Abstract
Phenotypic and genotypic diversity of the flagellin gene (fliC) of Clostridium difficile was studied in 47 isolates from various origins belonging to the serogroups A, B, C, D, F, G, H, I, K, X, and S3. Electron microscopy revealed 17 nonflagellated strains and 30 flagellated strains. PCR and reverse transcription-PCR demonstrated that the flagellin gene was present in all strains and that the fliC gene was expressed in both flagellated and nonflagellated strains. Southern blotting showed the presence of only one copy of the gene and three different hybridization patterns. DNA sequence analysis of fliC from the strains belonging to serogroups C, D, and X, representative of each profile, disclosed great variability in the central domain, whereas the N- and C-terminal domains were conserved. The variability of the flagellin gene fliC was further studied in the isolates by PCR-restriction fragment length polymorphism (RFLP) analysis. Nine different RFLP groups were identified (I to IX), among which three (I, VII, and VIII) corresponded to numerous serogroups whereas the six others (II, III, IV, V, VI, and IX) belonged to a single serogroup. Flagellin gene RFLP analysis could constitute an additional typing method employable in conjunction with other typing methods currently available.
Clostridium difficile is the major etiological agent of pseudomembranous colitis and antibiotic-associated diarrhea. In addition to the two major toxins, A and B, which represent the major virulence factors (7), a number of other putative accessory virulence factors have been described. These include adhesins mediating adherence to mucosa (15, 23, 48), fimbriae, and capsule and tissue-degradative enzymes (6). However, in some bacterial species, flagella may also be a virulence factor and play a role in colonization of the gastrointestinal tract. The flagellar structure plays a role in internalization of Campylobacter jejuni (18), Salmonella enterica serovar Typhi (26), and Proteus mirabilis (31) into cultured epithelial cells. Motility is an important factor in the virulence of Vibrio cholerae (39) and Vibrio anguillarum (30). Flagella are also involved in chemotaxis and have been implicated in mucus-cell adherence and colonization by Pseudomonas aeruginosa (1, 40), Helicobacter pylori (14), and Burkholderia pseudomallei (8). Since flagella are believed to constitute one of the virulence factors of various infectious bacteria, the flagellin gene could be considered a useful genetic marker for epidemiological and phylogenetic studies (20, 52).
One aspect of C. difficile that we have studied is its interaction with target cells (15, 23, 48). Adhesion to and colonization of target tissues by bacteria are frequently important first steps in establishing infection. It is likely that C. difficile is unable to colonize without attachment and will be quickly removed by nonspecific host defense mechanisms.
Our laboratory is interested in finding out whether flagella play a role in C. difficile intestinal colonization. Few studies concerning C. difficile flagella have been performed; Delmée et al. established that flagella were involved in cross-reactions of serogroups (11). In a previous study we characterized the 39-kDa flagellin protein (45). The flagellin gene was cloned and sequenced, and the recombinant protein was characterized.
The aim of this work was to study the phenotypic and genotypic variability of the flagellin gene (fliC) and its correlation with serogroups in C. difficile isolates from different origins. Strains were investigated by electron microscopy (EM). The presence of the fliC gene was verified by PCR amplification, and the expression of the flagellin gene was studied by reverse transcription (RT)-PCR. In order to investigate the flagellin gene structure, Southern analysis with serogroup reference strains and sequencing of fliC genes from three strains were performed. PCR amplification of flagellin genes combined with restriction fragment length polymorphism (RFLP) analysis were used in an attempt to study the variability among C. difficile isolates.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Forty-seven isolates belonging to 12 different serogroups (serogroups A1, A10, B, C, D, F, G, H, I, K, S3, and X) were selected at the Microbiology Unit of the Catholic University of Louvain, Brussels, Belgium, with care taken to choose strains isolated from several geographical locations. The 10 reference strains for specific serogroups were A (ATCC 43594), B (ATCC 43593), C (ATCC 43596), D (ATCC 43597), F (ATCC 43598), G (ATCC 43599), H (ATCC 43600), I (ATCC 43601), K (ATCC 43602), and X (ATCC 43603). Clostridium sordellii (Institut Pasteur, Paris, France) was used as a negative control, and C. difficile 79-685 was used as a positive control for the flagellin gene (Table 1).
TABLE 1.
C. difficile isolates studiedc
| Strain | Serogroup | Origina | Toxin A in vitro | Toxin B in vitro | Flagella shown by EM | PCR | RT-PCR | RFLP group |
|---|---|---|---|---|---|---|---|---|
| ATCC 43594b | A1 | Brussels 3, Belgium | + | + | + | + | NDd | VII |
| EX482 | A1 | Brussels 2, Belgium | + | + | + | + | ND | VII |
| 24573 | A1 | Charleroi, Belgium | + | + | + | + | ND | VII |
| SE810 | A10 | Annecy, France | − | − | + | + | ND | I |
| TO005 | A10 | Toronto, Canada | + | + | + | + | ND | IV |
| 55787 | A10 | Brussels 1, Belgium | − | − | + | + | ND | V |
| EX560 | B | Mauscron, Belgium | − | − | − | + | + | I |
| CO086 | B | Jouy en Josas, France | − | − | + | + | ND | III |
| CO109 | B | Jouy en Josas, France | − | − | + | + | ND | III |
| ATCC 43593b | B | Brussels 1, Belgium | − | − | + | + | ND | VII |
| ATCC 43596b | C | Namur, Belgium | + | + | − | + | + | I |
| 54637 | C | Brussels 4, Belgium | + | + | − | + | + | I |
| 54828 | C | Brussels 1, Belgium | + | + | − | + | + | I |
| 51936 | C | Sambreville, Belgium | + | + | − | + | + | I |
| 1075 | C | Brussels 1, Belgium | − | − | − | + | + | I |
| BR058 | D | Brussels 1, Belgium | − | − | + | + | ND | VIII |
| ATCC 43597b | D | Brussels 1, Belgium | − | − | − | + | + | VIII |
| 55944 | D | Brussels 1, Belgium | − | − | − | + | + | VIII |
| ATCC 43598b | F | Brussels 1, Belgium | − | + | + | + | ND | II |
| 5168 | F | Brussels 1, Belgium | − | + | + | + | ND | II |
| 6058 | F | Brussels 1, Belgium | − | + | − | + | + | II |
| 6100 | F | Brussels 1, Belgium | − | + | + | + | ND | II |
| 54126 | G | St. Ode, Belgium | + | + | + | + | ND | VII |
| 51187 | G | Brussels 1, Belgium | + | + | + | + | ND | VII |
| ATCC 43599b | G | Brussels 3, Belgium | + | + | + | + | ND | VII |
| SE956 | G | Annecy, France | + | + | − | + | + | VII |
| ATCC 43600b | H | Brussels 3, Belgium | + | + | − | + | + | VII |
| 50673 | H | Tournai, Belgium | + | + | + | + | ND | VII |
| 53444 | H | Brussels 1, Belgium | − | − | + | + | ND | VIII |
| ATCC 43601b | I | Brussels 1, Belgium | − | − | − | + | + | I |
| 54823 | I | Brussels 1, Belgium | − | − | − | + | + | I |
| 56026 | I | Brussels 1, Belgium | − | − | + | + | ND | I |
| 55684 | I | Brussels 1, Belgium | − | − | − | + | + | I |
| 52356 | K | Brussels 1, Belgium | + | + | − | + | + | I |
| 51659 | K | Soignies, Belgium | + | + | + | + | ND | VII |
| 48515 | K | Brussels 1, Belgium | + | + | + | + | ND | VII |
| SE752 | K | Annecy, France | + | + | + | + | ND | VII |
| ATCC 43602b | K | Brussels 1, Belgium | − | − | + | + | ND | VII |
| 79685 | S3 | Strasbourg, France | + | + | + | + | ND | I |
| 57207 | S3 | Brussels 1, Belgium | − | + | − | + | + | I |
| 37561 | S3 | Charleroi, Belgium | + | + | + | + | ND | VII |
| EX596 | S3 | Brussels 3, Belgium | + | + | + | + | ND | VII |
| 35962 | S3 | Brussels 1, Belgium | − | + | + | + | ND | VIII |
| 36678 | X | Brussels 1, Belgium | − | + | + | + | ND | I |
| 12934 | X | Verviers, Belgium | − | − | + | + | ND | VI |
| 20356 | X | Brussels 1, Belgium | − | + | − | + | + | VI |
| ATCC 43603b | X | Brussels 1, Belgium | − | − | + | + | ND | IX |
Brussels 1, Brussels 2, etc., represent different units or different hospitals in Brussels.
Serogroup reference strain.
+, positive result; −, negative result.
ND, not determined.
Clostridium strains were grown under anaerobic conditions on agar plates (Oxoid Ltd., Basingstoke, Hampshire, England) supplemented with 7% horse blood (BioMérieux, Marcy l'Etoile, France) or in TGY (tryptone glucose yeast infusion broth) (Difco Laboratories, Detroit, Mich.) for 48 h.
EM.
The strains of C. difficile were grown overnight on agar plates supplemented with 7% horse blood as described above. A bacterial suspension was made in 100 μl of phosphate-buffered saline. Copper grids (Touzard et Matignon, Paris, France) were placed facedown on the cell suspension for 5 min and then negatively stained with a 2% phosphotungstic acid solution (pH 7.2). The grids were air dried and observed under a transmission electron microscope (EM 301; Philips).
DNA extraction and Southern blotting.
DNA was extracted from 10 ml of overnight culture according to the protocol provided in the Puregene DNA gram-positive bacteria and yeast DNA extraction kit (Gentra Systems, Minneapolis, Minn.).
Southern blotting was carried out with 5 μg of DNA digested with 10 U of HindIII for 3 h under the conditions recommended by the provider (Life Technologies, Cergy Pontoise, France). Products of digestion were separated by electrophoresis on a 0.8% (wt/vol) agarose gel. The fragments were transferred onto a positively charged nylon membrane (Roche, Mannheim, Germany) using a vacuum blotter (Appligene-Oncor, Illkirch, France). The amplified fliC gene of the C. difficile 79-685 strain (45) was used as a C. difficile flagellin gene-specific probe. The DNA probe was labeled and detected by using the ECL direct nucleic acid labeling and detection system (Amersham-Pharmacia Biotech, Les Ulis, France). Hybridization and washing of membranes were carried out at low stringency (0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at 50°C).
PCR amplification and PCR-RFLP.
For amplification of the fliC gene from various C. difficile isolates, the specific primers used were Nter (5′-ATGAGAGTTAATACAAATGTAAGTGC-3′) and Cter (5′-CTATCCTAATAATTGTAAAACTCC-3′) corresponding to the 5′- and 3′- end sequences of the fliC gene of the C. difficile 79-685 strain. DNA amplification by PCR was performed in a reaction volume of 100 μl consisting of 1 μl of a bacterial suspension washed twice with phosphate buffer, primer Nter (1 mM), primer Cter (1 mM), deoxynucleoside triphosphates (0.2 mM), MgCl2 (2 mM), 1 U of Taq polymerase, and 1× polymerase buffer (Promega, Madison, Wis.). The reaction mixture was overlaid with mineral oil. Initial denaturation was carried out at 94°C for 5 min. Thirty-five cycles of amplification were performed in a Thermocycler 480 (Perkin-Elmer, Norwalk, Conn.). Each cycle consisted of three steps: denaturation at 94°C (30 s), annealing at 55°C (30 s), and extension at 72°C (1 min). An additional step of extension for 10 min at 72°C was performed at the end of the amplification to complete the extension of the primers. Samples (5 μl) of amplified products were digested with the restriction enzymes HindIII, DraI, HpaI, PvuII, HincII, HinfI (Amersham-Pharmacia Biotech), and RsaI (Life Technologies) according to the vendor's recommendations. The digested amplified products were analyzed by electrophoresis in a 1.2% (wt/vol) agarose gel with a 100-bp ladder (Amersham-Pharmacia Biotech) as the molecular size marker.
DNA sequencing.
PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). DNA sequencing was carried out with the BigDye terminator DNA sequencing kit (PE Applied Biosystems, Warrington, England). The samples were analyzed with the automated DNA sequencer ABI PRISM 310 genetic analyzer (Perkin-Elmer). The sequencing was initiated on both strands with the primers Nter and Cter and was finished with internal primers designed from the sequences obtained.
RNA extraction and RT-PCR.
RNA was extracted from 10 ml of 8-h C. difficile anaerobic culture. Bacteria were harvested by centrifugation at 5,000 × g for 5 min at 4°C and then resuspended in 0.5 ml of cold TE buffer (10 mM HCl, 1 mM EDTA, pH 7.4) and kept on ice. Glass beads (0.6 g; 425 μm < diameter < 600 μm; Sigma Chemical Co., St. Louis, Mo.) were added in a solution containing 0.17 ml of 4% (wt/vol) Bentone rheological additive (Rheox Ltd., Livingston, Scotland), 0.5 ml of acid phenol (Sigma Chemical Co.), and 0.05 ml of 10% (wt/vol) sodium dodecyl sulfate solution. The solution was mixed three times by vortexing for 1 min each, interrupted by 1-min pauses. The aqueous phase was recovered by centrifugation at 12,000 × g at 4°C for 15 min and then extracted three times with a phenol-chloroform (1:1, vol/vol) solution and precipitated with ethanol. The RNA pellet was washed with 75% (vol/vol) cold ethanol, vacuum dried, and resuspended in 50 μl of TE buffer. The RNA was treated with DNase I (Amersham-Pharmacia Biotech) and stored at −20°C.
The RT-PCR was carried out with the SuperScript one-step RT-PCR system (Life Technologies) in a 50-μl mixture containing 1 μg of RNA template, a 1 mM concentration each of primers Nter and Cter, 1.2 mM MgSO4, and the reaction cocktail according to the manufacturer's instructions. The RNA of C. difficile 79-685 was used as a positive control, and the RNA of C. sordellii was used as a negative control. The reaction mixture was overlaid with mineral oil. The cDNA synthesis step was performed at 50°C for 30 min, and a predenaturation step was performed at 94°C for 2 min. Thirty cycles of amplification were performed in a Thermocycler 480 (Perkin-Elmer). Each cycle consisted of three steps as described above. The amplified products were subjected to electrophoresis on a standard 1% (wt/vol) agarose gel.
Serogrouping and toxigenicity.
Serogroups were determined by slide agglutination with rabbit antisera (12) and were confirmed by typing by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (13). Toxin A production was determined by the C. difficile toxin A test (Oxoid). In vitro cytotoxin (toxin B) determination was performed with HeLa cells cultured in minimum Eagle medium with Earle's salts (Life Technologies) supplemented with 10% fetal calf serum (Life Technologies), 1% nonessential amino acids (Life Technologies), and 200 mM l-glutamine (Life Technologies) in microtiter plates (3 × 104 cells per well). Fivefold serial dilutions of filtrates of 48-h TGY liquid cultures of C. difficile were incubated for 18 h with the cells at 37°C in a 5% CO2 atmosphere. After fixation and coloration of the culture cells with methylene blue, cytotoxic effect was observed by inverse microscopy.
Computer analyses.
Nucleotide and protein sequence alignments were performed with DNA Strider software and the Multalin program (9).
Nucleotide sequence accession numbers.
The nucleotide sequence of the fliC locus of strains 79-685, 545, 3232, and 5036, corresponding to serogroups S3, C, D, and X, respectively, were assigned GenBank numbers AF065259, AF095236, AF095237, and AF095238.
RESULTS
Detection of fliC gene and flagella by EM and fliC gene expression.
As observed by EM, 30 out of 47 strains showed visible flagella, whereas 17 were nonflagellated (Table 1). All strains from serogroup A were flagellated; in contrast, no strain from serogroup C carried flagella. In other serogroups both flagellated and nonflagellated strains were observed. The number and length of flagella also varied considerably among strains.
PCR amplification using fliC-specific oligonucleotide primers Nter and Cter was employed to investigate the presence of the gene in C. difficile isolates. The amplification gave a single product in all 47 C. difficile strains studied (not shown), whereas no amplified product was obtained from the negative control C. sordellii strain. An 870-bp fragment was obtained from 46 strains, whereas the serogroup X reference strain revealed an 850-bp amplified fragment.
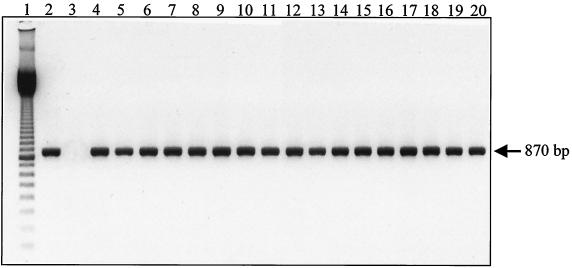
The lack of flagella on the bacterial surface could be due to the absence of transcription of the fliC gene. Therefore, to investigate expression of the flagellin gene in flagellated and nonflagellated strains, DNA transcription was investigated by detecting flagellin mRNA by RT-PCR. The results (Fig. 1) show that a single 870-bp amplified fragment was obtained from all the 17 nonflagellated strains, including four nonflagellated serogroup C, D, H, and I reference strains.
FIG. 1.
RT-PCR products obtained with primers Nter and Cter and RNA isolated from nonflagellated C. difficile strains. Lane 1, 100-bp ladder (Amersham-Pharmacia Biotech); lane 2, strain 79-685 (positive control); lane 3, RNA from C. sordellii (negative control); lane 4, EX560; lane 5, ATCC 43596 (serogroup reference C); lane 6, 54637; lane 7, 54828; lane 8, 51936; lane 9, 1075; lane 10, ATCC 43597 (serogroup reference D); lane 11, 55944; lane 12, 6058; lane 13, SE956; lane 14, ATCC 43600 (serogroup reference H); lane 15, ATCC 43601 (serogroup reference I); lane 16, 54823; lane 17, 55684; lane 18, 52356; lane 19, 57207; lane 20, 20356.
The absence of genomic DNA contamination in the RNA samples was verified by PCR using Nter and Cter primers. Furthermore, a counterexperiment was carried out with an RNA sample treated with RNase and subjected to an RT-PCR as described above. No amplified products were detected in these two control experiments, thus confirming the purity of the RNA preparation and the specificity of the target RNA.
Detection and copy number of fliC in C. difficile isolates.
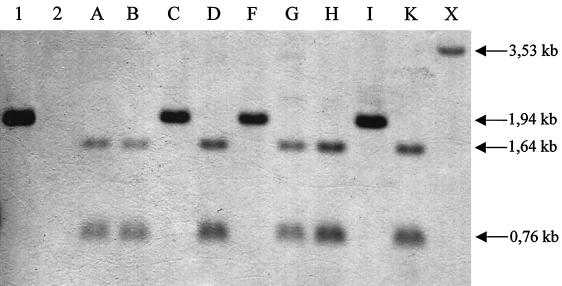
In some bacteria, fliC can be present in multiple copies on the bacterial chromosome. To assess whether fliC is present in mono- or multicopy, the amplified DNA from strain 79-685 was used as a probe in Southern hybridization of chromosomal DNA of strain 79-685 and the 10 reference serogroups (A, B, C, D, F, G, H, I, K, and X). Hybridization under low-stringency conditions showed that DNA of all isolates hybridized with the fliC-specific probe. Only one copy of the gene was present in each strain. Some strains carry a HindIII site and therefore show the presence of two bands (Fig. 2). The presence of a HindIII site was confirmed by subjecting the amplified fliC gene product to HindIII digestion. The digestion with HindIII allows the classification of the strains in three groups: the first group exhibits a single 1.94-kb single band (79-685, C, F, and I), and the second group displays two bands of 1.64 kb and 0.76 kb (A, B, D, G, H, and K), while there is a single 3.53-kb band for the serogroup X reference strain (Fig. 2).
FIG. 2.
Southern blot of chromosomal DNA isolated from C. difficile reference strains belonging to Delmée serogroups hybridized under low stringency with the fliC probe. DNA was digested with HindIII, electrophoresed, and transferred to a nylon membrane. Lane 1, strain 79-685 (positive control); lane 2, C. sordellii (negative control); lane A, ATCC 43594 (serogroup A); lane B, ATCC 43593 (serogroup B); lane C, ATCC 43596 (serogroup C); lane D, ATCC 43597 (serogroup D); lane F, ATCC 43598 (serogroup F); lane G, ATCC 43599 (serogroup G); lane H, ATCC 43600 (serogroup H); lane I, ATCC 43601 (serogroup I); lane K, ATCC 43602 (serogroup K); lane X, ATCC 43603 (serogroup X). The two bands in lanes A, B, D, G, H, and K are produced by the HindIII site near base 560 of the flagellin gene.
DNA sequence analysis.
In order to confirm that the 870-bp amplified product described above was the flagellin gene, PCR fragments obtained from the 10 reference serogroup strains were partially sequenced with the Nter primer. Sequencing revealed an N-terminal sequence identical to the fliC gene of the C. difficile 79-685 strain in all PCR products (data not shown). To investigate the conservation of the fliC gene coding region in strains representing the different profiles obtained by Southern blotting, the fliC gene of the serogroup C, D, and X reference strains was amplified by PCR using specific primers (Nter and Cter) as described in Materials and Methods. DNA and deduced amino acid sequence analysis revealed an open reading frame composed of 873 nucleotides (290 amino acids) for the 79-685 strain and our C and D reference strains, while the open reading frame was 846 nucleotides, corresponding to 281 amino acids, for the serogroup X reference strain. The latter strain carried a short deletion in the central region of flagellin (Fig. 3).
FIG. 3.
Sequence alignment of the deduced amino acid sequence of FliC of C. difficile strains 79-685, serogroup reference C, D and X strains. Identical residues are indicated with an asterisk (+). Deletions are indicated with a dash (—). The alignment was performed with the MULTALIN program (10).
The degree of the identity between the deduced amino acid sequences for the four strains was analyzed and showed 100% identity between FliC of 79-685, the gene of which was previously sequenced by us (45), and the serogroup C reference strain, while the identity was 90 and 77% between serogroup C and serogroup D and X reference strains, respectively, and 76% between serogroup D and X reference strains.
At the protein level, the identity between FliC of the serogroup C reference strain and of the serogroup D and X reference strains was 86 and 72%, respectively, and was 71% between serogroup D and X reference strains. It is evident from the analysis that flagellin proteins of C. difficile strains exhibit conservation in the N and C termini, while the central region is more diverse. The presence of a number of conserved alanine residues suggests a secondary structure in α-helical conformation (alanine is a helix-forming residue); this conservation could reflect the functional importance of the terminal regions in forming the tertiary structure of the flagellin protein monomers.
RFLP analysis of flagellin genes.
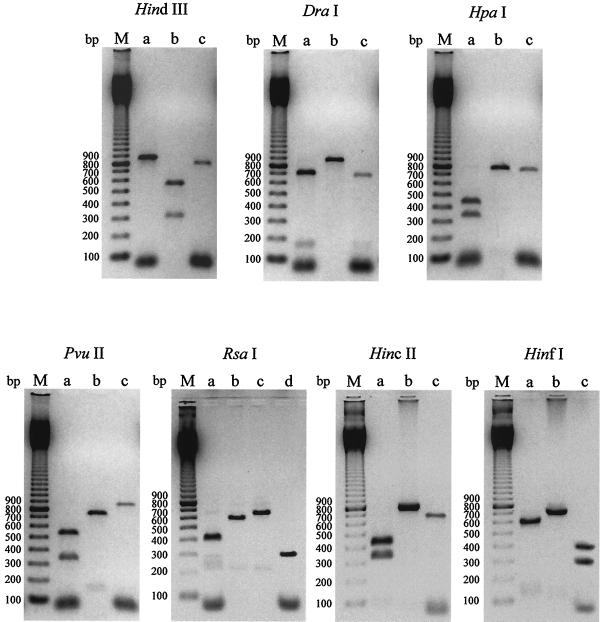
As sequence analysis of fliC from three strains suggested significant variability of gene structure, we decided to examine gene structure by PCR-RFLP analysis in all the isolates studied. The amplified fliC gene was digested with HindIII, DraI, HpaI, PvuII, RsaI, HincII, and HinfI. According to the restriction map (not shown), these restriction sites are distinct and therefore can be used to perform RFLP analysis.
The results (Fig. 4) show the different restriction patterns obtained from the C. difficile strains. Three different restriction profiles were obtained with HindIII, DraI, HpaI, PvuII, HincII, and HinfI enzymes, and four restriction profiles (designated a, b, c, and d) were obtained with RsaI endonuclease. As far as the serogroup X reference strain is concerned, the fliC RFLP analysis revealed a unique and different profile with each enzyme: profile c with HindIII, DraI, HpaI, PvuII, HincII, and HinfI restriction enzymes and profile d with RsaI.
FIG. 4.
RFLP patterns of PCR-amplified flagellin genes. The amplified fliC gene of C. difficile isolates was digested with HindIII, DraI, HpaI, PvuII, RsaI, HincII, and HinfI. The different restriction profiles obtained with each enzyme were designated a, b, c, and d. Lanes M, 100-bp ladder (Amersham-Pharmacia Biotech); lanes a, profile a; lanes b, profile b; lanes c, profile c; lanes d, profile d.
The strains can be classified into nine RFLP groups (Table 2). The most frequent RFLP types are I (15 strains) and VII (16 strains). RFLP type I encompasses mostly nonflagellated strains that are either toxin positive or negative, whereas RFLP type VII encompasses mostly flagellated strains that are toxin positive; types V and IX are rare (one of each) and include only toxin-negative strains. In addition, types III, VI, and VIII are also characterized by toxin-negative strains. The single isolate TO005 is the only one to be characterized as RFLP profile IV.
TABLE 2.
Classification of restriction enzyme patterns into RFLP groupgroups
| RFLP group | Restriction enzyme pattern generated by digestion witha:
|
||||||
|---|---|---|---|---|---|---|---|
| HindIII | DraI | HpaI | PvuII | RsaI | HincII | HinfI | |
| I | a | a | a | a | a | a | a |
| II | a | a | b | a | a | b | a |
| III | a | a | a | a | a | b | b |
| IV | a | a | b | b | b | b | b |
| V | a | a | b | b | c | b | b |
| VI | b | b | a | a | a | a | a |
| VII | b | b | b | b | b | b | b |
| VIII | b | b | b | b | c | b | b |
| IX | c | c | c | c | d | c | c |
Restriction enzyme patterns are shown in Fig. 4.
Serogroups C, D, F, G, and I are homogeneous in terms of RFLP profile, with all strains tested here displaying the same pattern. In contrast, profiles of serogroups, A, B, H, K, S3, and X are variable. Serogroup A is divided in four RFLP groups (I, IV, V, and VII), serogroup B is divided in three groups (I, III, and VII), serogroup H is divided in two groups (VII and VIII), serogroup K is divided in two groups (I and VII), serogroup S3 is divided in three groups (I, VII, and VIII), and serogroup X is divided in three groups (I, VI, and IX). The serogroup X reference strain has a separate profile and forms the RFLP group IX (Table 3).
TABLE 3.
Repartition of serogroups into the nine RFLP groups
| Serogroup | No. of strains studied | No. of strains belonging to the following RFLP group:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | ||
| A | 6 | 1 | 1 | 1 | 3 | |||||
| B | 4 | 1 | 2 | 1 | ||||||
| C | 5 | 5 | ||||||||
| D | 3 | 3 | ||||||||
| F | 4 | 4 | ||||||||
| G | 4 | 4 | ||||||||
| H | 3 | 2 | 1 | |||||||
| I | 4 | 4 | ||||||||
| K | 5 | 1 | 4 | |||||||
| S3 | 5 | 2 | 2 | 1 | ||||||
| X | 4 | 1 | 2 | 1 | ||||||
DISCUSSION
For numerous pathogens, the capacity to adhere, invade, and destroy the colonic mucosal cells appears to be an essential aspect of the first stage of their pathogenicity. C. difficile is responsible for the most-frequent hospital-acquired infection consequent to antibiotic therapy. It causes diarrheal disease, which can lead to an intense acute response: pseudomembranous colitis. C. difficile releases two exotoxins into the colon which are responsible for the disease: toxin A and toxin B. Toxin A (an enterotoxin) elicits fluid secretion, mucosal damage, and intestinal inflammation; toxin B (a cytotoxin) is completely devoid of enterotoxicity.
Before these events take place, C. difficile entering the gut must find pathways to reach suitable epithelial cells, which are naturally protected by a layer of dense mucus. C. difficile can adhere to the mucous layer (23). Afterwards the bacterium could penetrate the mucous layer with the aid of its proteases (38) and flagella; finally, C. difficile attaches to enterocytes by means of specific adhesins (23, 48); the role of flagella in this process has yet to be defined. At these different steps, the presence of complex and specialized chemotaxis and flagellar systems may play a role.
The studies undertaken here revealed that with EM certain strains of C. difficile are characterized by their inability to produce in vitro visible flagella. We have shown by PCR that the flagellin gene was present in both flagellated and nonflagellated C. difficile strains. The specificity of the amplification was confirmed by the fact that the flagellin gene of C. sordellii, highly flagellated and genetically very close to that of C. difficile, was not amplified with these primers. Gene amplification by PCR has been frequently used as a rapid method for detection and identification of pathogenic bacteria including Clostridium perfringens (16), Campylobacter spp. (36), Listeria monocytogenes (19), and Bordetella bronchiseptica (22). Therefore, amplification of the fliC gene could be used as a rapid method to detect and identify C. difficile.
We showed here that the flagellin gene was expressed in flagellated and nonflagellated C. difficile strains by detection of flagellin mRNA with the one-step method RT-PCR. According to Macnab (27) and Manson (28), the Escherichia coli flagellin gene was only expressed when both the basal body and the hook of the flagellum were fully formed through the membrane of bacterium. From that, the flagellin, the cap protein, and the junction hook-flagellum proteins were synthesized and assembled to achieve flagellar filament formation from the external membrane (5, 42). In nonflagellated C. difficile strains, the cap and/or junction hook-flagellum proteins could be inactivated by mutation, thus preventing transport of flagellin subunits through the bacterial membrane and polymerization. To confirm this hypothesis, it would be interesting to verify the presence of the flagellin protein in nonflagellated strains.
The nonflagellated C. difficile strains possess a cryptic flagellin gene. We cannot rule out, however, that in vivo all strains could be flagellated, and we intend to study the in vivo expression of fliC in our mouse model. Cryptic genes have been characterized in nonmotile bacteria. Indeed, the expression of surface flagella in some pathogenic bacteria may be induced only by factors related to a specific biological microenvironment or under certain in vitro growth conditions. Holt and Chaubal (21) showed that the carbon source, the viscosity of the medium, and the temperature of incubation can induce the motility of S. enterica Pullorum, thought to be nonmotile and nonflagellated. Shigella spp. are described as nonflagellated and nonmotile organisms. However, Giron (17) detected motility and flagella by EM in four strains and two clinical isolates, depending on the culture conditions under which temperature, salt, glucose, oxygen, or agar concentrations were modified. Tominaga et al. (46) showed the presence of intact cryptic flagellin genes in nonflagellated Shigella flexneri and Shigella sonnei strains. These genes produced normal-type flagella in an E. coli ΔfliC strain. Their results suggested a loss of the expression potential of flagellar genes, probably by various mutations and/or gene rearrangements. It would be interesting to investigate the role of mucus as an inducing factor for flagellal expression.
In order to study the variability of flagellin genes, the fliC gene was sequenced in three strains representing different profiles obtained by Southern blotting. Sequencing showed high conservation in the N-terminal and C-terminal regions and pronounced variability in the central domain of the flagellin protein (Fig. 3). The N- and C-terminal parts are responsible for secretion and polymerization of flagella, whereas the central region constitutes the surface-exposed antigenic part of the flagellar filament as described by Winstanley and Morgan (52), but flagellin may vary considerably among species in both amino acid sequence and size (49, 52). The deletion of amino acids in the variable domain of the serogroup X reference strain suggests, analogous to other bacteria, that the central region plays no role in the structure of flagella since this strain possesses flagella. Mutations of the flagellin gene do not account for the absence of flagella since the flagellin gene of the flagellated 79-685 strain is strictly identical to that of the nonflagellated serogroup C reference strain.
Different methods have been developed to study the epidemiology of C. difficile or to identify or type strains. Analysis of restriction patterns of DNA of clinical isolates has been used for investigations of epidemiology and typing of C. difficile-associated diarrhea (24). Pantosti et al. (37) used the electrophoretic patterns of extracted proteins to characterize C. difficile strains from various sources and showed correlation between certain electrophoretic patterns and virulence. Delmée et al. (13) compared serogrouping of C. difficile by slide agglutination with rabbit antisera and polyacrylamide gel electrophoresis of whole-cell proteins, permitting correlation between the two typing systems and establishment of a single classification. Recently, new molecular techniques have been developed to type C. difficile strains based on DNA polymorphism. DNA pattern profiles have been obtained by PCR amplification of a specific chromosomal region such as the rRNA gene (4) or the 16S-23S rRNA gene intergenic spacer region (44). Another molecular method, based on DNA polymorphism, has been found to be useful to distinguish strains of C. difficile, namely, arbitrary primed PCR, also called random amplified polymorphic DNA analysis. Arbitrary primed PCR or random amplified polymorphic DNA analysis has been used as an efficient discriminative method for investigation of nosocomial outbreaks of C. difficile-associated diarrhea (2, 3, 5; F. Barbut, N. Mario, J. Frottier, and J. C. Petit, Letter, Eur. J. Clin. Microbiol. Infect. Dis. 12:794–795, 1993).
In our study, we have used the PCR-RFLP method to study genetic diversity among C. difficile strains. With this molecular technique, correlation between RFLP groups and serogroups was clear for certain serogroups. Serogroups C and I are represented by a single RFLP group, group I, and serogroups D, F, and G are represented by RFLP groups VIII, II, and VII, respectively. We noticed that serogroup F exclusively possesses pattern II, which is not shared by any other strain in this study. Similar data were shown by Rupnik (41) concerning the toxinotype of strains belonging to serogroup F. Other serogroups (A, B, H, K, S3, and X) were subdivided into several RFLP groups. However, six RFLP groups, corresponding each to a single serogroup, could be used to confirm some strains. Indeed, RFLP groups II, III, IV and V, and VI and IX were correlated to serogroups F, B, A, and X, respectively. Some conclusions could also be drawn concerning the state of flagellation and toxigenesis, but a larger number of strains need to be investigated to draw more-definitive conclusions.
The study of flagellin gene diversity has been also carried out with other bacteria, such as C. jejuni (33, 35, 43), P. aeruginosa (32, 50), S. enterica (10), Vibrio parahaemolyticus (29), H. pylori (34), or Burkholderia cepacia (47, 51). The results have clearly demonstrated the pronounced genetic diversity of the flagellar gene of various bacteria. The PCR-RFLP method has sometimes been used with success to differentiate several bacterial flagellal types from isolates. However, in certain cases, this procedure does not appear sufficient to type bacterial species.
The flagellin genes are excellent biomarkers with which to study strain diversity. The particular structure of the flagellin gene, with terminal conserved regions allowing gene amplification by PCR, allows analysis by RFLP and sequencing to study the variations in the central region. The PCR-RFLP procedure is rapid, highly specific, and reproducible. If a vaccine is to be developed for C. difficile disease based on different proteins, the preparations should include a mixture of flagellin proteins from major RFLP groups to allow the best possible protection. C. difficile flagella could play a role in intestinal colonization during the first stage of pathogenesis. Colonization is induced in response to environmental conditions. It is likely that production of flagella could be under the control of a system which may be turned on or off by various factors in the gut environment. Important questions remain to be explored as to the identity of these factors and what role flagella and motility play in the pathogenic scheme.
ACKNOWLEDGMENTS
This work was supported in part by the FAIR Programme of the European Union (contract CT95-0433); program ACC-SV6 (Actions Concertées Coordonnées des Sciences du Vivant) of the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche of France; and a Medical Research Council Programme grant (G9122850).
We thank Michel Lemullois and Danielle Jaillard (Université de Paris-Sud, Orsay, France) for helping us with EM.
REFERENCES
- 1.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and characterization of Pseudomonas aeruginosa fliF, necessary for flagella assembly and bacterial adherence to mucin. Infect Immun. 1996;64:2130–2136. doi: 10.1128/iai.64.6.2130-2136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbut F, Mario N, Delmee M, Gozian J, Petit J C. Genomic fingerprinting of Clostridium difficile isolates by using a random amplified polymorphic DNA (RAPD) assay. FEMS Microbiol Lett. 1993;114:161–166. doi: 10.1111/j.1574-6968.1993.tb06567.x. [DOI] [PubMed] [Google Scholar]
- 3.Barbut F, Mario N, Meyohas M C, Binet D, Frottier J, Petit J C. Investigation of a nosocomial outbreak of Clostridium difficile-associated diarrhoea among AIDS patients by random amplified polymorphic DNA (RAPD) assay. J Hosp Infect. 1994;26:181–189. doi: 10.1016/0195-6701(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 4.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit J C. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 5.Blair D F, Dutcher S K. Flagella in prokaryotes and lower eukaryotes. Curr Opin Genet Dev. 1992;2:756–767. doi: 10.1016/s0959-437x(05)80136-4. [DOI] [PubMed] [Google Scholar]
- 6.Borriello S P. 12th C. L. Oakley Lecture. Pathogenesis of Clostridium difficile infection of the gut. J Med Microbiol. 1990;33:207–215. doi: 10.1099/00222615-33-4-207. [DOI] [PubMed] [Google Scholar]
- 7.Borriello S P, Davies H A, Kamiya S, Reed P J, Seddon S. Virulence factors of Clostridium difficile. Rev Infect Dis. 1990;12(Suppl. 2):S185–S191. doi: 10.1093/clinids/12.supplement_2.s185. [DOI] [PubMed] [Google Scholar]
- 8.Brett P J, Mah D C, Wood D E. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect Immun. 1994;62:1914–1918. doi: 10.1128/iai.62.5.1914-1919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dauga C, Zabrovskaia A, Grimont P A. Restriction fragment length polymorphism analysis of some flagellin genes of Salmonella enterica. J Clin Microbiol. 1998;36:2835–2843. doi: 10.1128/jcm.36.10.2835-2843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmée M, Avesani V, Delferriere N, Burtonboy G. Characterization of flagella of Clostridium difficile and their role in serogrouping reactions. J Clin Microbiol. 1990;28:2210–2214. doi: 10.1128/jcm.28.10.2210-2214.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmée M, Homel M, Wauters G. Serogrouping of Clostridium difficile strains by slide agglutination. J Clin Microbiol. 1985;21:323–327. doi: 10.1128/jcm.21.3.323-327.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmée M, Laroche Y, Avesani V, Cornelis G. Comparison of serogrouping and polyacrylamide gel electrophoresis for typing Clostridium difficile. J Clin Microbiol. 1986;24:991–994. doi: 10.1128/jcm.24.6.991-994.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton K A, Suerbaum S, Josenhans C, Krakowa S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eveillard M, Fourel V, Barc M C, Kerneis S, Coconnier M H, Karjalainen T, Bourlioux P, Servin A. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT29 cells in culture. Mol Microbiol. 1993;7:371–381. doi: 10.1111/j.1365-2958.1993.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 16.Fach P, Delbart M O, Schlachter A, Poumeyrol M, Popoff M R. Use of the polymerase chain reaction for the diagnostic of the Clostridium perfringens food borne diseases. Med Mal Infect. 1993;23:70–77. [Google Scholar]
- 17.Giron J A. Expression of flagella and motility by Shigella. Mol Microbiol. 1995;18:63–75. doi: 10.1111/j.1365-2958.1995.mmi_18010063.x. [DOI] [PubMed] [Google Scholar]
- 18.Grant C C, Konkel M E, Cieplak W J, Tompkins L S. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray D I, Kroll R G. Polymerase chain reaction amplification of the flaA gene for the rapid identification of Listeria spp. Lett Appl Microbiol. 1995;20(1):65–68. doi: 10.1111/j.1472-765x.1995.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 20.Hales B A, Morgan A W, Hart C A, Winstanley C. Variation in flagellin genes and proteins of Burkholderia cepacia. J Bacteriol. 1998;180:1110–1118. doi: 10.1128/jb.180.5.1110-1118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt P S, Chaubal L H. Detection of motility and putative synthesis of flagellar proteins in Salmonella pullorum cultures. J Clin Microbiol. 1997;35:1016–1020. doi: 10.1128/jcm.35.4.1016-1020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hozbor D, Fouque F, Guiso N. Detection of Bordetella bronchiseptica by the polymerase chain reaction. Res Microbiol. 1999;150:333–341. doi: 10.1016/s0923-2508(99)80059-x. [DOI] [PubMed] [Google Scholar]
- 23.Karjalainen T, Barc M C, Collignon A, Trollé S, Boureau H, Cotte-Laffite J, Bourlioux P. Cloning of a genetic determinant from Clostridium difficile involved in adherence to tissue culture cells and mucus. Infect Immun. 1994;62:4347–4355. doi: 10.1128/iai.62.10.4347-4355.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuijper E J, Oudbier J H, Stuifbergen W N, Jansz A, Zanen H C. Application of whole-cell DNA restriction endonuclease profiles to the epidemiology of Clostridium difficile-induced diarrhea. J Clin Microbiol. 1987;25:751–753. doi: 10.1128/jcm.25.4.751-753.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemann F, Chambon C, Barbut F, Gardin C, Briere J, Lambert-Zechovsky N, Branger C. Arbitrary primed PCR rules out Clostridium difficile cross-infection among patients in a haematology unit. J Hosp Infect. 1997;35:107–115. doi: 10.1016/s0195-6701(97)90099-2. [DOI] [PubMed] [Google Scholar]
- 26.Liu S L, Ezaki T, Miura H, Matsui K, Yabuuchi E. Intact motility as a Salmonella typhi invasion-related factor. Infect Immun. 1988;56:1967–1973. doi: 10.1128/iai.56.8.1967-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 28.Manson M D. Bacterial motility and chemotaxis. Adv Microb Physiol. 1992;33:277–346. doi: 10.1016/s0065-2911(08)60219-2. [DOI] [PubMed] [Google Scholar]
- 29.Marshall S, Clark C G, Wang G, Mulvey M, Kelly M T, Johnson W M. Comparison of molecular methods for typing Vibrio parahaemolyticus. J Clin Microbiol. 1999;37:2473–2478. doi: 10.1128/jcm.37.8.2473-2478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGee K, Horstedt P, Milton D L. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mobley H L T, Belas R, Lockatell V, Chippendale G, Trifillis A L, Johnson D E, Warren J W. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1996;64:5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan J A, Bellingham N F, Winstanley C, Ousley M A, Hart C A, Saunders J R. Comparison of flagellin genes from clinical and environmental Pseudomonas aeruginosa isolates. Appl Environ Microbiol. 1999;65:1175–1179. doi: 10.1128/aem.65.3.1175-1179.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura M, Nukina M, Kuroki S, Obayashi H, Ohta M, Ma J J, Saida T, Uchiyama T. Characterization of Campylobacter jejuni isolates from patients with Guillain-Barre syndrome. J Neurol Sci. 1997;153:91–99. doi: 10.1016/s0022-510x(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 34.Ohta-Tada U, Takagi A, Koga Y, Kamiya S, Miwa T. Flagellin gene diversity among Helicobacter pylori strains and IL-8 secretion from gastric epithelial cells. Scand J Gastroenterol. 1997;32:455–459. doi: 10.3109/00365529709025080. [DOI] [PubMed] [Google Scholar]
- 35.Owen R J, Leeton S. Restriction fragment length polymorphism analysis of the flaA gene of Campylobacter jejuni for subtyping human, animal and poultry isolates. FEMS Microbiol Lett. 1999;176:345–350. doi: 10.1111/j.1574-6968.1999.tb13682.x. [DOI] [PubMed] [Google Scholar]
- 36.Oyofo B A, Rollins D M. Efficacy of filter types for detecting Campylobacter jejuni and Campylobacter coli in environmental water samples by polymerase chain reaction. Appl Environ Microbiol. 1993;59:4090–4095. doi: 10.1128/aem.59.12.4090-4095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantosti A, Cerquetti M, Gianfrilli P M. Electrophoretic characterization of Clostridium difficile strains isolated from antibiotic-associated colitis and other conditions. J Clin Microbiol. 1988;26:540–543. doi: 10.1128/jcm.26.3.540-543.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poilane I, Karjalainen T, Barc M C, Bourlioux P, Collignon A. Protease activity of Clostridium difficile strains. Can J Microbiol. 1998;44:157–161. [PubMed] [Google Scholar]
- 39.Richardson K. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and characterization phenotypic of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro L. The bacterial flagellum: from genetic network to complex architecture. Cell. 1995;80:525–527. doi: 10.1016/0092-8674(95)90505-7. [DOI] [PubMed] [Google Scholar]
- 43.Slater E, Owen R J. Subtyping of Campylobacter jejuni Penner heat-stable (HS) serotype 11 isolates from human infections. J Med Microbiol. 1998;47:353–357. doi: 10.1099/00222615-47-4-353. [DOI] [PubMed] [Google Scholar]
- 44.Stubbs S L, Brazier J S, O'Neill G L, Duerden B I. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37:461–463. doi: 10.1128/jcm.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tasteyre A, Barc M C, Karjalainen T, Dodson P, Hyde S, Bourlioux P, Borriello P. A Clostridium difficile gene encoding flagellin. Microbiology. 2000;146:957–966. doi: 10.1099/00221287-146-4-957. [DOI] [PubMed] [Google Scholar]
- 46.Tominaga A, Mahmoud M A, Mukaihara T, Enomoto M. Molecular characterization of intact, but cryptic, flagellin genes in the genus Shigella. Mol Microbiol. 1994;12:277–285. doi: 10.1111/j.1365-2958.1994.tb01016.x. [DOI] [PubMed] [Google Scholar]
- 47.Tungpradabkul S, Wajanarogana S, Tunpiboonsak S, Panyim S. PCR-RFLP analysis of the flagellin sequences for identification of Burkholderia pseudomallei and Burkholderia cepacia from clinical isolates. Mol Cell Probes. 1999;13:99–105. doi: 10.1006/mcpr.1999.0221. [DOI] [PubMed] [Google Scholar]
- 48.Waligora A J, Barc M C, Bourlioux P, Collignon A, Karjalainen T. Clostridium difficile cell attachment is modified by environmental factors. Appl Environ Microbiol. 1999;65:4234–4238. doi: 10.1128/aem.65.9.4234-4238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson D R, Beveridge T J. Bacterial flagellar filaments and their component flagellins. Can J Microbiol. 1993;39:451–472. doi: 10.1139/m93-066. [DOI] [PubMed] [Google Scholar]
- 50.Winstanley C, Coulson M, Wepner B, Morgan A, Hart C. Flagellin gene and protein variation amongst clinical isolates of Pseudomonas aeruginosa. Microbiology. 1996;142:2145–2151. doi: 10.1099/13500872-142-8-2145. [DOI] [PubMed] [Google Scholar]
- 51.Winstanley C, Hales B A, Morgan J A, Gallagher M J, Puthucheary S D, Cisse M F, Hart C A. Analysis of fliC variation among clinical isolates of Burkholderia cepacia. J Med Microbiol. 1999;48:657–662. doi: 10.1099/00222615-48-7-657. [DOI] [PubMed] [Google Scholar]
- 52.Winstanley C, Morgan J A. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology. 1997;143:3071–3084. doi: 10.1099/00221287-143-10-3071. [DOI] [PubMed] [Google Scholar]