first cleavage removes the N-terminal MTS and a second cleavage separates the Arg6 and Arg5 enzymes (Figure 1F).
The Mitochondrial processing peptidase cleaves Arg5,6 in an internal mitochodrial targeting sequence–like sequence
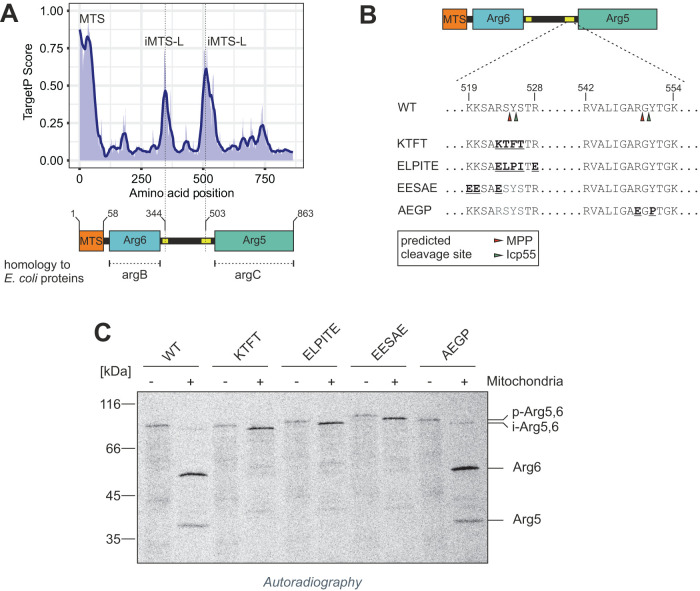
In Atp25, the internal MPP cleavage sites coincide with a sequence stretch that structurally mimics the properties of an N-terminal MTS. We subjected the amino acid sequence of Arg5,6 to an in silico prediction of such internal MTS-like sequences (iMTS-Ls), using an adapted version of the TargetP algorithm (Boos, Mühlhaus, et al., 2018). In fact, in addition to the N-terminal MTS, two internal regions with high TargetP score were detected around amino acid positions 344 and 503 (Figure 2A). A cleavage at the latter would produce polypeptides whose molecular masses matched those of the Arg6 and Arg5 proteins. To determine the site of processing more precisely, we used both TargetP (Emanuelsson et al., 2007; Almagro Armenteros, Salvatore, et al., 2019) and MitoFates (Fukasawa et al., 2015) to predict potential MPP cleavage sites within the second iMTS-L of Arg5,6. We found two sites that matched the classical MPP consensus motif R-2 with an arginine residue at the -2 position and an aromatic residue at the +1 position of the putative cleavage site (Taylor et al., 2001; Vögtle et al., 2009): RSY at 523–525 and RGY at 549–551. We generated mutated versions of Arg5,6 where either the RSY or the RGY motif was changed by point mutations (Figure 2B). We synthesized these mutant proteins in vitro and incubated the radiolabeled precursors with isolated mitochondria. All variants were imported into mitochondria, as they were protected from externally added protease after the import reaction and exhibited a small size shift, indicative of the removal of their N-terminal presequence. However, none of the RSY mutants was further processed, and the intermediate form of Arg5,6 accumulated, while the RGY mutant was efficiently cleaved into separate polypeptides whose sizes matched those of wild-type Arg6 and Arg5 (Figure 2C). Thus, MPP processes Arg5,6 in an internal MTS-like sequence and the cleavage site matches the classical MPP consensus motif at positions 523–525. Interestingly, MitoFates predicted additional processing of the newly formed N-terminus of Arg5 by one amino acid through the mitochondrial aminopeptidase Icp55 (Figure 2B). Obviously, the internal cleavage of Arg5,6 adheres to the same rules and is carried out by the same machinery as that of N-terminal processing of mitochondrial presequences.
FIGURE 2:
The internal MPP cleavage site in Arg5,6 is flanked by an iMTS-L and adheres to the R-2 motif. (A) Arg5,6 was subjected to TargetP profiling. High values indicate regions within the protein that structurally resemble mitochondrial presequences. (B) Putative cleavage sites for mitochondrial processing proteases in the second iMTS-L region of Arg5,6 were predicted with MitoFates. Mutations in the predicted recognition motif were introduced by site-directed mutagenesis. (C) Arg5,6 variants with and without the mutations displayed in B were synthesized as radiolabeled precursors and incubated with isolated mitochondria, followed by removal of nonimported material by PK treatment. Processing of the imported proteins was analyzed by SDS–PAGE, Western blotting, and autoradiography.
Arg5 and Arg6 can be imported into mitochondria separately and complement the arg5,6 deletion mutant
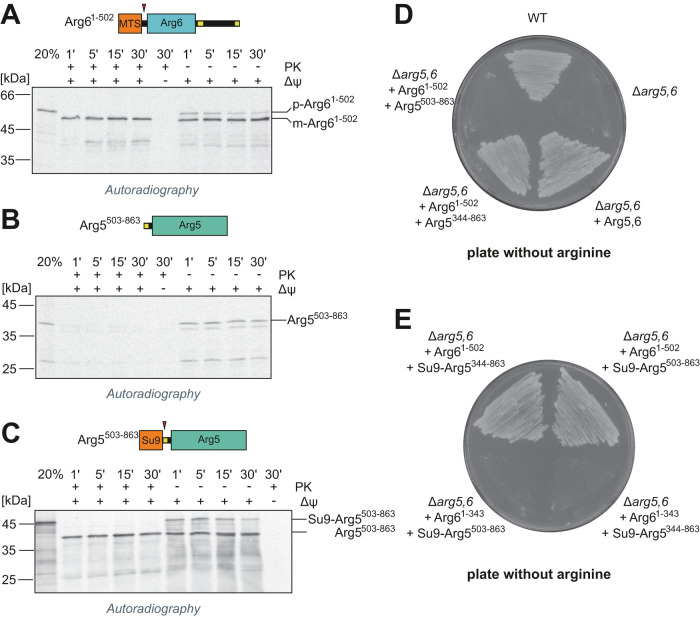
The unusual biogenesis of Arg5,6 prompted us to ask whether Arg6 and Arg5 can also be imported separately. Therefore, we created truncated versions of the ARG5,6 gene which contained only the N-terminal Arg6 with its MTS (Arg61–502) or only the C-terminal Arg5, starting at the first (Arg5344–863) or the second iMTS-L (Arg5503–863). For the latter two, we also generated variants that additionally carried the well-characterized presequence of ATP synthase subunit 9 from Neurospora crassa (Su9-Arg5344–863 and Su9-Arg5503–863).
Radiolabeled proteins were synthesized in vitro and incubated with isolated mitochondria to test their import competence. As expected, Arg61–502 was efficiently imported and its MTS was cleaved (Figure 3A). The shorter Arg5 variant (Arg5503–863) did not reach a protease-protected compartment and thus was not imported into amitochondria (Figure 3B). However, N-terminal fusion of the Su9 presequence completely restored import of Arg5503–863 (Figure 3C). Hence, import of Arg5 and Arg6 into mitochondria is in principle possible for separated polypeptides also, at least in the in vitro assay used here.
FIGURE 3:
Arg5 and Arg6 can be imported separately in vitro and in vivo. (A-C) Radiolabeled precursor proteins of Arg61–502, Arg5503–862, and Su9-Arg5503–862 were incubated with isolated mitochondria for the indicated times and analyzed by SDS–PAGE and autoradiography. Nonimported material was digested with proteinase K (left half). Twenty percent of the total lysate used per import lane is loaded for control. The membrane potential (Δψ) was depleted with VAO. Red arrowheads indicate processing sites. p, precursor, m, mature. (D, E) Yeast cells that lack endogenous Arg5,6 (Δarg5,6) were transformed with plasmids for expression of the indicated Arg5,6 variants and streaked out on plates containing minimal growth medium without arginine.
We next tested whether Arg6 and Arg5 can be imported separately in vivo and function in arginine biosynthesis. The deletion of the ARG5,6 gene renders yeast cells auxotrophic for arginine. We expressed either the full-length Arg5,6 precursor or combinations of separate Arg6 and Arg5 variants in a Δarg5,6 deletion mutant. If Arg6 and Arg5 make their way into mitochondria and acquire a functional conformation, arginine prototrophy should be restored. When we streaked out these cells on plates with minimal growth medium lacking arginine, we observed growth for the wild type and the Δarg5,6 mutant complemented with full-length Arg5,6, but not for Δarg5,6 carrying only an empty plasmid, as expected (Figure 3D).
The mutant expressing both Arg61–502 and the shorter Arg5503–863 variant was not able to grow without arginine (Figure 3D). However, when the presequence of Su9 was fused to the short Arg5503–863, cells regained arginine prototrophy (Figure 3E). All strains grew on plates containing arginine, showing that the Arg5503–863 protein has no toxic gain-of-function effect when residing in the cytosol (Supplemental Figure 1, C and D). Growing the strains in liquid medium lacking arginine confirmed the results obtained on plates and additionally demonstrated that the growth rate of the strain expressing Arg61–502 and Su9-Arg5503–863 is comparable to that of the wild type (Supplemental Figure 1E), at least under the overexpression conditions used here. This indicates that separate expression of Arg6 and Arg5 is possible, in principle, without adverse effects on cellular fitness.
A truncated version of Arg6 (Arg61–343) did not complement the deletion mutant with any of the Arg5 variants, even though this variant contains the entire region of homology to the bacterial acetyl glutamate kinase argB from E. coli (Figure 3E). Apparently, amino acids 344 to 502 are not merely a spacer or linker between Arg5 and Arg6, but relevant to the activity, the structure, and/or the assembly of the Arg5–Arg6 complex.
Taken together, Arg6 and Arg5 can be imported separately into mitochondria in vitro and in vivo and are functional in arginine biosynthesis, as long as mitochondrial localization is conferred by appropriate N-terminal targeting signals.
Internal processing of Arg5,6 by mitochondrial processing peptidase requires an N-terminal presequence
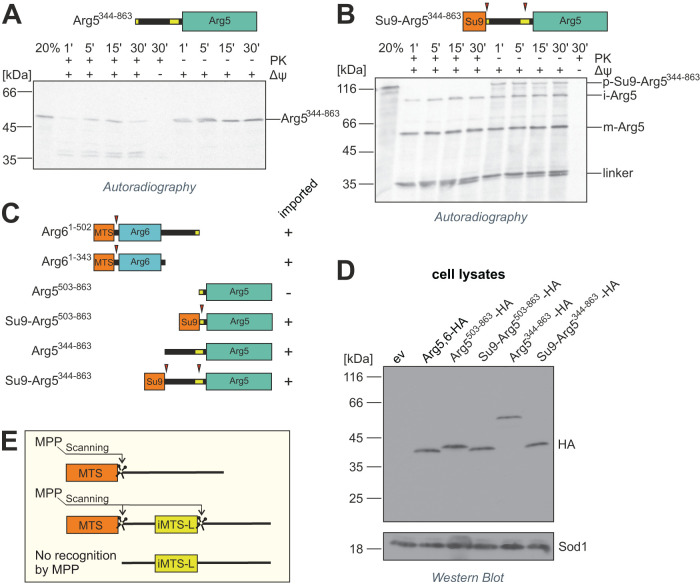
Surprisingly, cells expressing Arg61–502 and the longer Arg5344–863 could grow without arginine, indicating that this longer Arg5 variant can be imported even without fusion of an additional presequence (Figure 3D). Indeed, Arg5344–863 was imported into isolated mitochondria, albeit with low efficiency (Figure 4A). Addition of proteinase K to this import reaction resulted in the appearance of lower-running bands that were absent without protease, indicating that a portion of the precursor translocated only partially across the outer membrane. This is in agreement with earlier observations that iMTS-Ls can confer targeting to mitochondria when presented at the N-terminus, but are not necessarily able to drive complete translocation (Baker and Schatz, 1987; Backes, Hess, et al., 2018). As expected, fusion of the Su9 presequence to Arg5344–863 resulted in more efficient translocation (Figure 4, B and C).
FIGURE 4:
MPP requires a strong N-terminal MTS for internal processing of precursor proteins. (A, B) Radiolabeled precursor proteins of Arg5344–862 and Su9-Arg5344–862 were incubated with isolated mitochondria for the indicated times and analyzed by SDS–PAGE and autoradiography. Nonimported material is digested with proteinase K (left half). Twenty percent of the total lysate used per import lane is loaded for control. The membrane potential (Δψ) was depleted with VAO. Red arrowheads indicate processing sites. p, precursor, i, intermediate, m, mature. (C) Overview of truncated Arg5,6 variants and their import competence. Su9, presequence of N. crassa subunit 9. (D) Yeast cells expressing indicated variants of Arg5,6, all carrying a C-terminal HA tag, were lysed and protein extracts were analyzed by SDS–PAGE and immunoblotting directed against the HA epitope or Sod1 as a loading control. ev, empty vector. (E) MPP cleaves the Arg5,6 precursors at its internal processing site only if they carry a bona fide N-terminal presequence. Presumably, MPP recognizes its substrates primarily at their N-terminus and then scans the downstream polypeptide for internal cleavage sites.
Interestingly, even the fully imported Arg5344–863 was not processed inside mitochondria, while for Su9-Arg5344–863, prominent lower-running bands were observed. Their sizes fit those expected for the intermediate form after removal of the Su9 presequence (around 60 kDa), the completely matured Arg5 after cleavage at the second MPP site (around 40 kDa), and the cleaved “linker” between the Su9 presequence and Arg5 (around 25 kDa). This suggests that an internal MPP cleavage at an iMTS-L requires a bona fide N-terminal MTS.
To test whether this holds true also in vivo, we expressed HA-tagged Arg5,6 as well as Arg5 variants in yeast cells and analyzed cell lysates by immunoblotting against the HA epitope. In agreement with the in vitro import experiments, both Su9-Arg5344–863 and Su9-Arg5503–863 were processed and yielded a band at the same molecular weight as full-length Arg5,6. In contrast, Arg5344–863 and Arg5503–863 were exclusively present in their unprocessed forms (Figure 4D). Hence, even though Arg5344–863 is imported into mitochondria, its iMTS-L is not recognized by MPP. Obviously, internal MPP cleavage sites are dependent not only on the presence of a particular motif and its immediate context within the amino acid sequence, but also on more distant features of the polypeptide, such as the N-terminal presequence. In contrast to the in vivo and in organello situation, purified MPP was able to cleave radiolabeled Arg5344–863 in vitro, both with and without the Su9 presequence, demonstrating that the lack of internal cleavage is not an intrinsic problem of this peculiar precursor (Supplemental Figure 1F). In this minimal reconstituted in vitro system, MPP obviously exhibits a certain degree of promiscuity that is not observed inside mitochondria. These results indicate that in the physiological environment of the mitochondrial matrix, MPP recognizes its substrates primarily at their N-terminal and then scans the precursors for downstream internal processing sites (Figure 4E).
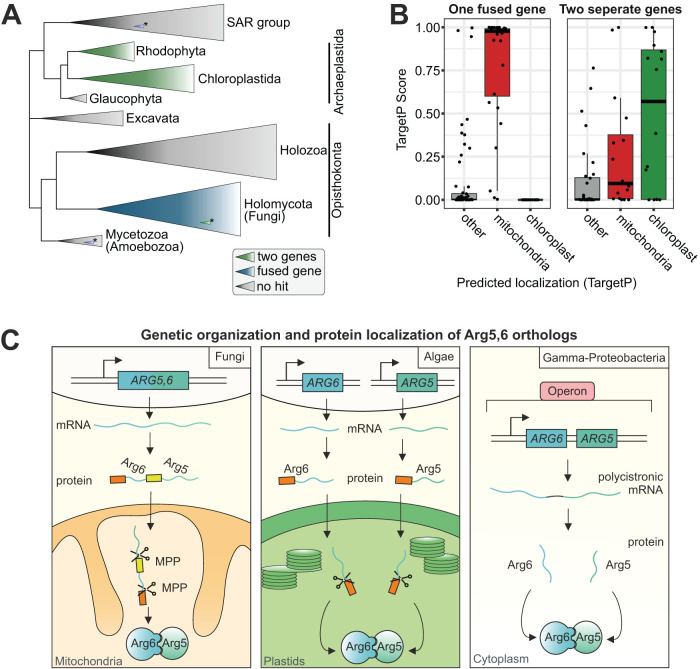
The ARG5,6 genes are fused in fungi, separated in algae, and encoded polycistronically in gamma-proteobacteria
Tandem organization of functionally related genes is rare in eukaryotes. In contrast, genomic colocalization of functionally related genes is pervasive in prokaryotes. The E. coli proteins argB and argC are homologous to Arg6 and Arg5, respectively, and are organized in an operon, i.e. they are transcribed polycistronically (Piette et al., 1982). We used the sequences of argB and argC of E. coli K-12 as query sequences to search a prokaryotic database of 5655 organisms (Supplemental Table 1A). Three thousand six hundred sixty-six species were identified that encoded exactly one copy each of argB and argC (Supplemental Table 1B), mainly distributed among proteobacteria, firmicutes, actinobacteria, and cyanobacteria. Among the 3666 prokaryotes, 882 strains (848 of which were gamma-proteobacteria) encoded the two open reading frames in immediate proximity, and hence presumably expressed them on a polycistronic mRNA (Supplemental Table 1C).
Even though we cannot directly trace back the evolutionary history of Arg5,6 of S. cerevisiae, these findings support the idea that this eukaryotic two-gene cluster evolved from syntenic genes in the eukaryotic ancestor. Nevertheless, Arg5,6 represents an exceptional case. Only very few proteins with a similar fusion structure are known in baker’s yeast, and for none of them do the separate proteins function in the same pathway (Ozkaynak et al., 1987; Finley et al., 1989; Woellhaf et al., 2016). However, the Arg5,6 homolog arg-6 in Neurospora crassa also encodes both acetyl glutamate kinase and acetyl glutamyl phosphate reductase, which are separated by proteolytic cleavage inside mitochondria (Gessert et al., 1994; Parra-Gessert et al., 1998). We therefore asked whether the composite structure of Arg5,6 is generally conserved across eukaryote species. To this end, we searched a dataset of 150 eukaryotes for homologues of the yeast composite Arg5,6 precursor and of its posttranslationally processed proteins Arg5 and Arg6 (Supplemental Table 1D). While 82 genomes returned no hits, we identified specific patterns within the diamond BLASTp hits of 36 eukaryotes (Figure 5A; Supplemental Figure 2; Supplemental Table 1E). In 28 species, all three query sequences hit the same subject, indicating the presence of an ARG5,6 gene fusion homolog. With the sole exception of Phytophthora sojae, the fusion homologues were exclusively found among fungi. In eight species, Arg5 and Arg6 each hit a single independent sequence, while Arg5,6 hit both of those sequences, which indicates that separate genes encode for Arg5 and Arg6. Again with a single exception (Fusarium graminearum PH-1), all of these organisms were algae, comprising both green and red algae. In 32 species we identified mixed blast patterns (Supplemental Table 1F), which were not further investigated. We also predicted the intracellular localization of all proteins via TargetP. Strikingly, mitochondrial localization was assigned to almost all fusion proteins of fungi, whereas most separate proteins in algae were predicted to be imported into chloroplasts (Figure 5B).
FIGURE 5:
Tandem organization and mitochondrial localization of Arg5,6 is conserved in fungi, whereas algae synthesize two separate proteins that localize to their plastids. (A) A database of 150 eukaryotes was searched for homologues of full-length Arg5,6 and the separate Arg5 and Arg6 proteins of S. cerevisiae. Clades in which Arg5,6 homologues were encoded as a single fusion protein are colored in blue; clades in which two separate Arg6 and Arg5 genes were found are colored in green. Gray, no Arg5,6 homologues were found in these clades. (B) In species that encode Arg5,6 as fusion protein, TargetP predicts the protein to be localized to mitochondria. In species with separate genes for Arg5 and Arg6, localization is predicted to be plastidal. (C) In fungi, Arg5,6 is a fusion protein that is matured by MPP into two separate enzymes in the mitochondrial matrix. In contrast, algae encode two separate proteins that localize to their plastids. Gamma-proteobacteria encode Arg5 and Arg6 polycistronically.
In summary, in fungi, acetyl glutamate kinase and acetyl glutamyl-phosphate reductase are generally encoded as a fusion protein, which is imported into mitochondria and processed twice by MPP to remove its presequence and gives rise to two functional enzymes. In contrast, algae encode two separate proteins that are individually imported into chloroplasts. Gamma-proteobacteria express the genes from one polycistronic RNA (Figure 5C).
Internal precursor processing by MPP is conserved among eukaryotes
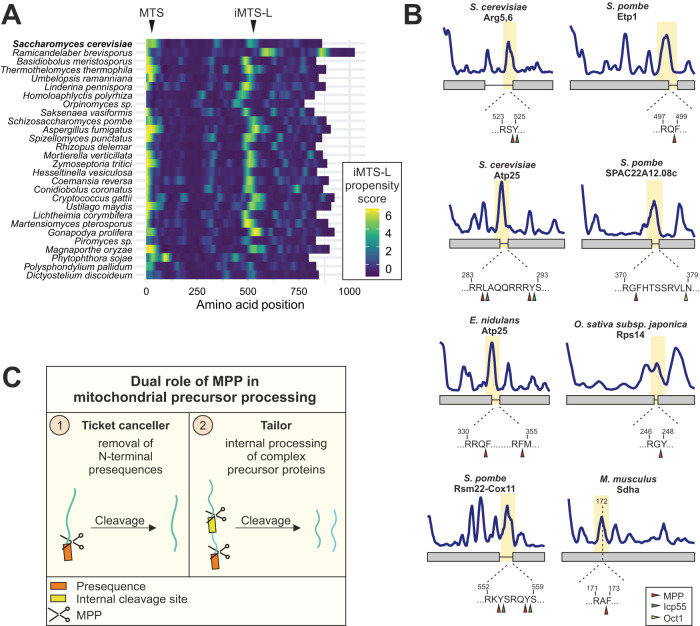
Because mitochondrial localization correlates well with the tandem structure of Arg5,6 homologues, we wondered whether the intramitochondrial processing into two separate enzymes by MPP might also be conserved in these species. To address this, we calculated the iMTS-L profiles of all fusion homologues of Arg5,6. In fact, we observed a common pattern across nearly all species: Besides the N-terminal MTS, our algorithm detected one conserved iMTS-L around amino acid position 500 (Figure 6A). Hence, all these fusion proteins harbor a potential MPP cleavage site precisely at the junction between the Arg5 and Arg6 parts.
FIGURE 6:
MPP not only functions as a presequence peptidase, but also has a conserved processing activity at internal cleavage sites of composite precursor proteins. (A) The iMTS-L at which MPP cleaves the Arg5,6 precursor in S. cerevisiae is conserved among species that encode Arg5,6 as fusion protein. Shown are iMTS-L propensity profiles along the sequence of Arg5,6 fusion protein homologues. (B) Previously described mitochondrial polyproteins from different species harbor an iMTS-L at the position of the junction between the fused polypeptides. Arrowheads indicate potential cleavage sites for MPP (red) and the downstream processing peptidases Icp55 (green) or Oct1 (yellow) as predicted by MitoFates. Sdha is no polyprotein, but an alternative N-terminus (dashed line) was recently identified, presumably the result of a posttranslational cleavage (Calvo et al., 2017). (C) In addition to its canonical role in presequence removal from mitochondrial precursor proteins, MPP has a second conserved function. It recognizes internal cleavage sites and processes complex precursor proteins into separate polypeptides.
This remarkable conservation of protein structure and processing inspired us to ask whether MPP could be responsible for internal cleavage of other mitochondrial proteins. Apart from Arg5,6 and its homologues, some other mitochondrial proteins with composite structure have been described in different species. These include proteins from different fungi (Atp25 from S. cerevisiae and Emericella nidulans and Etp1, Rsm22-Cox11, and the uncharacterized SPAC22A12.08c from Schizosaccharomyces pombe) and plants (RPS14 from Oryza sativa japonica; Oshima et al., 2005; Khalimonchuk et al., 2006; Woellhaf et al., 2016). We subjected these mitochondrial fusion proteins to our iMTS-L profiling analysis and to a cleavage site prediction and indeed found both prominent iMTS-Ls and MPP recognition motifs at each of their junction sites (Figure 6B).
DISCUSSION
Organization as fusion protein is an elegant solution to conferring mitochondrial targeting of two enzymes that reside in the same compartment and even act in consecutive steps of a biochemical pathway. It is still remarkable that this organization of Arg5,6 was retained during evolution even in distantly related organisms, indicating that there exists a strong constraint that maintained this organization for more than a billion years of evolution. To our knowledge, this is the only example of a fusion of two functionally related proteins whose organization is so widely conserved across eukaryote species. Eukaryotic genomes typically strongly disfavor even “milder” variants of physical coupling of genes, such as an operonlike organization, which is pervasively present in prokaryotes but basically absent in most eukaryotes. Interestingly, events of horizontal gene transfer from bacteria to eukaryotes were also accompanied by progressive loss of the polycistronic organization of the genes (Kominek et al., 2019). What might be the reason that the peculiar tandem structure of Arg5,6 survived several million years of evolution?
Because Arg6 and Arg5 form a complex, it is conceivable that the tandem structure of the precursor might facilitate their assembly. Many cytosolic protein complexes assemble cotranslationally, and this early-onset interaction between the partner subunits is crucial for function and, more generally, for maintenance of proteostasis (Shiber et al., 2018; Schwarz and Beck, 2019). Cotranslational assembly of nuclear-encoded mitochondrial proteins is hindered by the additional translocation step across two mitochondrial membranes. Therefore, coupling the assembly with the import of proteins into the mitochondrial matrix might represent the closest approximation to cotranslational assembly that is physically possible.
Besides its canonical role as a “ticket canceller” that clips targeting signals, the MPP obviously also possesses a “tailor” activity for internal processing of several precursor proteins (Figure 6C). This property is conserved across the fungi, plant, and potentially also animal kingdoms. Internal MPP cleavage requires a proximal recognition motif, which appears to be an iMTS-L, but remarkably also a strong N-terminal presequence. This suggests that to access internal cleavage sites, MPP has to be loaded onto a precursor as soon as it emerges from the TIM23 channel. MPP might then scan the still unfolded polypeptide for cleavage sites. A protein with a strong presequence will efficiently recruit MPP, which enables subsequent internal cleavage at an iMTS-L before this is buried by protein folding. In analyses of the N-proteome from yeast, mouse, and human mitochondria, a surprising variability in the N-termini of many proteins was observed, sometimes more than 100 amino acids downstream of the annotated start (Vögtle et al., 2009; Vaca Jacome et al., 2015; Calvo et al., 2017). Some of these isoforms might be generated by “leaky” internal MPP cleavage. For instance, the mouse protein Sdha has an alternative N-terminus at position 172 that coincides with an iMTS-L and an MPP cleavage site (Figure 6B). It will be exciting to elucidate the biogenesis and physiological role of such protein isoforms in future research. Its unusual biogenesis might render Arg5,6 a valuable model substrate to explore the mechanisms that confer specificity of internal MPP cleavage.
MATERIALS AND METHODS
Yeast strains and plasmids
All yeast strains used in this study were based on the WT strains BY4742 (Winston et al., 1995) or YPH499 (Sikorski and Hieter, 1989) and are listed in Supplemental Table 2. The mas1ts strain was kindly provided by Nora Vögtle (Poveda-Huertes et al., 2020). Unless indicated differently, strains were grown on synthetic medium (0.17% yeast nitrogen base and 0.5% (NH4)2SO4) containing 2% glucose.
The Arg5,6-coding region or a fragment of it was amplified by PCR and cloned into pGEM4 (Promega, Madison, WI) using the EcoRI and BamHI restriction sites. For construction of the Arg5,6-HA, Arg5344–863-HA, and Arg5503–863-HA versions, the corresponding sequence without stop codon was cloned into the expression plasmid pYX142, which harbors a constitutive TPI promoter upstream and the sequence of a hemagglutinin (HA) tag downstream of the multiple cloning site. For expression of Arg61–343-HA and Arg61–502-HA, pYX122 vectors were used, which differ from pYX142 in the selectable marker. The sequence of the Su9 presequence was amplified from a Su9-DHFR plasmid and cloned into the EcoRI site of the pYX142 plasmid carrying the ARG5 inserts to yield the Su9-Arg5344–863-HA and Su9-Arg5503–863-HA variants.
Site-directed mutations of putative MPP cleavage sites were introduced into the ARG5,6 open reading frame by Gibson Assembly (Gibson et al., 2009) using NEBuilder HiFi DNA Assembly. The mutated versions were inserted into the EcoRI and BamHI sites of the pGEM4 plasmid.
Isolation of yeast mitochondria
Isolation of mitochondria was performed essentially as described by Saladi, Boos, et al. (2020). Yeast cells were grown in YPGal medium (1% yeast extract, 2% peptone, 2% galactose) to an OD600 of 0.7–1.3, harvested, washed with water, and resuspended in MP1 buffer (10 mM DTT, 100 mM Tris). Cells were incubated for 10 min at 30°C, pelleted (5 min at 4000 × g at RT), and washed with 1.2 M sorbitol. The cell was digested for 1 h in MP2 buffer (1.2M sorbitol, 20 mM KH2PO4, pH 7.4) supplemented with 3 mg zymolyase/g wet weight (Seikagaku Corporation) at 30°C (23°C for mitochondria isolated from the mas1ts strain and its corresponding wild-type control). The spheroplasts were harvested, resuspended in ice-cold homogenization buffer (0.6 M sorbitol, 1 mM EDTA pH 8, 1 mM phenylmethanesulfonyl fluoride, 10 mM Tris-HCl pH 7.4, 0.2% fatty acid-free BSA), and lysed by douncing 10 times in a cooled potter homogenizer.
The homogenate was centrifuged for 5 min at 3500 × g at 4°C to separate cell debris and nuclei from organelles. The mitochondrial fraction was isolated by centrifugation of the supernatant from the previous step for 12 min at 12,000 × g at 4°C. The crude mitochondrial pellet was gently resuspended in SH buffer (0.6M sorbitol, 20 mM HEPES/KOH pH 7.4), centrifuged for 5 min at 4000 × g at 4°C, recovered from the supernatant by centrifugation for 12 min at 12,000 × g at 4°C, and finally resuspended in SH buffer. The protein concentration of the mitochondrial suspension was determined by a Bradford assay and mitochondria were diluted to a final concentration of 10 mg/ml protein with ice-cold SH buffer, aliquoted, frozen in liquid nitrogen, and stored at –80°C.
Import of radiolabeled proteins into isolated mitochondria
Proteins were synthesized from pGEM4 plasmid templates and radiolabeled with 35S methionine in a cell-free translation system (TNT Quick Coupled Transcription and Translation kit, Promega).
Import reactions were essentially performed as described previously (Peleh et al., 2015) in the following import buffer: 500 mM sorbitol, 50 mM HEPES, 80 mM KCl, 10 mM magnesium acetate, and 2 mM KH2PO4, pH 7.4. Mitochondria were energized by addition of 2 mM ATP and 2 mM NADH before radiolabeled precursor proteins were added. To dissipate the membrane potential, a mixture of 1 μg/ml valinomycin, 8.8 μg/ml antimycin, and 17 μg/ml oligomycin was added to the mitochondria. Precursor proteins were incubated with mitochondria for different times at 25°C before nonimported protein was degraded by addition of 100 μg/ml proteinase K. For import assays into mas1ts mitochondria, mitochondria were incubated in import buffer for 20 min at 37°C to inactivate MPP before radiolabeled precursor proteins were added to the reaction. The import was also performed at 37°C.
Growth assays
Growth curves were performed automated in a 96-well plate in technical triplicates using the ELx808 absorbance microplate reader (BioTek). Precultures of 100 μl were inoculated at an OD600 of 0.1 in microtiter plates and sealed with an air-permeable membrane (Breathe-Easy; Sigma-Aldrich, St. Louis, MO). The growth curves started at OD600 0.1 with incubation at 30°C for 72 h under constant shaking. The OD600 was measured every 10 min.
Antibodies
The antibodies against Sod1 and Ilv5 was raised in rabbits using purified recombinant protein. The secondary antibody was ordered from Biorad (goat Anti-rabbit IgG (H+L)-HRP conjugate #172-1019). The horseradish peroxidase-coupled HA antibody was ordered from Roche (Anti-HA-peroxidase, high affinity [3F10], #12 013 819 001). Antibodies were diluted in 5% (wt/vol) nonfat dry milk-TBS (Roth T145.2) with the following dilutions: Anti-Sod1 1:1,000, Anti-Ilv5 1:5,000, Anti-HA 1:500, Anti-rabbit 1:10,000.
Mitochondrial processing peptidase purification
The E. coli strain expressing histidine-tagged MPP subunits Mas1 and Mas2 from an expression plasmid was a gift of Vincent Géli (Luciano et al., 1997). The cells were grown at 37°C to an OD600 of 1 and induced with 0.5 mM IPTG overnight at 30°C. The bacteria were harvested and resuspended in buffer A (250 mM NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol, 0.2% NP-40, lysozyme), incubated at room temperature for 15 min, and snap-frozen in liquid nitrogen. After thawing, DNAseI was added and the cells were sonified 20 times for 1 s at a 60% duty level with a Branson sonifier 250. The lysate was cleared by centrifugation, loaded on buffer A-equilibrated Ni-NTA Sepharose resin (Aminitra, Expedeon, San Diego, CA), and washed with buffer A without detergent. A second wash was performed with buffer A adjusted to 1 M NaCl. The enzyme was eluted in buffer C (250 mM NaCl, 300 mM imidazole, 5% glycerol) and stored at –80°C.
Mitochondrial processing peptidase in vitro cleavage assay
The in vitro cleavage reactions were performed in 150 mM NaCl, 10% glycerol, 100 μM MnCl2, and 50 mM Tris, pH 7.5, in the presence of 250 μg of purified MPP and 5% reticulocyte lysate containing the substrate. If not otherwise stated, the incubation time was 1.5 h at 30°C. To inhibit MPP, the same reaction was performed in the presence of 2.5 mM EDTA and without addition of MnCl2.
Data reporting
Unless indicated otherwise, all experiments were performed in three independent biological replicates. For Western blots, import assays, and growth experiments, representative results are shown.
Prediction of iMTS-Ls
iMTS-Ls were predicted essentially as described (Backes, Hess, et al., 2018; Boos, Mühlhaus, et al., 2018). Briefly, multiple truncated sequences were generated by sequentially removing amino acids, one by one, N-terminally from the protein of interest. These sequences were submitted to TargetP with appropriate choice of “plant” or “nonplant” organism and without cutoffs. The mTP scores obtained were plotted against the corresponding amino acid position. A Savitzky–Golay filtering step with a window size of 21 (the expected value of the length distribution of known MTSs) was used to smooth the iMTS-L profile. The iMTS-L profiles can also be calculated using the iMTS-L predictor service online tool iMLP (http://imlp.bio.uni-kl.de/).
Prediction of mitochondrial processing peptidase cleavage sites
To detect putative cleavage sites for MPP, sequences were N-terminally truncated so that they started at the amino acid with the highest iMTS-L score. They were then submitted to MitoFates with appropriate choice of “fungus,” “metazoan,” or “plant” as organism (Fukasawa et al., 2015).
Arg5,6 homologues, intracellular localization and genomic distance
Amino acid sequences of yeast Arg5,6 and both posttranslationally processed proteins Arg5 and Arg6 were retrieved from (Boonchird et al., 1991a) and used as query sequences for a Diamond BLASTp (Buchfink et al., 2015) against a dataset of 150 eukaryotic genomes (Supplemental Table 1D). Subject organisms that exhibited hits (at least 25% local identity and a maximum E-value of 1E-10) were grouped by the number and pattern of the identified homologues, corresponding to encoding Arg5,6 either as one gene or as two genes (Supplemental Table 1E), by a python script. The intracellular localization of all homologues was checked via TargetP 2.0 (Almagro Armenteros, Salvatore, et al., 2019). Results were plotted on a eukaryotic reference tree generated in a previous analysis (Brueckner and Martin, 2020). For the identification of prokaryotic homologues, argB and argC amino acid sequences of E. coli K-12 were used as queries to search our dataset of 5655 complete prokaryotic genomes (Refseq, Supplemental Table 1A) (O’Leary et al., 2016). Query sequences were identified as homologues to yeast Arg5 and Arg6 via diamond BLASTp. Only prokaryotic subject organisms exhibiting exactly one homolog to each of the two query sequences (at least 25% local identity and a maximum E-value of 1E-10) were further investigated, and the nucleotide distance between the subject genes was calculated (Supplemental Table 1C). Sequence pairs with a maximum distance of 30 nucleotides were suspected to be encoded polycistronically. All genomic distances in nucleotides were derived from Refseq genome feature tables. Sequence pairs with overlapping start and end positions were given a distance of one nucleotide.
Supplementary Material
Acknowledgments
We thank Sabine Knaus, Andrea Trinkaus, Vera Nehr, Alexander Grevel, and Thomas Becker for assistance with the experiments, and Abdussalam Azem and Katja Hansen for helpful discussions and critical reading of the manuscript. We thank Nora Vögtle and Daniel Poveda-Huertes for providing the mas1 ts strain and for helpful discussions. This project was funded by grants from the Deutsche Forschungsgemeinschaft (DIP MitoBalance and 2803/10-1 to J.M.H. and 267205415–SFB 1208 to S.B.G.), the Volkswagen Stiftung (Life) to S.B.G., the Forschungsinitiative Rheinland-Pfalz BioComp to J.M.H. and F.B., the Minerva Stiftung to J.F., and the Joachim Herz Stiftung to F.B.
Abbreviations used:
- HA
hemagglutinin
- iMTS-L
internal MTS-like sequence
- MPP
mitochondrial processing peptidase
- MTS
mitochondrial targeting sequence
- PK
proteinase K
- TIM23
translocase of the inner mitochondrial membrane
- TOM
translocase of the outer mitochondrial membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-08-0524) on September 30, 2020.
REFERENCES
Boldface names denote co–first authors.
- Abadjieva A, Pauwels K, Hilven P, Crabeel M (2001). A new yeast metabolon involving at least the two first enzymes of arginine biosynthesis: acetylglutamate synthase activity requires complex formation with acetylglutamate kinase. J Biol Chem 276, 42869–42880. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, Nielsen H (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2, e201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S, Hess S, Boos F, Woellhaf MW, Godel S, Jung M, Mühlhaus T, Herrmann JM (2018). Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J Cell Biol 217, 1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Schatz G (1987). Sequences from a prokaryotic genome or the mouse dihydrofolate reductase gene can restore the import of a truncated precursor protein into yeast mitochondria. Proc Natl Acad Sci USA 84, 3117–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Song J, Pfanner N (2019). Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell Biol 29, 534–548. [DOI] [PubMed] [Google Scholar]
- Boonchird C, Messenguy F, Dubois E (1991a). Characterization of the yeast ARG5,6 gene: determination of the nucleotide sequence, analysis of the control region and of ARG5,6 transcript. Mol Gen Genet 226, 154–166. [DOI] [PubMed] [Google Scholar]
- Boonchird C, Messenguy F, Dubois E (1991b). Determination of amino acid sequences involved in the processing of the ARG5/ARG6 precursor in Saccharomyces cerevisiae. Eur J Biochem 199, 325–335. [DOI] [PubMed] [Google Scholar]
- Boos F, Mühlhaus T, Herrmann JM (2018). Detection of internal matrix targeting signal-like sequences (iMTS-Ls) in mitochondrial precursor proteins using the TargetP prediction tool. Bio-protocol 8, e2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw RA, Brickey WW, Walker KW (1998). N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem Sci 23, 263–267. [DOI] [PubMed] [Google Scholar]
- Brueckner J, Martin WF (2020). Bacterial genes outnumber archaeal genes in eukaryotic genomes. Genome Biol Evol 12, 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60. [DOI] [PubMed] [Google Scholar]
- Burkhart JM, Taskin AA, Zahedi RP, Vögtle FN (2015). Quantitative profiling for substrates of the mitochondrial presequence processing protease reveals a set of nonsubstrate proteins increased upon proteotoxic stress. J Proteome Res 14, 4550–4563. [DOI] [PubMed] [Google Scholar]
- Bykov YS, Rapaport D, Herrmann JM, Schuldiner M (2020). Cytosolic events in the biogenesis of mitochondrial proteins. Trends in Biochemical Sciences. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Julien O, Clauser KR, Shen H, Kamer KJ, Wells JA, Mootha VK (2017). Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol Cell Proteomics 16, 512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009). Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A (1989). The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401. [DOI] [PubMed] [Google Scholar]
- Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T (2006). The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics 5, 2336–2349. [DOI] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K (2015). MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SG, Gould SB (2016). The role of charge in protein targeting evolution. Trends Cell Biol 26, 894–905. [DOI] [PubMed] [Google Scholar]
- Gemayel R, Yang Y, Dzialo MC, Kominek J, Vowinckel J, Saels V, Van Huffel L, van der Zande E, Ralser M, Steensels J, et al. (2017). Variable repeats in the eukaryotic polyubiquitin gene ubi4 modulate proteostasis and stress survival. Nat Commun 8, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert SF, Kim JH, Nargang FE, Weiss RL (1994). A polyprotein precursor of two mitochondrial enzymes in Neurospora crassa. Gene structure and precursor processing. J Biol Chem 269, 8189–8203. [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Ott M, Funes S, Ostermann K, Rodel G, Herrmann JM (2006). Sequential processing of a mitochondrial tandem protein: insights into protein import in Schizosaccharomyces pombe. Eukaryot Cell 5, 997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominek J, Doering DT, Opulente DA, Shen XX, Zhou X, DeVirgilio J, Hulfachor AB, Groenewald M, McGee MA, Karlen SD, et al. (2019). Eukaryotic acquisition of a bacterial operon. Cell 176, 1356–1366 e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichel B, Falke S, Hilgenfeld R, Redecke L, Uetrecht C (2020). Processing of the SARS-CoV pp1a/ab nsp7–10 region. Biochem J 477, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P, Geoffroy S, Brandt A, Hernandez JF, Geli V (1997). Functional cooperation of the mitochondrial processing peptidase subunits. J Mol Biol 272, 213–225. [DOI] [PubMed] [Google Scholar]
- Luciano P, Tokatlidis K, Chambre I, Germanique JC, Geli V (1998). The mitochondrial processing peptidase behaves as a zinc-metallopeptidase. J Mol Biol 280, 193–199. [DOI] [PubMed] [Google Scholar]
- Minet M, Jauniaux JC, Thuriaux P, Grenson M, Wiame JM (1979). Organization and expression of a two-gene cluster in the arginine biosynthesis of Saccharomyces cerevisiae. Mol Gen Genet 168, 299–308. [DOI] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Hoss P, Feuerstein R, Gebert M, et al. (2017). Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 19, 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossmann D, Vögtle FN, Taskin AA, Teixeira PF, Ring J, Burkhart JM, Burger N, Pinho CM, Tadic J, Loreth D, et al. (2014). Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab 20, 662–669. [DOI] [PubMed] [Google Scholar]
- Müller UC, Deller T, Korte M (2017). Not just amyloid: physiological functions of the amyloid precursor protein family. Nat Rev Neurosci 18, 281–298. [DOI] [PubMed] [Google Scholar]
- Naamati A, Regev-Rudzki N, Galperin S, Lill R, Pines O (2009). Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J Biol Chem 284, 30200–30208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Yamasaki E, Ogishima T, Kadowaki K, Ito A, Kitada S (2005). Recognition and processing of a nuclear-encoded polyprotein precursor by mitochondrial processing peptidase. Biochem J 385, 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A (1987). The yeast ubiquitin genes: a family of natural gene fusions. EMBO J 6, 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Varshavsky A (1984). The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature 312, 663–666. [DOI] [PubMed] [Google Scholar]
- Parra-Gessert L, Koo K, Fajardo J, Weiss RL (1998). Processing and function of a polyprotein precursor of two mitochondrial proteins in neurospora crassa. J Biol Chem 273, 7972–7980. [DOI] [PubMed] [Google Scholar]
- Pauwels K, Abadjieva A, Hilven P, Stankiewicz A, Crabeel M (2003). The N-acetylglutamate synthase/N-acetylglutamate kinase metabolon of Saccharomyces cerevisiae allows co-ordinated feedback regulation of the first two steps in arginine biosynthesis. Eur J Biochem 270, 1014–1024. [DOI] [PubMed] [Google Scholar]
- Peleh V, Ramesh A, Herrmann JM (2015). Import of proteins into isolated yeast mitochondria. Methods Mol Biol 1270, 37–50. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Warscheid B, Wiedemann N (2019). Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol 20, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J, Cunin R, Boyen A, Charlier D, Crabeel M, Van Vliet F, Glansdorff N, Squires C, Squires CL (1982). The regulatory region of the divergent argECBH operon in Escherichia coli K-12. Nucleic Acids Res 10, 8031–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda-Huertes D, Matic S, Marada A, Habernig L, Licheva M, Myketin L, Gilsbach R, Tosal-Castano S, Papinski D, Mulica P, et al. (2020). An early mtUPR: redistribution of the nuclear transcription factor Rox1 to mitochondria protects against intramitochondrial proteotoxic aggregates. Mol Cell 77, 180–188.e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda-Huertes D, Mulica P, Vögtle FN (2017). The versatility of the mitochondrial presequence processing machinery: cleavage, quality control and turnover. Cell Tissue Res 367, 73–81. [DOI] [PubMed] [Google Scholar]
- Quiros PM, Langer T, Lopez-Otin C (2015). New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol 16, 345–359. [DOI] [PubMed] [Google Scholar]
- Saladi S, Boos F, Poglitsch M, Meyer H, Sommer F, Mühlhaus T, Schroda M, Schuldiner M, Madeo F, Herrmann JM (2020). The NADH Dehydrogenase Nde1 executes cell death after integrating signals from metabolism and proteostasis on the mitochondrial surface. Mol Cell 77, 189–202.e186. [DOI] [PubMed] [Google Scholar]
- Sato TK, Kawano S, Endo T (2019). Role of the membrane potential in mitochondrial protein unfolding and import. Sci Rep 9, 7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendzielorz AB, Schulz C, Lytovchenko O, Clancy A, Guiard B, Ieva R, van der Laan M, Rehling P (2017). Two distinct membrane potential-dependent steps drive mitochondrial matrix protein translocation. J Cell Biol 216, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz A, Beck M (2019). The benefits of cotranslational assembly: a structural perspective. Trends Cell Biol 29, 791–803. [DOI] [PubMed] [Google Scholar]
- Shiber A, Döring K, Friedrich U, Klann K, Merker D, Zedan M, Tippmann F, Kramer G, Bukau B (2018). Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Oyer PE (1967). The biosynthesis of insulin and a probable precursor of insulin by a human islet cell adenoma. Proc Natl Acad Sci USA 57, 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki CK, Suda K, Wang N, Schatz G (1994). Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264, 273–276. [DOI] [PubMed] [Google Scholar]
- Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J (2001). Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9, 615–625. [DOI] [PubMed] [Google Scholar]
- Vaca Jacome AS, Rabilloud T, Schaeffer-Reiss C, Rompais M, Ayoub D, Lane L, Bairoch A, Van Dorsselaer A, Carapito C (2015). N-terminome analysis of the human mitochondrial proteome. Proteomics 15, 2519–2524. [DOI] [PubMed] [Google Scholar]
- Varshavsky A (2011). The N-end rule pathway and regulation by proteolysis. Protein Sci 20, 1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veling MT, Reidenbach AG, Freiberger EC, Kwiecien NW, Hutchins PD, Drahnak MJ, Jochem A, Ulbrich A, Rush MJP, Russell JD, et al. (2017). Multi-omic mitoprotease profiling defines a role for Oct1p in coenzyme Q production. Mol Cell 68, 970–977.e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Prinz C, Kellermann J, Lottspeich F, Pfanner N, Meisinger C (2011). Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell 22, 2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C (2009). Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439. [DOI] [PubMed] [Google Scholar]
- von Heijne G (1986). Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 5, 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I, Arlt H, van Dyck L, Langer T, Neupert W (1994). Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J 13, 5135–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL (1995). Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Witte C, Jensen RE, Yaffe MP, Schatz G (1988). MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J 7, 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellhaf MW, Sommer F, Schroda M, Herrmann JM (2016). Proteomic profiling of the mitochondrial ribosome identifies Atp25 as a composite mitochondrial precursor protein. Mol Biol Cell 27, 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Ohta S, Schatz G (1985). A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J 4, 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost SA, Marcotrigiano J (2013). Viral precursor polyproteins: keys of regulation from replication to maturation. Curr Opin Virol 3, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 368, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.