Abstract
Bacteria of the Burkholderia cepacia complex consist of five discrete genomic species, including genomovars I and III and three new species: Burkholderia multivorans (formerly genomovar II), Burkholderia stabilis (formerly genomovar IV), and Burkholderia vietnamiensis (formerly genomovar V). Strains of all five genomovars are capable of causing opportunistic human infection, and microbiological identification of these closely related species is difficult. The 16S rRNA gene (16S rDNA) and recA gene of these bacteria were examined in order to develop rapid tests for genomovar identification. Restriction fragment length polymorphism (RFLP) analysis of PCR-amplified 16S rDNA revealed sequence polymorphisms capable of identifying B. multivorans and B. vietnamiensis but insufficient to discriminate strains of B. cepacia genomovars I and III and B. stabilis. RFLP analysis of PCR-amplified recA demonstrated sufficient nucleotide sequence variation to enable separation of strains of all five B. cepacia complex genomovars. Complete recA nucleotide sequences were obtained for 20 strains representative of the diversity of the B. cepacia complex. Construction of a recA phylogenetic tree identified six distinct clusters (recA groups): B. multivorans, B. vietnamiensis, B. stabilis, genomovar I, and the subdivision of genomovar III isolates into two recA groups, III-A and III-B. Alignment of recA sequences enabled the design of PCR primers for the specific detection of each of the six latter recA groups. The recA gene was found on the largest chromosome within the genome of B. cepacia complex strains and, in contrast to the findings of a previous study, only a single copy of the gene was present. In conclusion, analysis of the recA gene of the B. cepacia complex provides a rapid and robust nucleotide sequence-based approach to identify and classify this taxonomically complex group of opportunistic pathogens.
The Burkholderia cepacia complex is a very diverse group of bacteria (28). They are important opportunistic human pathogens that cause devastating infections in patients with cystic fibrosis (CF) (8, 13) and in other vulnerable individuals (25). The ability of B. cepacia to cause disease is not limited to the human host, as these bacteria are also important plant pathogens (7). In addition, B. cepacia complex bacteria may have commercially useful properties and have been used in agriculture as biocontrol agents and in the bioremediation of toxic agents (9, 14). Current taxonomic classification divides isolates previously classified as B. cepacia into five genomovars or discrete genomic species, all of which may be isolated from clinical infection (28). Strains of genomovar II have been proposed as the new species Burkholderia multivorans (28), and strains of genomovar V were found to be members of the proposed species Burkholderia vietnamiensis (6, 28). Strains within genomovar IV have recently been proposed as the new species Burkholderia stabilis (29). The remaining genomovars, I and III, await species designation if differential phenotypic tests can be found (28). Determination of the genomovar status of B. cepacia complex strains was based on a polyphasic taxonomic approach which utilized phenotypic tests such as whole-cell protein profile analysis and genotypic tests such as DNA-DNA hybridization (27, 28). A single test for identification of genomovar status is currently not available. Even conventional phenotypic identification of B. cepacia and its differentiation from closely related species are often not straightforward (3, 10, 24, 30). Incorrect diagnosis of infection, especially in patients with CF, may have serious clinical ramifications (13). A simple means of B. cepacia complex species identification will enhance our understanding of the pathogenesis and epidemiology of these opportunistic human pathogens.
Molecular diagnostic probes based on PCR provide a rapid and frequently highly discriminatory means of microbial identification (2, 15, 24, 30). In order to develop rapid tests to determine the genomovar status of B. cepacia complex isolates, we examined nucleotide sequence polymorphism in two genes, the 16S rRNA gene (16S rDNA) and recA gene. Although widely used for bacterial systematics, recent results demonstrate that 16S rDNA is limited in its ability to differentiate the B. cepacia complex (15, 23). Nucleotide sequence variation within this gene is not sufficient to enable all current genomovars within the complex to be easily identified (2, 15, 23). The B. cepacia complex recA (RecA is a protein essential for repair and recombination of DNA [5, 11]) was chosen as an alternative template gene for the following reasons. First, analysis of recA has proven very useful in previous studies of molecular systematics among closely related bacteria (5, 11). Second, although only a limited number of B. cepacia genes have been characterized, two recA sequences, each from a different strain, were available in the nucleotide sequence databases as a basis to initiate analysis (19, 31). Finally, in B. cepacia, recA has been shown to be diploid, with a single copy of the gene residing on each of the two large chromosomes present in B. cepacia strain ATCC 25416 (21). If both copies of recA are identical, they may form a more robust platform upon which to base diagnostic PCR probes than the multicopy rRNA genes, which are dispersed across the multiple replicons constituting the genome of B. cepacia and are potentially susceptible to genomic rearrangement (4, 12, 21). Diagnostic tests for the determination of B. cepacia complex genomovar status, developed using systematic genetic approaches, are described.
MATERIALS AND METHODS
Bacterial strains.
B. cepacia complex strains were cultured and biochemically identified as described previously (10, 16, 28). Bacteria were routinely grown on blood agar, Trypticase soy agar, or Luria-Bertani broth (LB; 23) at 35°C for 24 to 48 h until confluent growth was obtained (10, 16). Molecular identification approaches were developed using the 35 isolates listed in Table 1, of which 29 isolates were derived from a published panel of strains representative of each genomovar (18). The sources of the six additional strains initially screened in this study are also provided in Table 1. The latter 35 isolates and 68 further B. cepacia complex isolates examined (103 total) were recovered from various sources including patients with CF, patients with non-CF infection, and the natural environment. Strains were selected to be representative of the genetic diversity of B. cepacia complex from previous studies (10, 16–18, 28). Additional reference strains were obtained from the American Type Culture Collection (ATCC, Manassas, Va.). Related bacterial species commonly recovered from patients with CF and misidentified as B. cepacia were derived from previous studies (10, 16).
TABLE 1.
B. cepacia complex strains used to develop molecular diagnostic approaches
| Genomovar and straina | 16S rDNA DdeI RFLPb | Result of recA RFLP by digestion with:
|
recA GenBank accession no. | recA phylogenetic and PCR identification group | |
|---|---|---|---|---|---|
| HaeIIIc | MnlId | ||||
| B. cepacia genomovar I | |||||
| ATCC 25416T | 2 | D | d | AF143786 | |
| J1050e | 2 | D | d | I | |
| ATCC 17759 | 2 | E | e | AF143788 | I |
| CEP509 | 2 | E | e | AF143787 | I |
| LMG 17997 | 2 | E | e | I | |
| B. multivorans (formerly genomovar II) | |||||
| C5393 | 3 | F | a | AF143776 | B. multivorans |
| LMG 13010T | 3 | F | a | B. multivorans | |
| CF-A1-1 | 3 | F | a | B. multivorans | |
| C1576 | 3 | C | a | AF143774 | B. multivorans |
| HI-2308e | 3 | C | a | AF143777 | B. multivorans |
| JTC | 3 | F | a | AF143778 | B. multivorans |
| C1962 | 3 | F | a | B. multivorans | |
| ATCC 17616 | 3 | F | a | AF143775 | B. multivorans |
| 249-2 | 3 | F | a | B. multivorans | |
| B. cepacia genomovar III | |||||
| J2315 | 2 | G | f | III-A | |
| BC7 | 2 | G | f | III-A | |
| K56-2 | 2 | G | f | AF143779 | III-A |
| C5424 | 2 | G | f | AF143781 | III-A |
| C6433 | 2 | G | f | AF143780 | III-A |
| C4455e | 2 | G | f | AF143782 | III-A |
| C1394 | 2 | H | g | AF143783 | III-B |
| ATCC 17765 | 2 | H | g | III-B | |
| CEP511 | 2 | I | h | AF143785 | III-B |
| PC184 | 2 | J | i | AF143784 | III-B |
| B. stabilis (formerly genomovar IV) | |||||
| LMG 07000e | 2 | J | b | AF143789 | B. stabilis |
| LMG 14291e | 2 | J | b | AF143790 | B. stabilis |
| LMG 14294 | 2 | J | b | B. stabilis | |
| C7322 | 2 | J | b | B. stabilis | |
| LMG 14086 | 2 | J | b | B. stabilis | |
| LMG 18888 | 2 | J | b | B. stabilis | |
| B. vietnamiensis (formerly genomovar V) | |||||
| PC259 | 1 | A | c | AF143791 | B. vietnamiensis |
| LMG 16232 | 1 | A | c | B. vietnamiensis | |
| FC441 | 1 | A | c | B. vietnamiensis | |
| LMG 10929T | 1 | B | c | AF143793 | B. vietnamiensis |
| C2822e | 1 | B | c | AF143792 | B. vietnamiensis |
All strains except those indicated in footnote b below were derived from the B. cepacia complex strain panel where source and other data relevant to each are described (18).
Data are numerical 16S rRNA RFLP types as indicated in Fig. 1.
Letters correspond to alphabetical RFLP types as shown in Fig. 2A.
Letters correspond to alphabetical RFLP types as shown in Fig. 2B.
Strains not derived from the B. cepacia complex panel (18): J1050, isolated from non-CF patient within the U.K. and kindly provided by J. R. W. Govan; HI-2308, recovered from neonatal infection and kindly provided by J. J. LiPuma; C4455, recovered from a patient with CF (16); LMG 07000, recovered from a patient with septicaemia (20); LMG 14291, recovered from a patient with CF (20); and C2822, recovered from a patient with CF (16).
Genomovar status.
The genomovar status of B. cepacia complex strains used to develop the molecular identification strategies was determined by whole-cell protein profile analysis and a polyphasic approach as described previously (27, 28).
PCR analysis.
PCR was performed essentially as described previously (16, 17) however, PCR reagents were purchased from Qiagen Inc. Canada, and tests were performed in the presence of Qiagen Q solution, which enhances amplification of DNA templates which are rich in G+C content (Qiagen Inc. [http://www.qiagen.com]). Template DNA was prepared from fresh overnight cultures as previously described (17), with modification of the lysis buffer to include pronase at 0.5 mg/ml. DNA was quantitated by visualization on agarose gels, and approximately 20 ng was incorporated into 25-μl reactions which contained 1 U Taq DNA polymerase, 250 μM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, and 1× PCR buffer. Approximately 20 pmol of each appropriate oligonucleotide primer (Table 2) was added to each reaction, and amplification was carried out using a Gene Cycler (Bio-Rad, Mississauga, Ontario, Canada) for 30 cycles of 30 s at 94°C, 45 s at the appropriate annealing temperature (Table 2), and 60 s at 72°C; a final extension of 10 min at 72°C was applied to all thermal cycles. The sequences and specificity of all PCR primers used in this study are provided in Table 2.
TABLE 2.
Species-specific and nucleotide-sequencing PCR primers used in study
| Specificity and primer name | Sequence (5′ to 3′)a | Position | PCR annealing temperature (°C) | Product size |
|---|---|---|---|---|
| Universal 16S rDNA primer | ||||
| UNI2 | GACTCCTACGGGAGGCAGCAG | 336–356b | 60 | 1,020 bpc |
| UNI5a | CTGATCCGCGATTACTAGCGATTC | 1337–1360b | ||
| B. cepacia complex recA | ||||
| BCR1 | TGACCGCCGAGAAGAGCAA | 2–20d | 58 | 1,043 bpd |
| BCR2a | CTCTTCTTCGTCCATCGCCTC | 1044–1024d | ||
| recA sequencing primers | ||||
| BCR3 | GTCGCAGGCGCTGCGCAA | 513–530d | 58 | 532 bp,d 3′ half of B. cepacia complex recA gene in combination with primer BCR2 |
| BCR4a | GCGCAGCGCCTGCGACAT | 528–511d | 58 | 527 bp,d 5′ half of B. cepacia complex recA gene in combination with primer BCR1 |
| B. cepacia genomovar I | ||||
| BCRG11 | CAGGTCGTCTCCACGGGT | 112–129e | 62 | 492 bpe |
| BCRG12a | CACGCCGATCTTCATACGA | 603–585e | ||
| B. multivorans (Bm) | ||||
| BCRBM1 | CGGCGTCAACGTGCCGGAT | 321–339d | 62 | 714 bpd |
| BCRBM2a | TCCATCGCCTCGGCTTCGT | 1034–1016d | ||
| B. cepacia genomovar III-A | ||||
| BCRG3A1 | GCTCGACGTTCAATATGCC | 294–309f | 62 | 378 bpf |
| BCRG3A2a | TCGAGACGCACCGACGAG | 671–654f | ||
| B. cepacia genomovar III, RG-B | ||||
| BCRG3B1 | GCTGCAAGTCATCGCTGAA | 228–246g | 60 | 781 bpg |
| BCRG3B2a | TACGCCATCGGGCATGCT | 1008–991g | ||
| B. cepacia genomovar IV, RG-4 | ||||
| BCRG41 | ACCGGCGAGCAGGCGCTT | 361–378h | 64 | 647 bph |
| BCRG42a | ACGCCATCGGGCATGGCA | 100–990h | ||
| B. vietnamiensis, RG-BV | ||||
| BCRBV1 | GGGCGACGGCGACGTGAA | 84–101i | 62 | 378 bpi |
| BCRBV2a | TCGGCCTTCGGCACCAGT | 461–444i |
Base positions in primer sequence which were mismatched in other genomovars are underlined.
Position in relation to Escherichia coli 16S rRNA gene (GenBank accession number J01859).
Size in relation to B. cepacia 16S rRNA gene (GenBank accession number X87275).
Size and position in relation to B. multivorans ATCC 17616 recA sequence U70431.
Size and position in relation to B. cepacia genomovar I strain ATCC 17759 recA (Table 1).
Size and position in relation to B. cepacia genomovar III-A strain K56-2 recA (Table 1).
Size and position in relation to B. cepacia genomovar III-B strain C1394 recA (Table 1).
Size and position in relation to B. stabilis strain LMG14291 recA (Table 1).
Size and position in relation to B. vietnamiensis strain C2822 recA (Table 1).
For restriction fragment length polymorphism (RFLP) analysis, 5 to 10 μl of PCR product was combined with the appropriate restriction enzyme buffer and endonuclease as outlined by the manufacturer (New England BioLabs Inc., Mississauga, Ontario, Canada) and incubated at 37°C for 1 to 3 h. PCR and RFLP products were analyzed by agarose gel electrophoresis, with agarose concentrations adjusted between 1% and 2.5%, appropriate for the size range of DNA being analyzed, and using 0.5× Tris-borate-EDTA buffer (24). Molecular size markers of the appropriate size range were included on all gels (100-bp DNA ladder or 1-kb DNA ladder [Life Technologies GIBCO BRL Products, Burlington, Ontario, Canada]: 50-bp ladder [Pharmacia, Uppsala, Sweden]). RFLP patterns were analyzed as described previously (16, 17).
Typing of B. cepacia strains by random amplified polymorphic DNA (RAPD) PCR analysis was performed exactly as described previously (16). Detection by PCR of the B. cepacia epidemic strain marker (BCESM) and cable pilin subunit gene (cblA) was carried out as described previously (17).
Nucleotide sequence analysis.
Sequencing reactions were prepared using ABI PRISM DyeDeoxy Terminator cycle sequencing kits with AmpliTaq FS DNA polymerase according to the manufacturer's instructions and analyzed using either an ABI PRISM model 373 Stretch or a model 377 DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Raw sequences from both strands of the PCR products were then aligned, and a consensus sequence was derived using DNASTAR software (DNASTAR Inc., Madison, Wis.). Sequence identity was confirmed by analysis using the basic local alignment sequence tool (BLAST) (1) at the National Center for Biotechnology Information (NCBI, Bethesda, Md.).
Phylogenetic analysis.
Evolutionary relationships between recA genes were determined using the Data Analysis in Molecular Biology software (DAMBE; http://web.hku.hk/∼xxia/software/software.htm). After recA sequence determination, multiple sequence alignments were performed using CLUSTAL W (26). Phylogenetic trees were drawn from the resulting alignments using the genetic distance-based neighbor-joining algorithms of DAMBE. Paralinear and Jukes-Cantor-based algorithms were evaluated and found to demonstrate identical phylogenies for the strains examined (data not shown). Trees constructed using Jukes-Cantor distance matrices are presented in this report. Sequence input order was randomized, and 100 data sets were examined by bootstrapping resampling statistics for each analysis.
PFGE and Southern hybridization.
Macrorestriction and pulsed-field gel electrophoresis (PFGE) genomic fingerprinting of B. cepacia strains were performed as described previously (18). Multiple replicons which constitute the B. cepacia genome were separated as previously described (4, 21). Southern blot transfer of the DNA to nylon membranes was allowed to proceed for 48 h and probed with a digoxigenin-labeled recA PCR probe as previously described (17).
Nucleotide sequence accession numbers.
The complete recA nucleotide sequences were determined for the 20 B. cepacia complex strains listed in Table 1. Sequences were submitted to GenBank as an aligned set, and each was assigned an accession number as shown in Table 1.
RESULTS
Analysis of 16S rRNA genes of the B. cepacia complex and closely related species.
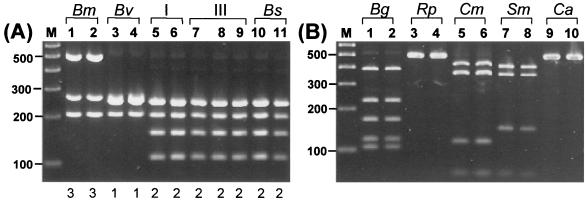
Amplified-rDNA restriction analysis (ARDRA; 23, 27) was performed to examine sequence polymorphism of the 16S rRNA gene in strains representative of the B. cepacia complex. The primer pair used for ARDRA analysis of the 16S rDNA in this study is novel (UNI2 and UNI5 [Table 2]) and has proven effective for amplification of the gene from all bacterial species tested to date (S. K. Byrne, unpublished data). To evaluate the efficacy of both the 16S rRNA gene (and recA [see below]) for systematic differentiation of the B. cepacia complex, a panel of 35 (Table 1) strains representative of all five genomovars of the B. cepacia complex (18) was initially examined. The 16S rDNA 1-kb amplicon was successfully amplified with primers UNI2 and UNI5 from all 35 strains. The identity of the amplified DNA was confirmed to encode 16S rDNA by direct nucleotide sequence analysis of PCR products (data not shown). To detect nucleotide sequence variation within the amplified gene, several restriction endonucleases expected to cleave frequently within bacterial rDNA were screened for their ability to reveal genomovar-specific RFLPs (data not shown). The enzyme DdeI was found to produce the most discriminatory RFLPs for the B. cepacia complex (Fig. 1A). Three 16S rDNA ARDRA patterns were found among the 35 strains (Table 1): type 1, B. vietnamiensis; type 2, B. cepacia genomovars I and III and B. stabilis; and type 3, B. multivorans (Fig. 1A). Polymorphism detected by ARDRA analysis of the 16S rDNA was consistent with the taxonomic classification of the new species B. multivorans and B. vietnamiensis (28) but was not sufficient to separate B. cepacia genomovars I and III (28) and the new species B. stabilis (29).
FIG. 1.
RFLP analysis of the 16S rRNA gene. (A) Analysis of the 16S rRNA gene of strains from the B. cepacia complex. Lanes: 1, LMG 13010T; 2, C1576; 3, LMG 10929T; 4, LMG 16232; 5, ATCC 25416; 6, ATCC 17759; 7, C5424; 8, C1394; 9, CEP511; 10, LMG 07000; and 11, LMG 14294. The genomovar status of each strain is indicated above the lane numbers, and the numerical 16S rRNA RFLP type is shown below each lane. Molecular size markers are shown in lane M (100-bp ladder). (B) Analysis of 16S rRNA genes of other bacterial species which may be misidentified as B. cepacia or grow on BCSA (10). Reference strains from Henry et al. (10): Lanes: 1, B. gladioli (Bg) CEP82; 2, B. gladioli CEP89; 3, R. picketti (Rp) ATCC 27511; 4, R. picketti ATCC 49129; 5, C. meningosepticum (Cm) FC113; 6, C. meningosepticum FC224; 7, S. maltophilia (Sm) CEP272; 8, S. maltophilia C4525; 9, C. acidovorans (Ca) FC77; 10, C. acidovorans CEP145. In panel A, the numerical 16S rRNA RFLP type of each isolate is shown below each lane.
Although the 16S rDNA ARDRA method was limited with regard to its differentiation of the B. cepacia complex, it is a versatile means of rapid identification and discrimination for other bacterial species which may be recovered from CF patient sputum (24). Burkholderia gladioli, Ralstonia picketti, Chryseobacterium meningosepticum, Stenotrophomonas maltophilia, and Commomonas acidovorans are examples of organisms which in clinical settings have been incorrectly identified as members of the B. cepacia complex (3, 10). ARDRA analysis of the 16s rDNA with DdeI of the latter species is shown in Fig. 1B. Each of these microbial species was easily distinguished from bacteria of the B. cepacia complex by ARDRA analysis (Fig. 1).
PCR amplification of B. cepacia complex recA gene.
Two recA sequences, each from a different strain of B. cepacia, were available within GenBank. The genomovar status of B. cepacia strain JN25, from which one recA sequence (GenBank accession no. D90120) was derived, was not known; it was an isolate of clinical origin characterized in Japan in 1990 (19). The second recA sequence (accession no. U70431) was derived from the B. cepacia reference strain ATCC 17616 (31); this strain has been classified as the new species B. multivorans (28). Each sequence was aligned using BLAST (1). PCR primers BCR1 and BCR2 (Table 2) were designed from homologous sequences at the 5′ and 3′ ends of the recA open reading frame. These primers amplified a single 1-kb amplicon from all 35 strains representative of the B. cepacia complex (Table 1). The identity of the 1-kb fragment was subsequently confirmed to be recA by direct nucleotide sequence analysis of PCR products (see below). A further 68 isolates, each with biochemical- (10) and genomovar-specific (28) properties characteristic of the B. cepacia complex, also tested positive with these recA primers. PCR with primers BCR1 and BCR2 failed to amplify PCR products from the following species: B. gladioli, R. picketti, C. meningosepticum, S. maltophilia, C. acidovorans, Escherichia coli, and Pseudomonas aeruginosa (data not shown).
RFLP analysis of the B. cepacia complex recA gene.
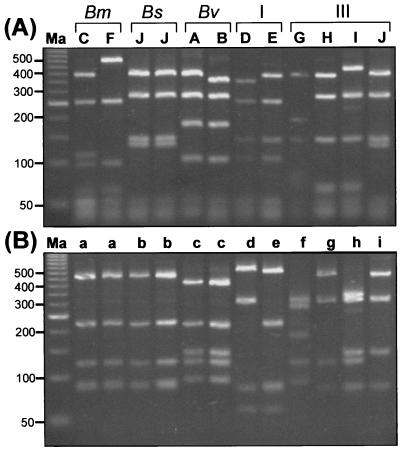
RFLP analysis with AluIII, BsaWI, MnlI, and HaeIII was investigated. RFLP types generated by digestion with HaeIII were the most discriminatory among the four restriction enzymes (data not shown). Ten distinct HaeIII RFLP patterns were found among the 35-isolate B. cepacia complex panel initially examined, and each pattern was assigned an alphabetical code (Fig. 2; Table 1). HaeIII RFLP analysis was capable of discriminating among all five genomovars (Fig. 2A) except for RFLP pattern J, which was shared by the genomovar III strain PC184 and all of the B. stabilis strains examined. To distinguish genomovar III and B. stabilis strains with this RFLP type, analysis with an additional enzyme (such as MnlI [Fig. 2B]) was required.
FIG. 2.
RFLP analysis of the B. cepacia complex recA amplified by PCR. (A) The alphabetical RFLP type is shown above each lane. Lanes: (left to right) C, C1576; F, ATCC 17616; J, LMG 14086; J, LMG 14294; A, FC441; B, LMG 10929T; D, ATCC 25416; E, ATCC 17759; G, K56-2; H, C1394; I, CEP511; and J, PC184. The genomovar status of each strain is indicated above the relevant lane. Molecular size standards (50-bp ladder) are in lane Ma. B. MnlI RFLP analysis of the recA gene. The designated Mnl RFLP type is shown above each lane. Molecular size standards (50-bp ladder) are in lane Ma.
RFLP analysis with MnlI revealed eight patterns among the 35 B. cepacia complex isolates (Table 1). Single-signature RFLP types were produced for the B. multivorans, B. vietnamiensis, and B. stabilis strains examined (Fig. 2B; Table 1). The B. stabilis and B. multivorans MnlI RFLP types (patterns a and b, respectively) were only slightly different, with the second largest RFLP fragment being consistently smaller in size among the B. stabilis strains examined (Fig. 2B). The same number of distinct patterns as observed with HaeIII were also obtained by MnlI digestion for the remaining genomovar I and III strains. Genomovar III strain PC184 produced a distinct MnlI recA RFLP type (pattern i [Fig. 2; Table 1]) which was easily distinguished from the MnlI RFLP type of B. stabilis (pattern b [Fig. 2; Table 1]). The remaining 68 B. cepacia complex isolates examined were all analyzed using HaeIII for RFLP analysis of their amplified recA genes (Table 3).
TABLE 3.
Evaluation of genomovar-specific primers on a collection of genetically diverse B. cepacia complex strains
| PCR primer set specificity | No. of isolates testing positive with each primer set | recA HaeIII RFLP type within positive isolates | No. of isolates within each RFLP type | No. of genetically heterogeneous strains within each RFLP type | 16s rDNA RFLP type (DdeI) |
|---|---|---|---|---|---|
| Genomovar I | 8 | D | 5 | 3 | 2 |
| E | 3 | 3 | 2 | ||
| B. multivorans | 20 | C | 3 | 3 | 3 |
| F | 17 | 16 | 3 | ||
| Genomovar III-A | 16 | G | 16 | 8 | 2 |
| Genomovar III-B | 24 | H | 4 | 3 | 2 |
| I | 10 | 8 | 2 | ||
| J | 10 | 7 | 2 | ||
| B. stabilis | 22 | J | 22 | 4 | 2 |
| B. vietnamiensis | 13 | A | 6 | 5 | 1 |
| B | 7 | 6 | 1 | ||
| Total | 103 | 10 | 103 | 66 |
Nucleotide sequence analysis of recA.
To confirm the sequence variation detected by RFLP analysis of the amplified recA gene and facilitate the design of PCR primers specific to each B. cepacia genomovar, complete nucleotide sequence analysis of the gene was performed for 20 strains representative of each recA RFLP type (Table 1). The complete recA gene was sequenced in two 500-bp segments using the combinations of PCR primers BCR1, BCR2, BCR3, and BCR4 described in Table 2. The sequence of each segment was combined to produce the full-length recA. Sequence analysis of the recA amplicon of B. multivorans ATCC 17616 was performed as a control for the sequencing strategy; the sequence determined in this study was identical to the published sequence for this strain (31). BLAST analysis (1) of each nucleotide sequence confirmed that each encodes RecA with homology to the published B. cepacia sequences (19, 31). Computational analysis of the HaeIII restriction sites within each nucleotide sequence matched those HaeIII sites determined by RFLP analysis of the complete recA amplicon for each strain. Sequence-based restriction mapping of recA from genomovar III strain PC184 and the B. stabilis strains demonstrated the presence of closely overlapping HaeIII sites. However, seven cleavage sites were present in PC184 and six in the B. stabilis recA, and each of these recA sequences was quite different (see phylogenetic analysis below). The minor differences in the HaeIII fragment sizes were not distinguished under the electrophoresis conditions used for RFLP analysis (Fig. 2A).
Phylogenetic analysis of recA.
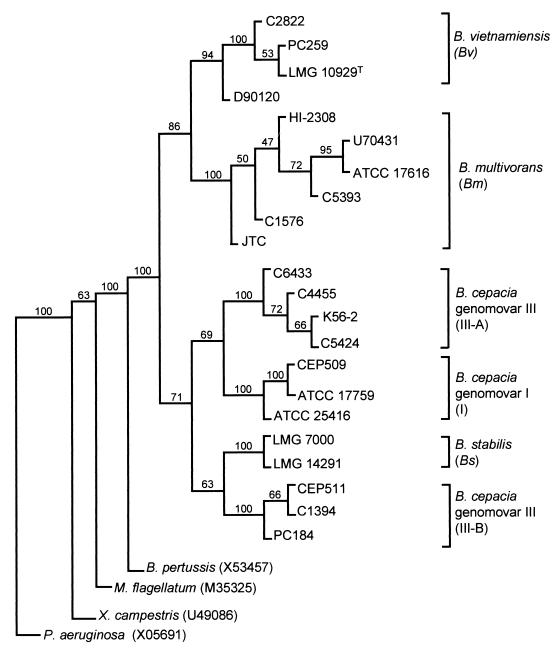
Construction of a recA phylogenetic tree of the B. cepacia complex was carried out as described in Materials and Methods. The Bordetella pertussis recA gene (accession no. X53457) was used to root the tree because previous molecular systematic analysis of bacterial recA genes had demonstrated that it was closely related to B. cepacia recA and had placed both bacteria within the β-subgroup of the Proteobacteria (5, 11). The P. aeruginosa (accession no. X05691), Methylobacillus flagellatum (accession no. M35525), and Xanthomonas campestris (accession no. U49086) recA sequences were also included in the phylogenetic analysis as indicators of the ability of the analysis to differentiate among unrelated species. The resulting nucleotide sequence-based phylogenetic tree is shown in Fig. 3. All five previously determined genomovars (28) formed distinct arms within the tree, consistent with their proposal as new species (28, 29) or unnamed genomic species awaiting formal binomial classification (28). In total, six distinct phylogenetic groups were present within the recA tree and were given the following designations as recA groups (Fig. 3): I, B. cepacia genomovar I; III-A and III-B, B. cepacia genomovar III recA clusters A and B, respectively; Bs, B. stabilis; Bm, B. multivorans; and Bv, B. vietnamiensis. These B. cepacia complex phylogenies were also distinct from the unrelated bacterial species chosen as out groups for the tree (Fig. 3). Phylogenetic trees based on the protein translation of each recA nucleotide sequence also demonstrated the same clustering of genomovars and new species (data not shown).
FIG. 3.
Phylogenetic tree of the B. cepacia complex based on the complete recA gene sequence. Multiple sequence alignment was performed on these genes and the published recA sequences from B. pertussis, M. flagellatum, X. campestris, and P. aeruginosa. The phylogenetic tree was rooted with the B. pertussis recA gene. Bootstrapping resampling statistics were applied to the tree (100 data sets), and bootstrap values are shown on each horizontal limb of the tree. The genomovar status of each B. cepacia complex strain is indicated on the right of the figure (the corresponding recA phylogenetic group is shown in brackets).
The separation of B. multivorans, B. vietnamiensis, and B. stabilis strains within the recA tree (Fig. 3) was consistent with their designation as new species (28, 29). The published sequence of strain JN25 (21) aligned closely with the B. vietnamiensis recA sequences. Separation of B. cepacia complex genomovars I and III was also apparent in the recA phylogenetic tree and consistent with their designation as distinct genomovars (28), except for the subdivision of isolates which had previously been identified as genomovar III by protein profile analysis (27, 28). RecA group III-A (Fig. 3) included epidemic CF strains from the cable pilus-encoding lineage (18, 22) and Vancouver outbreaks (16, 18). RecA group III-B (Fig. 3) contained epidemic CF strains from outbreaks in Cleveland, Ohio, in Manchester, England, and in Australia (18). B. cepacia genomovar III strain PC184, which shared the same recA HaeIII RFLP as B. stabilis (Fig. 2), did not align with the latter (Fig. 3), clearly demonstrating the presence of genomovar III-specific sequence variation in the complete recA gene.
Genomovar-specific PCR.
Using the nucleotide sequence alignment of the 20 novel recA genes determined above and the two within the databases (19, 31), specific PCR primers were designed to detect the six recA-derived clusters of the B. cepacia complex identified by phylogenetic analysis (see Fig. 3). Primer pairs for the identification of recA groups I, III-A, III-B, B. multivorans, B. stabilis, and B. vietnamiensis are listed in Table 2. Each primer pair was designed to match all sequences determined within each phylogenetic subgroup (see Fig. 3) and be mismatched at the 3′ base (and as many other bases as possible) with all other B. cepacia complex recA sequences (Table 2). Each primer set was tested on all 103 B. cepacia complex isolates. Amplification products of the correct size (Table 2) were obtained from strains of the appropriate recA group with each specific primer pair (data not shown). No products of the predicted size were seen in strains outside the target group for each primer pair (data not shown). The identity of each recA group-specific PCR product was confirmed to be the appropriate portion of the recA gene by direct sequence analysis of the PCR product for each of the following strains: B. vietnamiensis PC259, B. multivorans ATCC 17616, B. stabilis LMG 7000, B. cepacia ATCC 25416 (I), B. cepacia C5424 (III-A), and B. cepacia C1394 (III-B) (data not shown). The recA-specific PCR, HaeIII recA RFLP analysis, and 16S ARDRA results obtained from all 103 B. cepacia complex isolates are summarized in Table 3.
The majority of 103 strains examined were genetically heterogeneous (66 out of 103) and possessed unique RAPD or PFGE fingerprints (Table 3). Each recA primer set was 100% specific for isolates which possessed recA HaeIII RFLP types (Table 1) characteristic of the respective new species, conventional genomovar, or recA phylogenetic subgroup for which they were designed (Table 3). RFLP types and specific PCR results remained stable for sequential or multiple isolates of a given genetic strain type. ARDRA profiles obtained with the 16S rDNA primers also correlated as expected with the recA-specific primer results (Table 3).
Genomic location of recA.
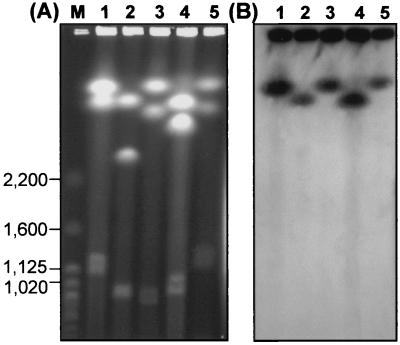
Given the multichromosomal structure of the B. cepacia complex genome (4, 12, 21), it was important to determine the copy number and genomic location of recA. Previous studies on B. cepacia genomovar I strain ATCC 25416 (Table 1) demonstrated the presence of two copies of the gene, one on each large chromosome (21). Electrophoretic separation of linear replicons from B. multivorans genomovars I, III-A, and III-B and B. stabilis strains is shown in Fig. 4A. Each genomovar possessed a multireplicon structure, with between two and four large replicons, ranging from approximately 0.6 to 3.8 Mb in size, being detected (data not shown for B. vietnamiensis). For each B. cepacia complex genomovar tested, only the largest chromosome produced a positive signal when probed with recA by Southern hybridization (Fig. 4B). Further analysis of the genomic location of the recA gene using RFLP approaches was also consistent with the presence of only one copy of the recA gene within the B. cepacia complex genome (data not shown).
FIG. 4.
Genomic location of the recA gene in strains of the B. cepacia complex. (A) Lanes: M, molecular size standards (S. cerevisiae chromosomes; relevant size bands are indicated in kilobases); 1, strain ATCC 25416 (genomovar I); 2, B. multivorans strain 17616; 3, strain C5424 (recA group III-A); 4, strain C1394 (recA group III-B); and 5, B. stabilis strain LMG 14291. (B) Southern hybridization of separated chromosomes from panel A with a recA gene probe from B. cepacia strain K56-2 (genomovar III-A). A positive hybridization signal was obtained from the largest chromosome in each strain examined.
DISCUSSION
Systematic analysis of two conserved genes, 16S rDNA and recA, in strains of the B. cepacia complex demonstrated that each may have a use in the identification and classification of this important group of opportunistic pathogens. Although widely used for bacterial identification, analysis of the 16S rDNA of the B. cepacia complex by an ARDRA approach demonstrated that this gene was useful for discrimination of the two new species, B. vietnamiensis and B. multivorans, but insufficient to separate genomovars I and III and B. stabilis. Because of these discriminatory limitations, a novel PCR-RFLP analysis was developed based on the B. cepacia recA gene. Nucleotide sequence polymorphism was identified in recA which was sufficient to identify all five current B. cepacia complex genomovars defined by a polyphasic approach (28). This study validates the use of nonribosomal housekeeping genes in the development of alternative molecular diagnostic strategies, especially among taxonomically complex species such as B. cepacia. A complete nucleotide sequence-based identification approach based on this conserved, stable, and single-copy gene was developed.
Four approaches based on the B. cepacia complex recA gene may be used to identify the genomovar status of clinical isolates and, depending on the resources of the diagnostic laboratory, they may be applied as individual tests or multiple complementary analyses. First, amplification of recA with primers BCR1 and BCR2 can be used as an initial means of placing an isolate within the B. cepacia complex since cross-reaction of these primers with other species commonly found in CF sputum was not detected. In the future, direct testing and application of these B. cepacia complex-specific primers to CF sputum, which contains multiple bacterial species, may also be possible. Second, after successful amplification of recA, RFLP with HaeIII and MnlI may be used to place the isolate within a specific genomovar or recA group (Table 1). Third, the design of recA group-specific primers capable of identifying all of the genomovars within the current B. cepacia complex (28, 29) provides a means of genomovar identification in a single test. With further development such recA-based primer sets may be employed in a single multiplex PCR for rapid genomovar detection. Finally, nucleotide sequence determination of recA provides a powerful means of both identification and classification of these poorly defined bacteria. Overall, the availability of several complementary tests based on a single diagnostic gene provides a robust approach to B. cepacia complex identification.
Phylogenetic analysis of recA from a range of bacteria has demonstrated that the gene may be very useful for the separation of closely related species and may define evolutionary trees that are consistent with those observed for rRNA genes (5, 11). Its application herein confirms these observations and illustrates the benefits of examining protein-encoding DNA when systematic analysis of ribonucleotide-encoding sequences fails to yield discriminatory classifications among closely related species. Analysis of a single genomic locus, recA, in this study was consistent with the polyphasic taxonomic approach (27) originally used to define the B. cepacia complex genomovars, with the exception of the phylogenetic subdivision of organisms classified as genomovar III by DNA-DNA hybridization experiments (28). The taxonomic significance of the subdivision of genomovar III-classified strains into recA groups III-A and III-B awaits further research. In addition B. cepacia complex strains possessing novel recA RFLPs, not reported in this study, have recently been identified (E. Mahenthiralingam and P. Vandamme, unpublished data); preliminary polyphasic analysis (27) of these strains indicates that they are members of novel genomic species within the complex (Mahenthiralingam and Vandamme, unpublished data). The ability to use recA analysis as a means to assist taxonomic classification of the B. cepacia complex was demonstrated in the recent proposal of B. stabilis (formerly genomovar IV) as a new species (29) and is also corroborated by the data presented in this study.
Analysis of recA also revealed new insights into the epidemiology and pathogenesis of the B. cepacia complex. Concerns about commercial use of these bacteria (9, 14) are confirmed by the recA phylogenetic analysis. Strains isolated from clinical infection and those recovered from the natural environment cluster within the B. multivorans, B. vietnamiensis, and B. cepacia genomovar I arms of the evolutionary tree (Fig. 3). In addition, among the 103 isolates screened, there were clinical and environmental strains of B. cepacia genomovar III-B which shared the same RFLP type (Table 3) and hence would cluster phylogenetically if nucleotide sequence analysis were subsequently performed. Only within recA group III-A and B. stabilis were no environmental isolates found in this study. The results of recA analysis corroborate the taxonomic findings of Vandamme et al. (28) which demonstrate that all genomovars within the B. cepacia complex can cause human opportunistic infection. Phylogenetic distinctions between environmental and clinical strains do not appear to exist, and all strains within the complex appear to possess conserved traits which enable them to cause infection in vulnerable individuals.
Determination of the phylogeny of the B. cepacia complex enabled the association of known epidemiological features with each recA subgroup to be examined. The cable pilus gene was primarily associated with genomovar III strains of the ET12 lineage (22), such as K56-2 and C5424, which clustered in recA group III-A (Table 1; Fig. 3). However, one unique strain recovered from an individual CF patient at a center in Australia, which possessed DNA homologous to cblA, was identified as recA HaeIII RFLP type E and B. cepacia genomovar I by specific PCR (Table 3). Therefore, possession of cblA still appears to be a rare feature for the B. cepacia complex (17), but it is not solely possessed by genomovar III strains of the ET12 lineage (22). The prevalence of the BCESM within the B. cepacia complex recA phylogeny also appears more widespread that originally observed (17). The marker was associated with strains that had spread among patients with CF when it was originally described (17); however, unlike cblA, no specific virulence trait has been associated with the BCESM locus. From the results of this study, DNA homologous to the BCESM appears to be primarily associated with epidemic CF strains belonging to recA groups III-A and III-B (all classified as genomovar III) (18). However, BCESM was also found in B. vietnamiensis strain ATCC 29424, but none of the other B. vietnamiensis strains examined (Table 3), suggesting that it may occasionally occur outside of the highly transmissible genomovar III strains (16–18). The recA probes described herein provide an additional means of identifying strains with an epidemiological precedent (16–18) for patient-to-patient spread among individuals with CF. The recA group III-A- and III-B-specific primers detect strains which have caused several outbreaks (16–18) and may ultimately prove to be better diagnostic probes than the not-yet fully-defined BCESM DNA (17). Early diagnosis of infection by B. cepacia recA PCR may in future facilitate rapid-enactment infection control measures and aggressive therapy to improve the poor outcome associated with B. cepacia infection in patients with CF (13).
In conclusion, nucleotide sequence analysis of recA is a rapid and reproducible means of identifying all of the current genomovars and new species within the B. cepacia complex. Rapid identification of bacteria recovered from patients with CF and from other vulnerable individuals is vital if we are to further understand the clinical risks posed by each genomovar and new species within the B. cepacia complex.
ACKNOWLEDGMENTS
We thank David Speert for his support and helpful discussion and for providing access to the Canadian B. cepacia Strain Repository. Deborah Henry is acknowledged for assistance in biochemical analysis and identification of B. cepacia strains. We thank Gary Probe, Richard Parkes, and Julie Fadden for excellent technical assistance.
This work was funded by grants from the Canadian Cystic Fibrosis Foundation and UK Cystic Fibrosis Trust (E.M., project grant PJ472) and the Fund for Scientific Research, Belgium (P.V.). E.M. acknowledges the British Columbia Lung association for provision of a Career Development Award and the British Columbia Research Institute for Children's and Women's Health for an Investigator Establishment Award.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D L. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Scheider I, Jungwirth R, Roller C. Discrimination of Burkholderia species detectable in cystic fibrosis patients by PCR. J Clin Microbiol. 1998;36:2748–2751. doi: 10.1128/jcm.36.9.2748-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdge D R, Noble M A, Campbell M E, Krell V L, Speert D P. Xanthomonas maltophilia misidentified as Pseudomonas cepacia in cultures of sputum from patients with cystic fibrosis: a diagnostic pitfall with major clinical implications. Clin Infect Dis. 1995;20:445–448. doi: 10.1093/clinids/20.2.445. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H-P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisen J A. The RecA protein as a model molecule for systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 7.Gonzalez C F, Pettit E A, Valadez V A, Provin E M. Mobilization, cloning and sequence determination of a plasmid encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas) cepacia. Mol Plant-Microbe Interact. 1997;10:840–851. doi: 10.1094/MPMI.1997.10.7.840. [DOI] [PubMed] [Google Scholar]
- 8.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aerugionosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govan J R W, Vandamme P. Agricultural and medical microbiology—a time for bridging gaps. Microbiology. 1998;144:2373–2375. doi: 10.1099/00221287-144-9-2373. [DOI] [PubMed] [Google Scholar]
- 10.Henry D A, Campbell M E, LiPuma J J, Speert D P. Identification of Burkholderia cepacia from patients with cystic fibrosis and a new selective medium for its isolation. J Clin Microbiol. 1996;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlin S, Weinstock G M, Brendel V. Bacterial classifications derived from RecA protein sequence comparisons. J Bacteriol. 1995;177:6881–6893. doi: 10.1128/jb.177.23.6881-6893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessie T G, Hendrickson W, Manning B D, Devereux R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 13.LiPuma J J. Burkholderia cepacia—management issues and new insights. Clin Chest Med. 1998;19:473–486. doi: 10.1016/s0272-5231(05)70094-0. [DOI] [PubMed] [Google Scholar]
- 14.LiPuma J J, Mahenthiralingam E. Commercial use of Burkholderia cepacia: are there additional threats? Emerg Infect Dis. 1999;5:5–6. doi: 10.3201/eid0502.990226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for the identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by random amplified polymorphic DNA (RAPD) fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic strains of Burkholderia cepacia recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahenthiralingam E, Coenye T, Chung J, Speert D P, Govan J R W, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakazawa T, Kimoto M, Abe M. Cloning, sequencing and transcriptional analysis of the recA gene of Pseudomonas cepacia. Gene. 1990;94:83–88. doi: 10.1016/0378-1119(90)90471-3. [DOI] [PubMed] [Google Scholar]
- 20.Revets H, Vandamme P, Van Zeebroeck A, De Boeck K, Streulens M J, Verhaegen J, Ursi J P, Verschaegen G, Franckx H, Malfroot A, Dab I, Lauwers S. Burkholderia (Pseudomonas) cepacia and cystic fibrosis: the epidemiology in Belgium. Acta Clin Belg. 1996;51:222–230. doi: 10.1080/22953337.1996.11718514. [DOI] [PubMed] [Google Scholar]
- 21.Rodley P D, Römmling U, Tümmler B. A physical genome map of the Burkholderia cepacia type strain. Mol Microbiol. 1995;17:57–67. doi: 10.1111/j.1365-2958.1995.mmi_17010057.x. [DOI] [PubMed] [Google Scholar]
- 22.Sajjan U S, Sun L, Goldstein R, Forstner J F. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning—a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speert D P, Bond M, Woodman R C, Curnutte J T. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J Infect Dis. 1994;170:1524–1531. doi: 10.1093/infdis/170.6.1524. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 29.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Pelt C, Verduin C M, Goessens W H F, Vos M C, Tümmler B, Segonds C, Reubsaet F, Verbrugh H, van Belkum A. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Waasbergen L G, Kidambi S P, Miller R V. Construction of a recA mutant of Burkholderia (formerly Pseudomonas) cepacia. Appl Microbiol Biotechnol. 1998;49:59–65. doi: 10.1007/s002530051137. [DOI] [PubMed] [Google Scholar]