Abstract
Hedgehog proteins govern crucial developmental steps in animals and drive certain human cancers. Before they can function as signaling molecules, Hedgehog precursor proteins must undergo N-terminal palmitoylation by Hedgehog acyltransferase (HHAT). Here we present cryo-electron microscopy (EM) structures of human HHAT in complex with its palmitoyl-CoA substrate and of a product complex with a palmitoylated Hedgehog peptide at 2.7-Å and 3.2-Å resolutions, respectively. The structures reveal how HHAT overcomes the challenges of bringing together substrates that have different physiochemical properties from opposite sides of the endoplasmic reticulum membrane within a membrane-embedded active site for catalysis. These principles are relevant to related enzymes that catalyze acylation of Wnt and the appetite-stimulating hormone ghrelin. The structural and mechanistic insights may advance the development of inhibitors for cancer.
One Sentence Summary:
Snapshots of the Hedgehog acyltransferase reaction reveal mechanisms of palmitoylation necessary for Hedgehog signaling.
Main text:
Hedgehog signaling is crucial for mammalian development. Downregulated in most adult tissues, aberrant Hedgehog activation has been implicated in pancreatic, breast, lung, and other malignancies (1). Mammals contain three Hedgehog proteins—Sonic, Desert, and Indian Hedgehog—each of which operates as a secreted morphogen for short- and long-range cell-to-cell signaling. Before they can initiate downstream signaling, Hedgehog precursor proteins must undergo a series of processing steps that culminate in the attachment of an acyl lipid by the enzyme Hedgehog acyltransferase (HHAT) (fig. S1A) (2, 3). The attached lipid is required for downstream signaling and is structurally recognized by the Hedgehog receptor Patched1 (4–6). Inhibition of HHAT is a new avenue for therapeutic intervention in Hedgehog-dependent cancers (7). HHAT is the only major component of the Hedgehog signaling pathway for which an atomic structure has not been reported.
HHAT is an endoplasmic reticulum (ER) membrane protein that catalyzes the transfer of a palmitoyl lipid from palmitoyl-CoA to the N-terminal cysteine of Hedgehog precursor proteins (2, 8). The enzyme displays exquisite selectivity for Hedgehog proteins, which are thought to be its only substrates (9). On the basis of low (< 15%) amino acid sequence similarity, HHAT can be classified as a member of the diverse family of membrane bound O-acyl transferase (MBOAT) enzymes (8, 10). Most MBOAT family members acylate lipids as part of lipid biosynthesis pathways (10–12). Three members—HHAT, GOAT, and Porcupine—are functionally distinct in that they acylate protein substrates. Like HHAT, the mature products of GOAT and Porcupine, the appetite-stimulating hormone ghrelin and the developmental and oncogenic regulator Wnt, respectively, operate as key secreted signaling molecules (13, 14). The members of this enzyme triumvirate are thought to obtain their hydrophobic acyl-CoA donor substrates from the cytosolic leaflet of the ER membrane and their hydrophilic protein substrates from the lumen of the ER. This localization suggests a remarkable mechanistic feat: HHAT, GOAT, and Porcupine obtain physiochemically diverse substrates from opposite sides of a membrane and bring them together within a membrane-embedded active site to achieve catalysis. Available three-dimensional structures of lipid-modifying MBOAT enzymes (ACAT1, DGAT1, and DltB) provide limited mechanistic insights into this process because those enzymes acylate small lipophilic molecules rather than hydrophilic proteins, and they have low sequence identity with protein-modifying enzymes (10–13 % with HHAT) (15–20). Additionally, no structure of any MBOAT enzyme in complex with an acceptor substrate or an acylated product has been determined.
To investigate its molecular mechanisms, we determined cryo-electron microscopy (EM) structures of human HHAT that represent snapshots of its reaction coordinate: a 2.7-Å resolution structure of a complex with palmitoyl-CoA, and a 3.2-Å resolution structure with a palmitoylated Hedgehog peptide product.
We expressed full length human HHAT in mammalian cells and purified it for functional and structural analyses (fig. S1B). Using an assay to measure the activity of the enzyme in microsomes and in purified form, we observe kinetic parameters similar to those previously reported (8), indicating that the sample used for cryo-EM analysis has full activity (fig. S2).
Size-exclusion chromatography suggested that HHAT is monomeric (fig. S1C). At merely 57 kDa, HHAT was too small for high-resolution single particle cryo-EM studies on its own. Consequently, we developed monoclonal antibodies that recognize conformational epitopes on human HHAT to use as fiducial markers. We identified two antibodies that were able to bind to the enzyme simultaneously and used these for cryo-EM analysis of HHAT in complex with its palmitoyl-CoA substrate. Single particle cryo-EM analysis of HHAT in complex with Fab fragments of these antibodies (denoted 1C06 and 3H02) and the palmitoyl-CoA substrate yielded a structure at 2.7-Å resolution (Fig. 1A, fig. S3). Density for the HHAT polypeptide (amino acids 1–491) is well resolved in the map (fig. S4A). The resulting atomic model has good stereochemistry and agreement with the map (fig. S4D, table S1). Density is well-resolved for the variable domains of the Fab fragments, which contact HHAT, but less well-ordered for the constant domains. The antibodies bind on opposite sides of the membrane. Neither antibody contacts the palmitoyl-CoA substrate (Fig. 1A, B).
Fig. 1. Overall structure of HHAT.
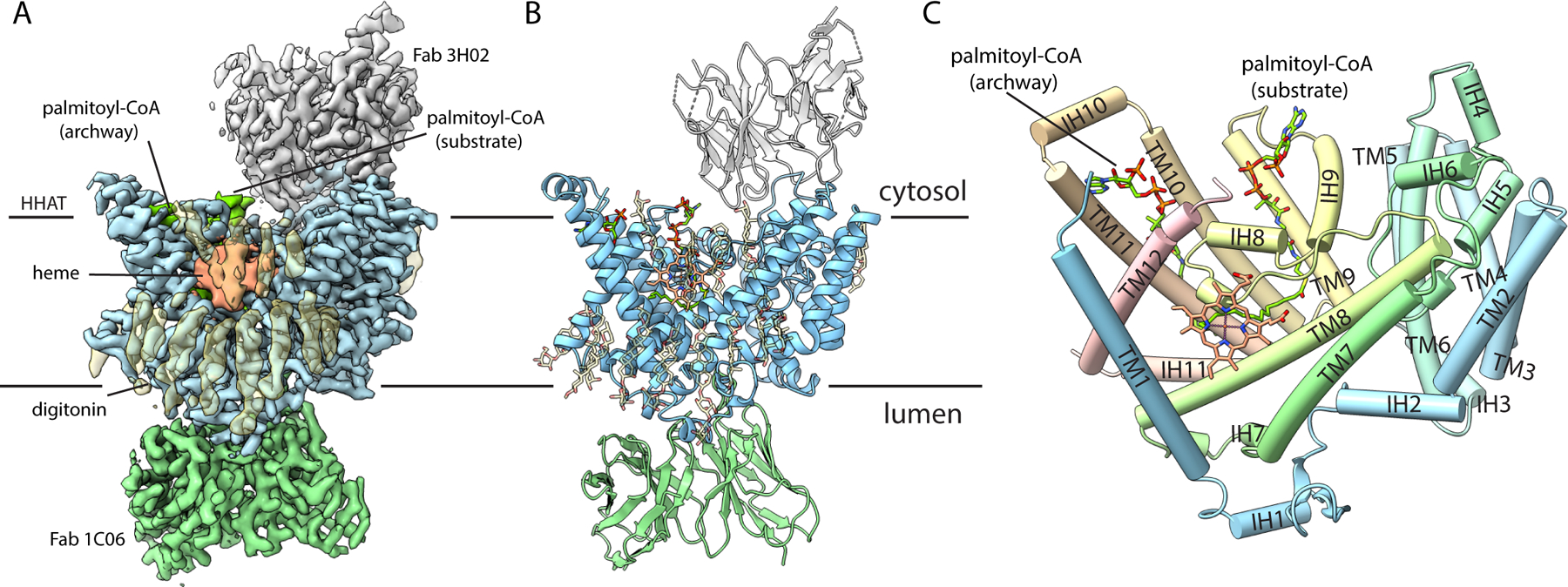
(A, B) Cryo-EM reconstruction (A) and cartoon representation (B) of HHAT in complex with palmitoyl-CoA and two Fab antibody fragments, viewed from the membrane. Variable domains of antibodies are shown. (C) Architecture of HHAT; helices drawn as cylinders.
The structure of HHAT reveals 12 transmembrane a helices (TM1-TM12), intervening a helical (IH1-IH11) and loop regions. The enzyme spans the ER membrane with only minimal regions extending into the cytosol or lumen (Fig. 1, fig. S1D). A structural “MBOAT fold” identified from structures of lipid-modifying MBOATs (15–20) is recognizable in the TM4-TM11 portion of HHAT (fig. S5, S6). This region contains a hallmark histidine residue (His379 in HHAT) that has been implicated in catalysis (10, 14, 21). Substantial differences in the MBOAT fold region create features particular to HHAT that constitute substrate binding and access sites (as described below). TM10 and TM11 are longer than their counterparts in other MBOAT structures (fig. S6); together with intervening cytosolic helix IH10 (also found only in HHAT), this motif forms a distinctive archway that appears to be involved in palmitoyl-CoA substrate access (Fig. 1C). The TM1-TM3 region, which has no structural counterpart in the lipid-modifying enzymes, forms a module (Fig. 1C, fig. S6) that is involved in Hedgehog substrate binding.
The molecular surface of HHAT contains invaginations from both sides of the membrane (Fig. 2). The cytosolic opening is occupied by well-defined density for palmitoyl-CoA (Fig. 2B). Its binding site comprises two distinct sections: one cavity for the acyl chain and one occupied by the CoA moiety (Fig. 2D). These cavities meet with an approximate right angle at the hallmark His379 near the middle of the membrane, thereby identifying the active site. A concomitant bend in the substrate occurs near its central carbonyl carbon, the site of transfer to hedgehog. Basic and hydrophilic residues on the enzyme’s cytosolic side coordinate the CoA moiety and its phosphate groups (Fig. 2B, fig. S7). The acyl chain adopts an extended conformation within its cavity, which is slender and roughly colinear with the midline of the membrane. The walls of acyl cavity are lined by hydrophobic amino acids that are conserved among HHAT orthologs (figs. S7, S8). The binding location of palmitoyl-CoA indicates that this substrate would access the active site from the cytosolic side of the membrane, consistent with where palmitoyl-CoA is synthesized (22).
Fig. 2. Palmitoyl-CoA substrate complex.
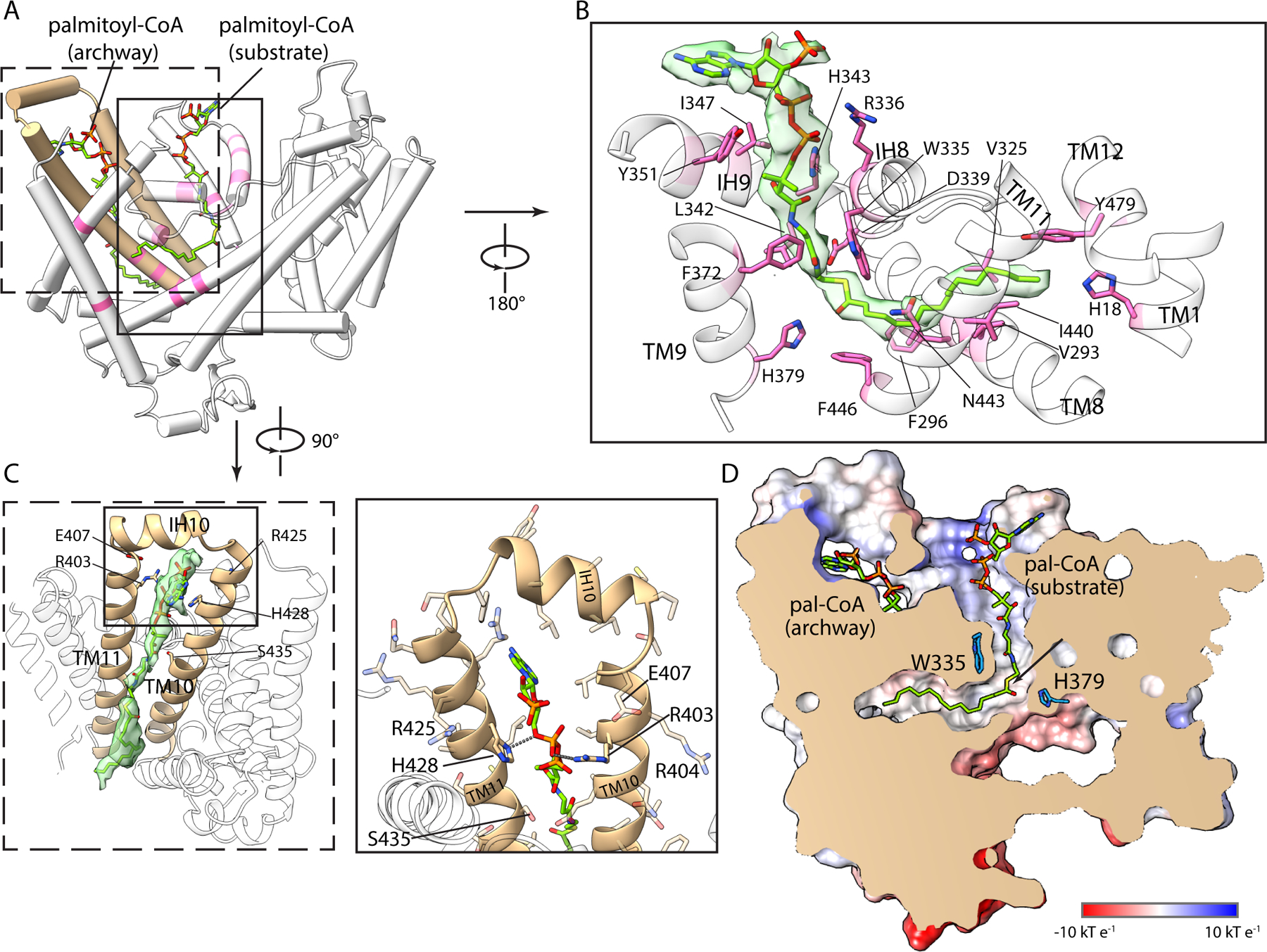
(A) Binding of two palmitoyl-CoA molecules (sticks with green carbon atoms): one in the active site and one in the archway. Regions of HHAT that contact these ligands are pink; the archway is tan. (B) Detailed view of palmitoyl-CoA substrate in the active site. Its density (green surface) and interacting amino acids (pink sticks) are shown. (C) Palmitoyl-CoA in the archway, shown from the membrane perspective. (D) Slice of the molecular surface of HHAT highlighting the active site cavities and bound ligands. An arrow indicates the carbonyl carbon of palmitoyl-CoA in its substrate binding site.
The archway, formed by the TM10-IH10-TM11 motif, connects the active site to the membrane and contains density for a second palmitoyl-CoA molecule (Fig. 2C). This palmitoyl-CoA molecule is oriented vertically with its CoA moiety near the cytosolic side and its acyl chain extending luminally along the enzyme’s transmembrane surface (Fig. 2A, C). The archway is the only membrane-exposed opening to the cytosolic leaflet large enough to accommodate a palmitoyl-CoA, and its chemical environment is apt for palmitoyl-CoA access to the active site: hydrophilic amino acids face the active site whereas hydrophobic ones face the membrane (Fig. 2C). The phosphate groups of the CoA moiety are within hydrogen bonding distance to Arg403 and His428 on the inside of the archway (Fig. 2C). Substitution of either of these residues with tryptophan, which would be expected to narrow the archway, reduce catalytic activity (fig. S9), corroborating the archway as the access route for palmitoyl-CoA to the active site.
On the surface of the membrane-spanning region of HHAT, nestled between a V-shaped arrangement of TM1 and TM7, resides density for a heme B prosthetic group (Fig. 1A, 3A). Although it was not previously noted as a component of HHAT or other MBOAT enzymes, biochemical analysis confirms that purified HHAT contains a stoichiometric amount of heme (fig. S10B). Its location and density for surrounding detergent molecules suggest that the heme would be directly exposed to membrane lipids. To evaluate this hypothesis, we performed an all-atom molecular dynamics simulation and observe that the heme interacts with surrounding membrane lipids (fig. S10C, and Movie S1). We found that mutation of Cys324, which coordinates the central iron (Fig. 3B), abolishes detectable catalytic activity (Fig. 4). A distance of 22 Å between the heme and the active site is presumably too far for a direct role in catalysis. Marked reduction in thermal stability of the mutant is consistent with a structural role (fig. S11). Further analysis is required to assess possible biochemical and physiological importance of the heme and whether these are regulatable by oxidation state.
Fig. 3. Heme prosthetic group.
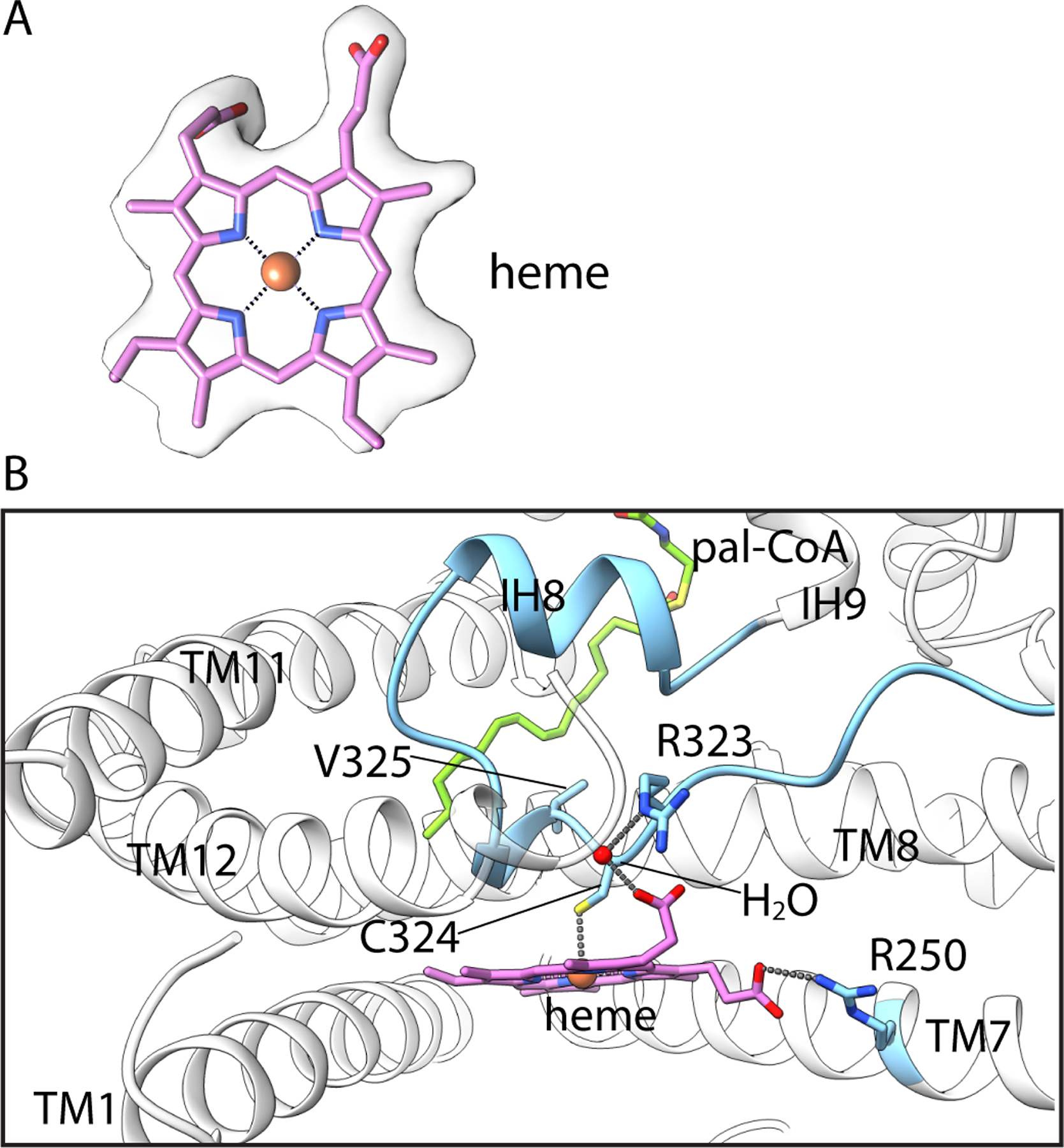
(A) Density for the heme. (B) Interactions with HHAT. Amino acids contacting the heme are drawn as sticks. Dashed lines indicate hydrogen bonds and interactions with Fe(III).
Fig. 4. Palmitoylated Hedgehog peptide product complex.
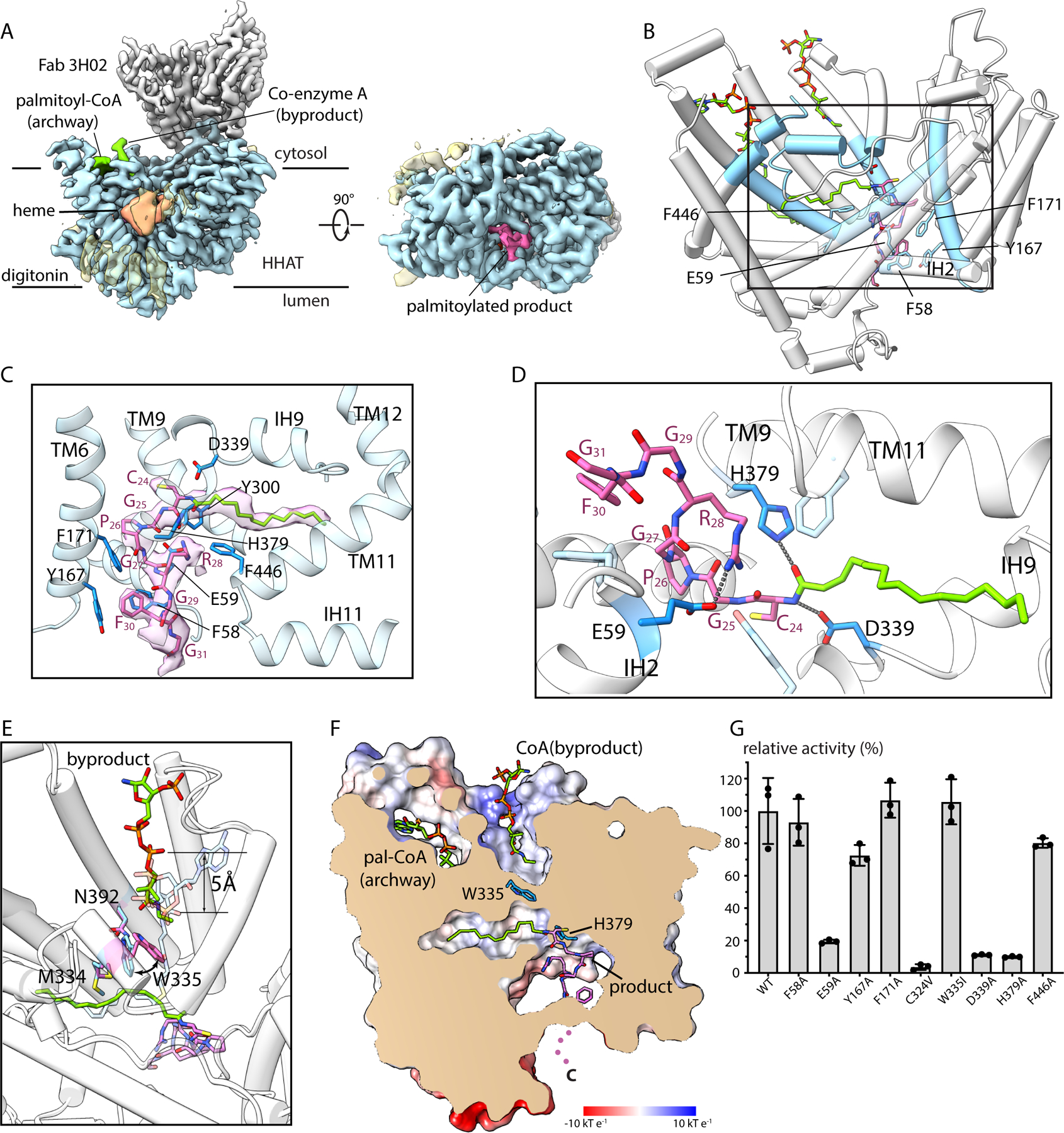
(A) Cryo-EM reconstruction, in orthogonal views. (B) Overall structure, highlighting the locations of ligands. Regions in contact with the product are colored blue. (C-D) Close up views of the product, showing cryo-EM density (C) and contacting HHAT residues. (E) A curved arrow indicates repositioning of Trp335 from the substrate complex (light blue) to the product complex (pink). Positioning of the CoA byproduct relative to the substrate is indicated. (F) Slice of the molecular surface showing the product in the reaction chamber and the CoA byproduct in the cytosolic cavity. Dots indicate the disordered C-terminal region of the Hedgehog peptide. (G) Relative enzymatic activity of indicated mutants.
We found that the luminal antibody (1C06) is a potent inhibitor of HHAT whereas the cytosolic antibody (3H02) has minimal effect on catalytic activity (fig. S2D). The inhibitory antibody partially occupies a V-shaped cleft that would be exposed to the ER lumen in a cellular setting (Fig. 1B). Suspecting that this cleft might comprise the binding site for the acceptor substrate, we pursued a structure using only the cytosolic antibody to obtain structural information for the interaction with Hedgehog.
To investigate Hedgehog binding, we pre-formed a complex of HHAT with the cytosolic antibody and added stoichiometric amounts of palmitoyl-CoA and a peptide substrate comprising the first 12 amino acids of partially processed human Sonic Hedgehog. Single particle cryo-EM analysis yielded a structure at 3.2 Å resolution (Fig. 4, figs. S3, S12). The structure contains well-ordered density for a palmitoylated Hedgehog peptide product within the active site (Fig. 4). Like in the other structure, density for the polypeptide of HHAT is well-resolved throughout. The structures superimpose with an RMSD of 0.46 Å. There are noteworthy side chain conformational differences that appear to be involved with the release of the CoA byproduct, but there are no domain-level differences or α-helical shifts between the structures. The absence or presence of the luminal antibody has no discernable effect on the structure of the enzyme, confirming that its inhibition is due to occlusion of the Hedgehog binding site.
Well-defined density for the peptide and palmitoyl group portions of the product, including density for the amide linkage to the N-terminal cysteine (C24) of Hedgehog, indicates that the palmitoylation reaction has occurred (Fig. 4C). The acyl chain is located in a position analogous to that observed in the substrate complex (Fig. 4). Density is also present for the CoA byproduct of the reaction (fig. S12), which is displaced by approximately 5 Å toward the cytosolic solution relative to its location in the substrate complex (Fig. 4E). With displacement of the CoA, the side chain of Trp335 swings in and seals off the acyl cavity from the cytosolic side of the enzyme (Fig. 4E–F). This and concomitant rearrangements of interacting residues Met334 and Asn392 (fig. S13) constitute the only marked conformational differences in HHAT between the structures. Mutation of Trp335 to Ile has no discernable effect on palmitoyl transfer activity, suggesting that Trp335 does not have a direct role in catalysis (fig. 4G). As before, a palmitoyl-CoA molecule occupies the archway, presumably poised to enter the active site chamber following release of the products.
Sonic, Desert, and Indian Hedgehog contain a conserved peptide sequence at their N-termini (24CGPGR28) that has been shown to underlie their ability to act as substrates of HHAT (9) (fig. S14). The consensus comprises an N-terminal cysteine (C24), a proline (P26), an arginine (R28), and intervening glycine residues. The first eight amino acids of the palmitoylated Hedgehog peptide product, containing this consensus sequence, are ordered within the luminal cavity of the reaction chamber (Fig. 4C). The peptide makes an S-shaped curve in the active site, with the palmitoylated C24 binding deepest within the enzyme. The tightly bent conformation of the peptide is facilitated by the flexibility of glycine residues and a turn that occurs at P26 (Fig. 4C). The peptide makes extensive van der Waals and hydrogen bonding interactions with HHAT. The residues involved emanate from a congregation of secondary structural elements that converge on the luminal side of the reaction chamber (Fig. 4B–D). Among these, the IH2 helix, within the TM1-TM3 structural element, shields the peptide from the membrane (Fig. 4B). Notable hydrogen bonds include one between His379 and the carbonyl oxygen of the amide bond at the palmitoylated N-terminus and one between Asp339 and the amide nitrogen (Fig. 4D). Mutation of either Asp339 or His379 nearly eliminates activity (fig. 4G), as has been observed previously for His379 (21). Several aromatic amino acids lining the peptide-binding site also contribute to the contour of the peptide binding site, but those we tested have only modest effects on activity when mutated (fig. 4G). The only ion pair interaction with the peptide portion of the product is between Glu59, a component of the TM1-TM3 element structural element, and R28. Corroborating a previous implication of R28 in substrate specificity (9), mutation of either Glu59 or R28 to alanine diminishes both kcat and kcat/Km (fig. S2B). The compacted shape of the peptide and the confines of the luminal cavity indicate that the reaction occurs in the middle of the membrane, shielded from both bulk solvent and the lipid bilayer.
The structures of substrate and product complexes indicate how HHAT overcomes an impressive challenge: bringing together two substrates—separated by a membrane and with divergent chemical properties—into a membrane-embedded active site to achieve catalysis (fig. S15). HHAT utilizes a remarkably efficient design to catalyze Hedgehog acylation. Palmitoyl-CoA accesses the active site by passing under the archway, which complements the substrate’s physiochemical properties and localization. It then proceeds to its substrate binding site. By contrast, the Hedgehog substrate reaches the active site from the aqueous environment of the lumen. Catalysis has been proposed to proceed through either a one step or a two-step mechanism(3, 8). Our work cannot definitively distinguish between these, but the following one-step mechanism seems more plausible: Asp339 acts as a general base to activate the amino terminal end of Hedgehog for nucleophilic attack on the carbonyl carbon, thereby releasing CoA as the leaving group and forming the amide linkage of the fully mature Hedgehog product (fig. S15B). Our work indicates that release of its products is directional. The CoA byproduct is released into the cytosol, as is evidenced by its binding location in the structure. Palmitoylated Hedgehog can only be released into the lumen for the simple reason that the Hedgehog polypeptide would be too large to fit through the narrow path exposed to the cytosol. The enzyme therein transfers one palmitoyl acyl group across the membrane per reaction cycle.
In vitro experiments indicate that in addition to the chemical modification of Hedgehog, HHAT can enable movement of an intact palmitoyl-CoA molecule from the cytosolic leaflet of the membrane to the luminal one (23). The physiological implications of this biochemical functionality are not known. However, the structure shows that this functionality is not involved in the palmitoylation of Hedgehog because the catalytically competent palmitoyl-CoA substrate within the active site is located on the cytosolic side, because the CoA moiety is released into the cytosol, and because the confines of the active site would prohibit simultaneous binding of both the Hedgehog substrate and palmitoyl-CoA in the luminal cavity.
Our work reveals marked differences from the structures of MBOAT enzymes that acylate lipophilic substrates. Unlike DGAT1 and ACAT1, which are catalytically active only as dimers or tetramers (16–20), the present study identifies that HHAT functions as a monomer. The dimer interfaces of those enzymes roughly correspond to the location of the heme in HHAT (fig. S16). The palmitoyl-CoA access archway is found exclusively in HHAT; DGAT1 and ACAT1 have a cleft between two transmembrane helices, roughly corresponding to TM9 and TM10 in HHAT, that may act as the access route for acyl-CoA substrates of those enzymes but is sealed in the structure of HHAT (fig. S16A–C). Different modes of acyl-CoA access may contribute to substrate specificities of the enzymes, with HHAT having preference for palmitoyl-CoA, whereas DGAT1 and ACAT1 utilize an array of acyl-CoA donors. The slender cavity of HHAT in which the palmitoyl moiety of the substrate binds is distinct from a bulging cavity observed in ACAT1/DGAT1 enzymes (fig. S16E–F) – a difference that is also in line with the acyl donor specificity profiles of the enzymes. With regard to acyl acceptor substrate binding, in contrast to the Hedgehog binding site—which is shielded from the membrane and exposed to the aqueous environment of the ER—large cavities in DGAT1 and ACAT1 are directly exposed to the luminal leaflet of the membrane by openings in their molecular surfaces that may function as entry and egress routes for the lipophilic acyl acceptor substrates and products (fig. S16D–G). In HHAT, protein substrate access and the release of fully mature Hedgehog occur via a membrane-embedded cavity that is nonetheless hydrophilic and exposed only to the lumen. These principles provide a structural framework for inhibitor development and may be relevant to the mechanisms of acylation of ghrelin and Wnt.
Supplementary Material
Acknowledgements:
We thank members of the S.B. Long and R.K. Hite laboratories at MSKCC for discussions; the staff of the New York Structural Biology Center Simons Electron Microscopy Center, M. J. de la Cruz of the MSKCC Cryo-EM facility, and Frances Weis-Garcia and staff of the Antibody Core Facility at MSKCC.
Funding:
This work was supported, in part, by a Geoffrey Beene Cancer Research Center Award (to S.B.L) and an NIH core facilities grant to MSKCC (P30 CA008748).
Footnotes
Competing interests: The authors declare no competing financial interests.
Data and materials availability: Atomic coordinates and EM maps of the substrate and product complexes have been deposited in the PDB (accession numbers 7MHY and 7MHZ) and EMDB (EMD-23836 and EMD-23837, respectively). All other data are available in the main text or the supplementary materials.
References and Notes:
- 1.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y, Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem Biol 24, 252–280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamoun Z et al. , Skinny Hedgehog, an acyltransferase required for palmitoylation and activity of the Hedgehog signal. Science 293, 2080–2084 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Mann RK, Beachy PA, Novel lipid modifications of secreted protein signals. Annual review of biochemistry 73, 891–923 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT, Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev 18, 641–659 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi XF, Schmiege P, Coutavas E, Wang JW, Li XC, Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128-+ (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi X, Schmiege P, Coutavas E, Li X, Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrova E, Matevossian A, Resh MD, Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene 34, 263–268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buglino JA, Resh MD, Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. The Journal of Biological Chemistry 283, 22076–22088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy RY, Resh MD, Identification of N-terminal residues of Sonic Hedgehog important for palmitoylation by Hedgehog acyltransferase. J Biol Chem 287, 42881–42889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann K, A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci 25, 111–112 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Chang CC et al. , Recombinant acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) purified to essential homogeneity utilizes cholesterol in mixed micelles or in vesicles in a highly cooperative manner. J Biol Chem 273, 35132–35141 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Hishikawa D et al. , Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc Natl Acad Sci U S A 105, 2830–2835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grzeschik KH et al. , Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nature Genetics 39, 833–835 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL, Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 132, 387–396 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Ma D et al. , Crystal structure of a membrane-bound O-acyltransferase. Nature 562, 286-+ (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan CC et al. , Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nature Communications 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long T, Sun Y, Hassan A, Qi X, Li X, Structure of nevanimibe-bound tetrameric human ACAT1. Nature 581, 339–343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian H et al. , Structural basis for catalysis and substrate specificity of human ACAT1. Nature 581, 333–338 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Sui X et al. , Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature 581, 323–328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L et al. , Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature 581, 329–332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buglino JA, Resh MD, Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog. PloS one 5, e11195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broadway NM et al. , The liver isoform of carnitine palmitoyltransferase 1 is not targeted to the endoplasmic reticulum. Biochem J 370, 223–231 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asciolla JJ, Resh MD, Hedgehog Acyltransferase Promotes Uptake of Palmitoyl-CoA across the Endoplasmic Reticulum Membrane. Cell Rep 29, 4608–4619 e4604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez M, Sabio L, Galvez N, Capdevila M, Dominguez-Vera JM, Iron Chemistry at the Service of Life. Iubmb Life 69, 382–388 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Baradaran R, Long SB, Structure and Reconstitution of an MCU-EMRE Mitochondrial Ca(2+) Uniporter Complex. J Mol Biol 432, 5632–5648 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhofer A et al. , Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–138 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Kawate T, Gouaux E, Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure (London, England : 1993) 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanov J et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebschner D et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goddard TD et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Williams CJ et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo S, Kim T, Im W, Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2, e880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Jo S, Lim JB, Klauda JB, Im W, CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys J 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J et al. , CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J Chem Theory Comput 15, 775–786 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Wu EL et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL, OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40, D370–376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J et al. , CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks BR et al. , CHARMM: the biomolecular simulation program. J Comput Chem 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraham MJ et al. , GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 48.Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Stone JE, Vandivort KL, Schulten K, paper presented at the Proceedings of the 8th International Workshop on Ultrascale Visualization - UltraVis ‘13, 2013.
- 50.Diver MM, Long SB, Mutational analysis of the integral membrane methyltransferase isoprenylcysteine carboxyl methyltransferase (ICMT) reveals potential substrate binding sites. Journal of Biological Chemistry 289, 26007–26020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez M, Sabio L, Galvez N, Capdevila M, Dominguez-Vera JM, Iron Chemistry at the Service of Life. Iubmb Life 69, 382–388 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


