Abstract
Tyrosine kinase inhibitors of anaplastic lymphoma kinase (ALK-TKIs) including alectinib have been the standard therapy against ALK fusion gene-positive non–small cell lung cancers (NSCLCs). Many ALK fusion variants have been identified in NSCLCs, and the predominant variants are echinoderm microtubule-associated protein-like 4-ALK (EML4-ALK) variant 1 (V1), V2 and V3a/b. However, there have been conflicting reports on the clinical responses of these variants to ALK-TKIs, and there are few reports on other less common ALK variants. To examine the influence of ALK variants on the efficacy of ALK-TKIs, we analyzed the sensitivity to alectinib of eight types of ALK variant: three major variants (V1, V2 and V3a) and five less common variants (V4; kinesin family member 5-ALK; kinesin light chain 1-ALK; striatin, calmodulin-binding protein-ALK; and tropomyosin-receptor kinase fused gene-ALK). Analysis was done by cell-free kinase assays using the recombinant proteins and by cell, growth assays using murine Ba/F3 cells expressing ALK variants. The kinase activity of each recombinant protein was significantly inhibited by alectinib. Intracellular ALK phosphorylation levels and its downstream signaling mediators, STAT3 and ERK, were suppressed by alectinib in each ALK variant-expressing Ba/F3 cell. Each cellular proliferation was markedly inhibited by alectinib treatment. There was no significant difference in the IC50 values between cells, with a <3.6-fold difference in responsiveness. In conclusion, these eight ALK variants had similar sensitivity to alectinib in vitro, indicating that it may not be possible to predict the response to alectinib just by determination of the ALK variant type in ALK fusion-positive NSCLCs.
Keywords: alectinib, ALK, EML4, fusion, KIF5B, KLC1, lung cancer, STRN, TFG, variant
Introduction
Genetic rearrangement in the anaplastic lymphoma kinase (ALK) gene occurs in approximately 8% of non–small cell lung cancers (NSCLCs) [1]. Patients with ALK-fusion-positive (ALK+) NSCLC have been found to have improved clinical outcomes when treated with ALK tyrosine kinase inhibitors (ALK-TKIs), which has led to the approval of agents such as crizotinib, ceritinib, alectinib, brigatinib and lorlatinib [2]. More than 90 distinct ALK fusion variants have been identified in ALK+ NSCLC [3]. The most common variants are echinoderm microtubule-associated protein-like 4 (EML4)-ALK variant 1 (V1), variant 2 (V2) and variants 3a and 3b (V3a/b), resulting from inversion at different breakpoints of EML4 fused to ALK on chromosome 2; these occur with frequencies of 42.7, 10.5 and 37.1%, respectively [4,5].
In previous clinical studies with crizotinib or lorlatinib, patients with V3a/b seem to have had significantly shorter progression-free survival (PFS) than did patients with V1 [6,7]. However, these results conflict with those of other studies with crizotinib or alectinib that report insignificant differences in PFS among patients with V1, V2 and V3a/b [5,8,9]. In addition, there is a very limited amount of clinical data available with which to examine whether or not minor uncommon ALK variants (accounting for about 10% of all variants other than V1, V2 and V3a/b) affect the response to ALK-TKIs.
In this preclinical study, we analyzed the effects of alectinib on major ALK variants as well as on minor variants. With reference to the Catalogue of Somatic Mutations in Cancer database version 81 released in 2017 (https://cancer.sanger.ac.uk/cosmic) and to the reports of Shaw et al. and Sasaki et al. [10,11], we selected eight different ALK fusion variants in ALK+ NSCLC: V1; V2; V3a; EML4-ALK variant 4 (V4); kinesin family member 5 (KIF5B)-ALK; kinesin light chain 1 (KLC1)-ALK; striatin, calmodulin-binding protein (STRN)-ALK; and tropomyosin-receptor kinase fused gene (TFG)-ALK.
Materials and methods
Reagents and cell line
Alectinib was synthesized in Chugai Pharmaceutical Co., Ltd. (Tokyo, Japan), and crizotinib was obtained from Selleck Chemicals (Houston, Texas, USA) [12]. They were dissolved in DMSO. The murine bone marrow-derived pro B cell line Ba/F3 was purchased from the Riken Bioresource Center (Ibaraki, Japan), and maintained in RPMI1640 (Sigma-Aldrich Co., Ltd., St. Louis, Missouri, USA) supplemented with 10% FBS (Sigma-Aldrich Co., Ltd.), 1 ng/mL mouse IL-3 protein (R&D Systems, Minneapolis, Minnesota, USA) at 37°C in 5 % CO2.
Expression constructs
Human cDNAs for the eight variants (V1, V2, V3a, V4, KIF5B-ALK, KLC1-ALK, STRN-ALK and TFG-ALK) were synthesized by GenScript (Tokyo, Japan), and were each cloned into the cloning site located downstream of the cytomegalovirus constitutive promoter in the pIRES plasmid vector (Takara Bio, Shiga, Japan). This vector also contains a neomycin resistance gene to select for stable integration of the ALK variant gene in Ba/F3 cells.
In vitro cell-free kinase assay
N-terminal glutathione S-transferase-tagged human recombinant proteins of all eight ALK variants expressed by baculovirus in Sf9 insect cells were obtained from Signalchem (Richmond, BC, Canada) (Table 1). Kinase activity following treatment with or without 1000 nM alectinib for 1 h was measured with an ADPsensor universal kinase activity assay kit (BioVision, Milpitas, California, USA) according to the manufacturer’s instructions.
Table 1.
ALK fusion variants in this study
| ALK variants | CDS reference | Catalog number of recombinantsc |
|---|---|---|
| V1 | AB274722.1a | A19-19G |
| V2 | AB275889.1a | A19-19HG |
| V3a | AB374361.1a | A19-19FG |
| V4 | AB374363.1a | A19-19GG |
| KIF5B-ALK | AB462413.1a | A19-19MG |
| KLC1-ALK | 22347464b | A19-19IG |
| STRN-ALK | 24475247b | A19-19JG |
| TFG-ALK | 18083107b | A19-19KG |
CDS of eight ALK variants in this study were referenced from RefSeq database
a and COSMIC database
b. Recombinant ALK variant proteins were obtained from Signalchem
c. CDS, coding sequence.
Establishment of ALK variant-expressing cells
Each of the vectors coding ALK fusion variants was individually transfected into Ba/F3 cells by NEPA21 electroporation (Nepa Gene, Chiba, Japan). After 2 days of transfection, the culture medium was replaced with medium containing 500 µg/ml G418 (Nacalai Tesque, Kyoto, Japan) for 13 days to select ALK-variant-expressing cells. An empty control vector was also transfected into Ba/F3 cells as described above.
Cell proliferation assay
Cells were seeded in 96-well plates in a culture medium without G418 and mouse IL-3, and the following day, alectinib was added at the indicated concentrations. After 4 days, viability was determined by quantitation of ATP present in the cell, which signals the presence of metabolically active cells, using CellTiter-Glo 3D cell viability assay (Promega, Madison, Wisconsin, USA).
RT-PCR assay
RNA was obtained from the cells by using a SuperPrep II Cell Lysis & RT kit for qPCR (Toyobo, Osaka, Japan), and the cDNA was synthesized using a PrimeScript RT reagent kit (Takara Bio). Real time-PCR was performed using the LightCycler 480 Instrument II (Roche Diagnostics, Basel, Switzerland) and Taqman probes for the neomycin resistance gene and mouse GAPDH (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The expression levels of the neomycin resistance gene normalized to mouse GAPDH were calculated using the LightCycler 480 software (Roche Diagnostics).
Western blotting assay
Cells were treated with or without 100 nM alectinib for 3 h. The same amount of protein lysate was loaded for each Western blotting assay using the Sally Sue capillary electrophoresis–based protein analysis system (ProteinSimple, Santa Clara, California, USA). Antibodies against ALK, phospho-ALK (Y1282/1283), ERK, phospho-ERK (T202/Y204), AKT, phospho-AKT (T450), STAT3, phospho-STAT3 (Y705) and β-actin (Cell Signaling Technology, Danvers, Massachusetts, USA) were used.
Statistical analysis
Analysis of statistical significance was performed using JMP version 15.0.0 software (SAS Institute, Tokyo, Japan). Significance of treatment with alectinib as compared with vehicle control in in vitro kinase assays was determined by Student’s t-test. Significant difference in all pairwise comparisons of neomycin resistance gene expression among all eight ALK-variant-expressing cells was determined by Tukey–Kramer’s HSD test. P < 0.05 indicates a statistically significant result and was labeled with asterisks.
Results
Inhibition of cell-free kinase activity of recombinant ALK variants by alectinib
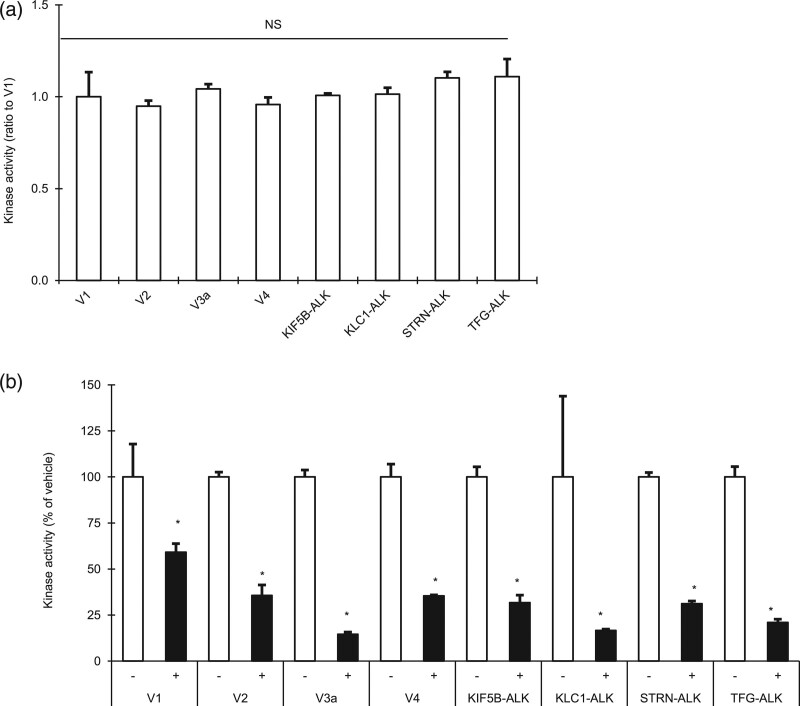
To examine the effect of alectinib on ALK variants identified in ALK+ NSCLC, we selected eight types of ALK variants (V1, V2, V3a, V4, KIF5B-ALK, KLC1-ALK, STRN-ALK and TFG-ALK). In cell-free kinase assay using recombinant proteins, none of the eight recombinant variants showed any significant difference in kinase activity (Fig. 1a), and the kinase activity of each was significantly inhibited by alectinib (Fig. 1b).
Fig. 1.
Effect of alectinib on recombinant ALK fusion variants in vitro. (a) The kinase activities of eight recombinant ALK variants were measured and their activities relative to that of V1 were calculated. NS means no significant difference in any pairwise comparison between each variant by Tukey–Kramer’s HSD test. (b) The kinase activity was measured with alectinib (+) or without alectinib (−), and the relative activity (percentage) in the presence of alectinib with respect to activities in the presence of vehicle alone was calculated. Asterisks indicate a significant difference between them at P<0.05 by Student’s t test. Each point represents the mean + SD of triplicates.
Inhibition of kinase activity of intracellular ALK variants by alectinib
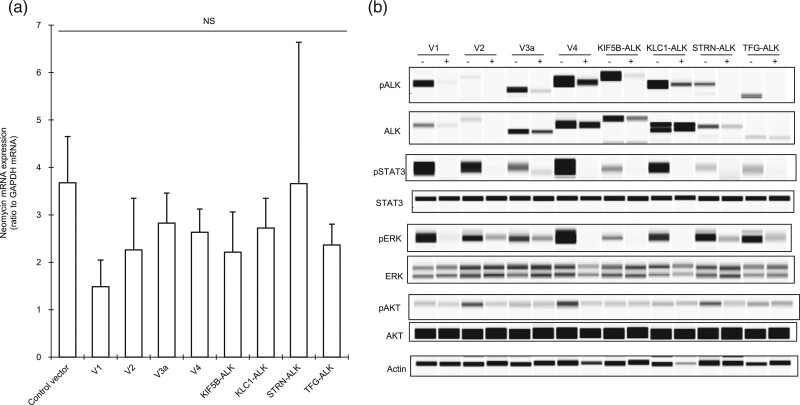
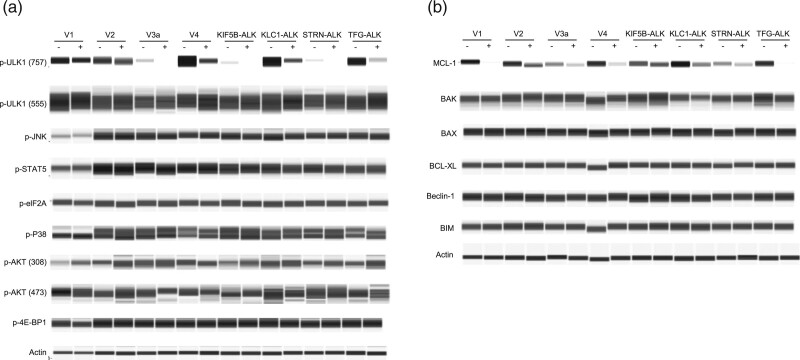
Next, to assess the effect of alectinib on the intracellular kinase activity of the ALK variants, we established eight types of stable cells, each expressing one of the eight types of ALK variant, by individually transfecting a vector containing each variant into the murine Ba/F3 cell line. We also established stable control cells by transfecting an empty vector. Since a neomycin resistance gene in the vector is also consistently transcribed by a constitutive promoter in the cell, the mRNA expression level of the neomycin resistance gene was thought to be a good marker with which to estimate the copy number of the vector as well as the ALK variant gene in each Ba/F3 cell after transfection. There was an insignificant difference in the mRNA level of the neomycin resistance gene between each of the nine cells (Fig. 2a), suggesting that the level of mRNA expression of each of the ALK variants would also be almost the same. Phosphorylation levels of the intracellular ALK variant and its downstream signaling mediators, STAT3 and ERK, were suppressed by alectinib in all eight ALK variant cells (Fig. 2b). In addition, the phosphorylation of Ser757 of ULK1 was suppressed, and the MCL-1 protein level was downregulated by alectinib in all variant cells (Fig. 3a,b). Decrease of ULK1 Ser757 phosphorylation promotes autophagy [13,14], and MCL-1 is one of the antiapoptotic Bcl-2 family members, indicating that alectinib could induce autophagy initiation as well as apoptosis in each variant cell regardless of ALK variant types. Therefore, not only major ALK variants V1, V2, and V3a but also minor variants V4, KIF5B-ALK, KLC1-ALK, STRN-ALK and TFG-ALK had constitutive kinase activity, which activates downstream oncogenic signaling via STAT3, ERK or ULK1. However, alectinib could inhibit these signaling pathways through suppression of ALK phosphorylation regardless of ALK variant types.
Fig. 2.
Effect of alectinib on intracellular ALK fusion variants. (a) RNA was extracted from stable Ba/F3 cells, and mRNA expression level of the neomycin resistance gene relative to that of GAPDH was measured by RT-PCR analysis. NS means no significant difference in any pairwise comparison between each ALK variant cell by Tukey–Kramer’s HSD test. Each point represents the mean + SD of quadruplicates. (b) Immunoblots of cell lysates from the ALK variant cells treated with (+) or without (−) alectinib.
Fig. 3.
Effect of alectinib on various signaling molecules of ALK variant cells. Immunoblots of cell lysates from the ALK variant cells treated with (+) or without (−) 100 nM alectinib for 3 h (a) or 24 h (b). Antibodies against phospho-JNK, phospho-P38, phospho-STAT5, phospho-eIF2A, phospho-4E-BP1, phospho-AKT (473), phospho-ULK1 (555 or 757), MCL-1, BCL-XL, BAK, Beclin-1, BIM (Cell Signaling Technology), BAX (SIGMA) and phospho-AKT (308) (Thermo Fisher Scientific) were used.
Inhibition of cell proliferation of ALK variant cells by alectinib
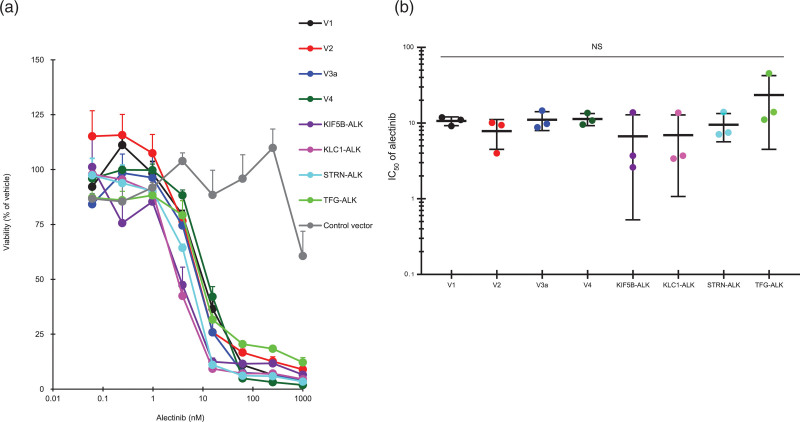
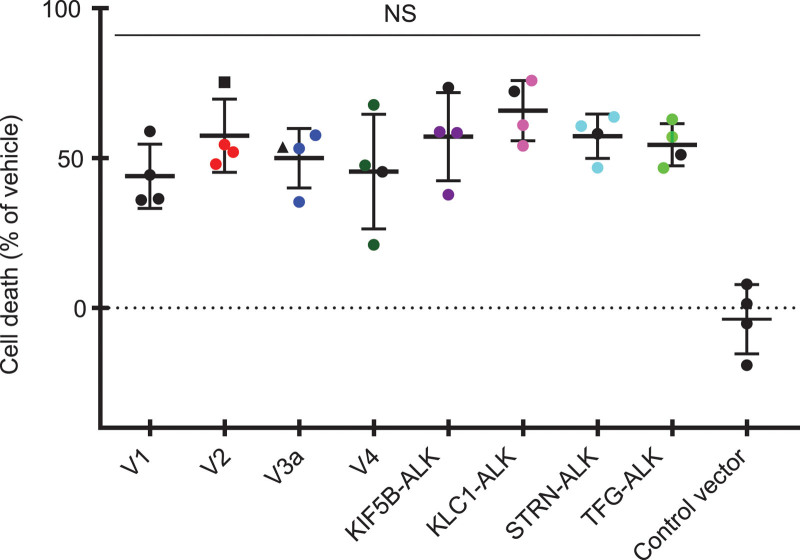
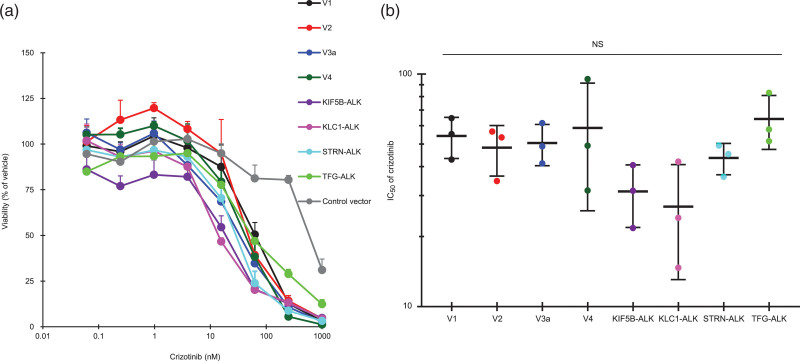
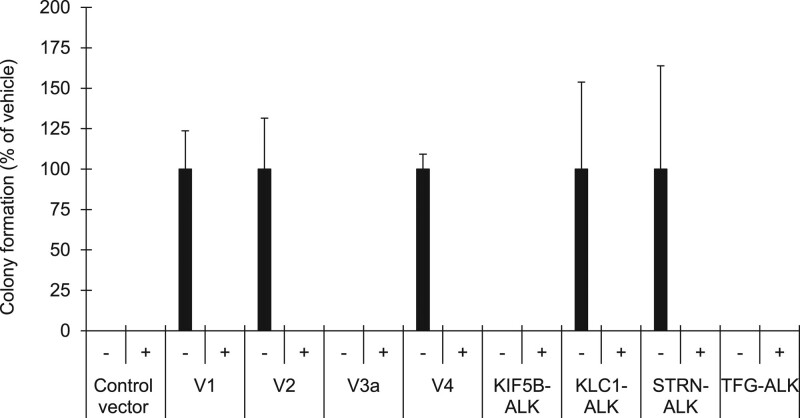
Finally, we examined the effect of alectinib on the growth of Ba/F3 cells expressing ALK variants. Proliferation of these ALK-variant-expressing cells was markedly suppressed by alectinib treatment for 4 days in a dose-dependent manner (Fig. 4a), and the IC50 values were not significantly different among them, with a <3.6-fold difference in responsiveness (Fig. 4b and Table 2). The suppression of proliferation of control cells was less than 40% at the maximum dose of 1000 nM alectinib (Fig. 4a). We investigated cell death by directly counting the numbers of viable and nonviable cells with double-fluorescent dye staining acridine orange and DAPI. The cell death ratio was not significantly different among all variant cells after treatment with alectinib at 10 nM for 4 days (Fig. 5). As was the case with alectinib, crizotinib treatment for 4 days also markedly suppressed cell proliferation of each ALK variant in a dose-dependent manner (Fig. 6a), and the IC50 values among them were not significantly different (Fig. 6b). Next, we investigated the effects of pretreatment of alectinib for 24 h on colony formation in semisolid agar media in a 6 day culture. Each colony of V1, V2, V4, KLC1-ALK, STRN-ALK variant cells was completely blocked by pretreatment with alectinib (Fig. 7). There was no colony in V3a, KIF5B-ALK, TFG-ALK variant cells with or without alectinib pretreatment. These results indicate that cell growth of all eight ALK-variant-expressing cells is immediately and similarly suppressed by treatment with ALK-TKIs regardless of ALK variant type.
Fig. 4.
Effect of alectinib on cell proliferation of ALK variant cells. (a) ALK-variant-expressing Ba/F3 cells or control Ba/F3 cells were cultured with or without alectinib, and their viability with alectinib relative to their viability with vehicle alone was measured. Each point represents the mean + SD of triplicates. (b) IC50 values of alectinib were calculated as described previously [20]. NS means no significant difference in any pairwise comparison between each ALK variant cell by Tukey–Kramer’s HSD test. Horizontal bars indicate the mean ± SD of triplicates.
Table 2.
IC50 of ALK variant-expressing cells
| ALK variant cells | IC50 (nM) |
|---|---|
| V1 | 10.7 ± 1.4 |
| V2 | 7.8 ± 3.3 |
| V3a | 11.1 ± 3.1 |
| V4 | 11.3 ± 2.1 |
| KIF5B-ALK | 6.7 ± 6.2 |
| KLC1-ALK | 7.0 ± 5.9 |
| STRN-ALK | 9.5 ± 3.9 |
| TFG-ALK | 23.5 ± 19.0 |
IC50 values of alectinib in ALK variant-expressing cells were calculated as described previously [18]. Each data represents the mean + SD of triplicates.
Fig. 5.
Effect of alectinib on cell death of ALK variant cells. ALK-variant-expressing Ba/F3 cells or control Ba/F3 cells were cultured with or without 10 nM alectinib for 4 days. Then, the cell numbers of viable and nonviable cells were counted with double-fluorescent dye staining acridine orange and DAPI of cell count & viability assay kit using Image Cytometer NucleoCounter NC-3000. The cell death ratio was calculated as follows: Cell death ratio % = 100−100× (viable cell numbers with alectinib treatment/viable cell numbers without treatment). NS means no significant difference in any pairwise comparison between each ALK variant cell by Tukey–Kramer’s HSD test. Horizontal bars indicate the mean ± SD of quadruplicates.
Fig. 6.
Effect of crizotinib on cell proliferation of ALK variant cells. (a) ALK-variant-expressing Ba/F3 cells or control Ba/F3 cells were cultured with or without crizotinib, and their viability with crizotinib relative to their viability with vehicle alone was measured. Each point represents the mean + SD of triplicates. (b) IC50 values of crizotinib were calculated as described previously [20]. NS means no significant difference in any pairwise comparison between each ALK variant cell by Tukey–Kramer’s HSD test. Horizontal bars indicate the mean ± SD of triplicates.
Fig. 7.
Effect of transient alectinib treatment on colony formation of ALK variant cells. ALK-variant-expressing Ba/F3 cells or control Ba/F3 cells were cultured with or without 100 nM alectinib for 24 h. After washing the cells, they were cultured in semisolid agar media for 6 days using CytoSelect 96-well in vitro tumor sensitivity assay kit (Cell Biolabs). Each point represents the mean + SD of triplicates.
Discussion
We found that the type of ALK fusion variant – V1, V2, V3a, V4, KIF5B-ALK, KLC1-ALK, STRN-ALK or TFG-ALK – did not affect the alectinib sensitivity of ALK-variant-transformed Ba/F3 cells, with the IC50 values indicting an insignificant <3.6-fold difference in responsiveness among all variants.
Two prior studies using a cellular proliferation assay for the same Ba/F3 cells as in our study; however, showed inconsistent results. Heuckmann et al. reported that cells expressing V3a showed >6.7-fold higher IC50 than did cells expressing V2 [15] and Woo et al. reported that cells expressing V3a showed >11.8-fold higher IC50 than did cells expressing V1 or V2 [16], suggesting that V3a-expressing cells are more resistant to alectinib than V1- or V2-expressing cells. Although the exact reason for the contradiction is unknown, considering that the amplification of ALK fusion genes has been reported to induce crizotinib resistance in ALK+ H2228 cells [17], it is possible that the results may have been affected by differences in the expression level of each ALK variant gene. In addition, it is also possible that the turnover rate, as well as the instability of intracellular ALK fusion protein, may differ markedly among V1, V2 and V3a variants [15].
Thus, to clarify the difference in alectinib sensitivity more precisely, we considered it is essential to not only adjust the level of each ALK variant protein but also the level of gene expression among all eight ALK variants-expressing cells. In this study, we electroporated each variant into Ba/F3 cells at the same time, and we succeeded in making eight ALK-variant-expressing cells with no significant difference between them in the amount of expression of each ALK variant gene, as estimated by measuring the expression of the neomycin resistance gene. If the expression level of each variant had been considerably unbalanced, it might have been possible to deduce that variants with high expression are less sensitive to alectinib than are variants with low expression, regardless of the type of ALK variant. This hypothesis may partly explain the discordance between their results and ours in the responses of V1, V2 and V3 to alectinib. However, the results of our in vitro model showed no significant difference in alectinib sensitivity among the eight types of ALK variant, including V1, V2 and V3a, which was consistent with the clinical response observed in a recent phase III trial of first-line treatment with alectinib that found that the objective response rate was not significantly different between patients with V1, V2 or V3a/b variants [5].
Nearly 90 types of ALK fusion variants have been identified in patients with ALK+ NSCLC, and the clinical responsiveness to alectinib of almost all variants other than the major V1, V2 and V3a/b variants are poorly understood. Approximately 17% of patients with ALK+ NSCLC do not respond to alectinib [5], and the causes of their poor response are also unknown. In the current preclinical study, not only did the three major variants not significantly affect alectinib or crizotinib sensitivity but neither did five uncommon variants (V4, KIF5B-ALK, KLC1-ALK, STRN-ALK and TFG-ALK). Therefore, these five variants at least might not contribute to any differences observed in the clinical response to alectinib in ALK+ NSCLC patients, and further studies with various ALK-variant-expressing cells other than the eight types tested here are needed before we can speculate on any correlation between clinical responses and ALK variants.
There were several limitations to our study. First, the number of ALK variants was limited. Since it was experimentally difficult to establish a large number of ALK variants-expressing Ba/F3 cells with nearly equivalent expression, we selected eight ALK variants in this study. Second, we did not directly measure ALK variant gene expression in the cells, because the stability of each ALK variant mRNA is unknown. However, a neomycin resistance gene driven by the same constitutive promoter is present in each of the eight ALK variant vectors. Thus, we indirectly gauged the expression of each ALK variant gene by measuring the expression level of the common neomycin resistance gene. Third, our ALK variant expressing cells were constructed with not a human cell line but a mouse cell line, Ba/F3, although we believe Ba/F3 cells are suitable for this study as they have been used in prior studies to examine fusion gene kinase activity such as with ALK or ROS1 or RET [15,16,18,19]. To overcome these limitations and make an entire map of sensitivity of all ALK fusion variants to alectinib in vitro, it will be useful to test as many ALK variants as possible using not only Ba/F3 cells but also human lung epithelial BEAS-2B cells.
In conclusion, the kinase activity of eight ALK variants (V1, V2, V3, V4, KIF5B-ALK, KLC1-ALK, STRN-ALK and TFG-ALK) was significantly inhibited by alectinib, and cells expressing each variant showed no significant difference in sensitivity to alectinib treatment. Our findings indicate that NSCLC patients with any of these eight ALK fusion variants might also show a similar response to alectinib regardless of the type of ALK variant, and the difference in these variants might have little impact on alectinib response in patients with ALK fusion-positive NSCLC. Furthermore, since only a very limited number of uncommon ALK fusion variants have been detected in the clinic, the preclinical models we constructed in this study may be useful tools with which to investigate the clinical responses of these minor variants to alectinib treatment.
Acknowledgements
The authors thank MK for their technical support in the experiments.
The study was funded by Chugai Pharmaceutical Co., Ltd., and no other specific funding was received for this study.
K.F. performed all experiments, analyzed and interpreted the data, and was a major contributor in writing the manuscript. N.H. and Y.Y. supported the interpretation and writing of the manuscript. All authors contributed to the final manuscript and approved it for submission.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of interest
All authors are employees of Chugai Pharmaceutical Co., Ltd., and there are no conflicts of interest.
References
- 1.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014; 311:1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia B, Nagasaka M, Zhu VW, Ou SI, Soo RA. How to select the best upfront therapy for metastatic disease? Focus on ALK-rearranged non-small cell lung cancer (NSCLC). Transl Lung Cancer Res. 2020; 9:2521–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ou S-HI, Zhu VW, Nagasaka M. Catalog of 5’ fusion partners in ALK-positive NSCLC circa 2020. JTO Clin Res Rep. 2020; 1:100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK variants: biological and molecular properties, and the implications for patients. Cancers (Basel). 2017; 9:E118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camidge DR, Dziadziuszko R, Peters S, Mok T, Noe J, Nowicka M, et al. Updated efficacy and safety data and impact of the EML4-ALK fusion variant on the efficacy of alectinib in untreated ALK-positive advanced non-small cell lung cancer in the global phase III ALEX study. J Thorac Oncol. 2019; 14:1233–1243 [DOI] [PubMed] [Google Scholar]
- 6.Yoshida T, Oya Y, Tanaka K, Shimizu J, Horio Y, Kuroda H, et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2016; 34:3383–3389 [DOI] [PubMed] [Google Scholar]
- 7.Lin JJ, Zhu VW, Yoda S, Yeap BY, Schrock AB, Dagogo-Jack I, et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J Clin Oncol. 2018; 36:1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei YY, Yang JJ, Zhang XC, Zhong WZ, Zhou Q, Tu HY, et al. Anaplastic lymphoma kinase variants and the percentage of ALK-positive tumor cells and the efficacy of crizotinib in advanced NSCLC. Clin Lung Cancer. 2016; 17:223–231 [DOI] [PubMed] [Google Scholar]
- 9.Mitiushkina NV, Tiurin VI, Iyevleva AG, Kholmatov MM, Filippova EA, Moiseyenko FV, et al. Variability in lung cancer response to ALK inhibitors cannot be explained by the diversity of ALK fusion variants. Biochimie. 2018; 154:19–24 [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013; 13:772–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki T, Rodig SJ, Chirieac LR, Jänne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010; 46:1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura Y, Kurasawa M, Yorozu K, Puig O, Bordogna W, Harada N. Antitumor activity of alectinib, a selective ALK inhibitor, in an ALK-positive NSCLC cell line harboring G1269A mutation: efficacy of alectinib against ALK G1269A mutated cells. Cancer Chemother Pharmacol. 2016; 77:623–628 [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011; 27:107–132 [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011; 13:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuckmann JM, Balke-Want H, Malchers F, Peifer M, Sos ML, Koker M, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012; 18:4682–4690 [DOI] [PubMed] [Google Scholar]
- 16.Woo CG, Seo S, Kim SW, Jang SJ, Park KS, Song JY, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol. 2017; 28:791–797 [DOI] [PubMed] [Google Scholar]
- 17.Courtin A, Smyth T, Hearn K, Saini HK, Thompson NT, Lyons JF, Wallis NG. Emergence of resistance to tyrosine kinase inhibitors in non-small-cell lung cancer can be delayed by an upfront combination with the HSP90 inhibitor onalespib. Br J Cancer. 2016; 115:1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama R, Kobayashi Y, Friboulet L, Lockerman EL, Koike S, Shaw AT, et al. Cabozantinib overcomes crizotinib resistance in ROS1 fusion-positive cancer. Clin Cancer Res. 2015; 21:166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kodama T, Tsukaguchi T, Satoh Y, Yoshida M, Watanabe Y, Kondoh O, Sakamoto H. Alectinib shows potent antitumor activity against RET-rearranged non-small cell lung cancer. Mol Cancer Ther. 2014; 13:2910–2918 [DOI] [PubMed] [Google Scholar]
- 20.Furugaki K, Fukumura J, Iwai T, Yorozu K, Kurasawa M, Yanagisawa M, et al. Impact of bevacizumab in combination with erlotinib on EGFR-mutated non-small cell lung cancer xenograft models with T790M mutation or MET amplification. Int J Cancer. 2016; 138:1024–1032 [DOI] [PubMed] [Google Scholar]