Abstract
Cyclin E is a G1 cyclin essential for S-phase entry and has a profound role in oncogenesis. Previously this laboratory found that cyclin E is overexpressed and present in lower-molecular-weight (LMW) isoforms in breast cancer cells and tumor tissues compared to normal cells and tissues. Such alteration of cyclin E is linked to poor patient outcome. Here we report that the LMW forms of cyclin E are hyperactive biochemically and they can more readily induce G1-to-S progression in transfected normal cells than the full-length form of the protein can. Through biochemical and mutational analyses we have identified two proteolytically sensitive sites in the amino terminus of human cyclin E that are cleaved to generate the LMW isoforms found in tumor cells. Not only are the LMW forms of cyclin E functional, as they phosphorylate substrates such as histone H1 and GST-Rb, but also their activities are higher than the full-length cyclin E. These nuclear localized LMW forms of cyclin E are also biologically functional, as their overexpression in normal cells increases the ability of these cells to enter S and G2/M. Lastly, we show that cyclin E is selectively cleaved in vitro by the elastase class of serine proteases to generate LMW forms similar to those observed in tumor cells. These studies suggest that the defective entry into and exit from S phase by tumor cells is in part due to the proteolytic processing of cyclin E, which generates hyperactive LMW isoforms whose activities have been modified from that of the full-length protein.
In the past decade, new findings in the fields of cell biology and molecular genetics of cancer have revealed a deregulation of the cell cycle as a critical event for the onset of tumorigenesis. Progression through the cell cycle, the sequence of events between two cell divisions, is governed by the actions of positive and negative regulators in the eukaryotic cell. The mammalian cell cycle is positively regulated by heterodimeric complexes of stable cyclin-dependent kinases (CDKs) and unstable regulatory cyclin subunits (1, 51). Mitogenic stimuli result in the phosphorylation and thereby activation of cyclin-CDK complexes by CDK-activating kinase (17, 22, 35). The activated cyclin-CDK complexes in turn phosphorylate substrates such as the retinoblastoma protein (pRb) throughout the cell cycle (16, 36, 51).
The connection between cyclins and cancer has been substantiated with G1-type cyclins (25–27, 55). Cyclin E, a G1 cyclin which forms complexes with CDK2, is essential for S-phase entry (41, 47) and has a profound role in oncogenesis (29, 31). In dividing cells, the expression of cyclin E increases to a maximum at the G1/S transition, with a peak expression level near the restriction point (13, 34). When coupled to CDK2, the active kinase follows cyclin D-CDK4 in progressively phosphorylating pRb, releasing it from members of the E2F family (15). As E2F is released it activates a number of S-phase genes, including cyclin E and E2F-1 (18, 40). This state of readiness to enter S phase requires cyclin E or cells will arrest at this point in the cell cycle (42, 57). For example, the functional knock out of cyclin E by injection of anti-cyclin E antibodies into fibroblast cells causes cell arrest in the G1 phase (42). Conversely, the overexpression of cyclin E protein causes acceleration through G1 along with a decreased cell size (42, 48). In addition to its requirement for DNA synthesis, cyclin E also plays a key role in senescence (14), development (7, 48), and modulation of downstream signals involving pRb (58) and E2F (18, 40). Normal expression and activity of cyclin E play a crucial role in cell proliferation, and any defects in its expression could have a critical effect in oncogenesis.
We and others have reinforced the linkage between oncogenesis and cyclin E by correlating the altered expression of cyclin E to the loss of growth control in breast cancer (8, 30, 32). Furthermore, several tumor cohort studies (reviewed in reference 55) have documented a strong correlation between cyclin E overexpression and poor patient outcome (31, 44) and lack of estrogen receptor expression (21, 39, 49). In addition, patients with high cyclin E levels in their tumors have a significantly increased risk of death and/or relapse from breast cancer even if they are node negative (39, 44). Additionally, examination of the oncogenic potential of cyclin E in transgenic mice under the control of the bovine β-lactoglobulin promoter revealed that lactating mammary glands of the transgenic mice overexpressing cyclin E contain hyperplasia and over 10% also develop mammary carcinomas (6). Lastly, constitutive overexpression of cyclin E (but not cyclin D1 or A) in both immortalized rat embryo fibroblasts and human breast epithelial cells results in chromosome instability (53). Collectively, these data provide strong support for the role of cyclin E in breast cancer tumorigenesis.
In normal cells, cyclin E is expressed precisely when needed and then is rapidly degraded. In tumor cells, cyclin E expression is different in several ways. First, amplification of the cyclin E gene and overexpression of cyclin E mRNA by 64-fold have been reported in a subset of breast cancer cell lines (8, 32). Second, the protein expression and associated kinase activity are no longer cell-cycle regulated (21, 50). Third, in addition to the typical 50-kDa protein, a pattern of low-molecular-weight (LMW) forms ranging in size from 49 to 33 kDa is also detected by immunoblotting (23, 31, 32). However, despite the tumor-specific expression of LMW forms of cyclin E and the compelling prognostic evidence, very little is known about what gives rise to these isoforms.
Previously we reported that although cyclin E is subject to extensive alternative splicing, these cyclin E mRNA variants do not account for the LMW forms of the protein observed in tumor cells (43). Here we show that the generation of these tumor-specific LMW forms of cyclin E is predominantly derived from proteolytic processing of the full-length cyclin E. Mass spectrometry analysis revealed that the full-length cyclin E, which is 50 kDa and is found predominantly in both normal and tumor cells, is the EL form (i.e., the 15-amino-acid [aa] elongated variant of cyclin E [42]). By using site-specific mutations and transient transfection, we have also identified two protease-sensitive domains in cyclin E. Four of the five LMW forms of cyclin E in tumor cells are accounted for by proteolysis at these two sites, with posttranslational modification of the two proteolytic forms creating closely migrating doublets. Mutation analysis of amino-acyl residues within the proteolytic cleavage domain suggests that cyclin E is a substrate for the elastase class of proteases. In addition to these proteolytically derived bands, a fifth band appears from an alternate translation start at M46 of cyclin EL. More importantly, the cyclin E LMW forms are hyperactive, as they display stronger activity toward their native substrates, such as histone H1 and GST-Rb, than the full-length form does. Additionally, these LMW isoforms of cyclin E are localized to the nucleus and can stimulate the cell to progress through the cell cycle more effectively than does the full-length form. The proteolysis of cyclin E described here is a newly discovered level of tumor-specific cyclin regulation distinct from the ubiquitin-proteasome pathway.
MATERIALS AND METHODS
Materials, cell lines, culture conditions, and transfections.
Serum was purchased from HyClone Laboratories (Logan, Utah), and cell culture medium was obtained from Life Technologies, Inc. (Grand Island, N.Y.). All other chemicals used were reagent grade. The culture conditions for the normal 70N and 81N cell strains, immortalized MCF-10A cell line, and breast cancer MCF-7, MDA-MB-157, MDA-MB-231, and MDA-MB-436 cell lines were described previously (21). The 76N-E6 cell line (gift from V. Band, Tufts Medical Institute, Boston, Mass.) was immortalized and cultured as described previously (1, 2). All cells were cultured and treated at 37°C in a humidified incubator containing 6.5% CO2 and maintained free of mycoplasma as determined by Hoechst staining (24). Transfection by electroporation was carried out on the tumor cell line MDA-MB-157 at 0.27 kV and 960 μF capacitance. For each condition, 1 × 107 cells were suspended in 0.5 ml of media (serum free), with 40 μg of the indicated plasmid in a 0.4-cm gap cuvette. Following transfection, cells were plated with complete medium and harvested 24 h posttransfection for analysis. Transfection with the FuGene Transfection Reagent was carried out in 70N, 81N, MCF-10A, 76NE6, and MDA-MB-436 cells according to the manufacturer's instructions (Roche Molecular Biochemicals, Indianapolis, Ind.). Cells were harvested for analysis 24 h posttransfection.
Sorting and analysis of transfected cells.
Following cotransfection of 81N cells with cyclin EL-FLAG and green fluorescent protein (GFP) constructs, cells (1 × 107) were harvested and resuspended in sterile ice-cold phosphate-buffered saline (PBS) and subjected to sorting with a Vantage fluorescence-activated cell sorter (FACS) from Becton Dickinson with a 488-nm argon ion laser. The GFP fluorescence was measured using a band-pass filter at 530/30 nm. Cell sorting was performed under sterile conditions. The sorting gates were set to sort the GFP-negative cells in the left tube and the GFP-positive cells in the right tube. Sorted cells were collected in a tube containing growth medium. The GFP transfection efficiency was 30%. GFP-positive cells were gated by separating nonfluorescent cells, as generated by mock transfection, from the fluorescent cells. On average, 3 × 106 cells were collected in the GFP-negative tube and 9 × 105 cells were collected in the GFP-positive tube. After sorting, an aliquot of the sorted cells was run on the FACS Vantage to check the purity of the two populations. A purity of >99% was noted for each group. The remaining cell suspensions for each condition were spun down and resuspended in ice-cold PBS and used for three different analyses. DNA content analysis was performed with 1 × 105 of the GFP-positive cells collected from each transfectant group. FLAG immunohistochemical analysis was performed on 5 × 104 GFP-positive and -negative cells collected from each transfectant group, following their plating on glass coverslips for 16 h. Lastly, Western blot analysis was performed on the remaining sorted and unsorted cells as described below.
For DNA content analysis, the GFP-positive sorted cells were centrifuged at 1,000 × g for 5 min, fixed by the gradual addition of ice-cold 70% ethanol (30 min at 4°C), and washed with PBS. Cells were then treated with RNase (10 μg/ml) for 30 min at 37°C, washed once with PBS, and resuspended and stained in 1 ml of 69 μM propidium iodide (PI) in 38 mM sodium citrate for 30 min at room temperature. The cell cycle phase distribution was determined by analytical DNA flow cytometry as described previously (29).
For Western blot analysis, cells were homogenized by sonication and high-speed centrifugation. The cleared cell lysate supernatant was assayed for total protein content and then subjected to Western blot analysis as described below.
Western blot, immunoprecipitation, and H1 kinase analysis.
Cell lysates were prepared and subjected to Western blot analysis as previously described (45). Briefly, 50 μg of protein from each condition was electrophoresed in each lane of a 10% (cyclin E, FLAG, cyclin D1, and actin) or 13% (CDK2, CDK4, p21, p27, and actin) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to Immobilon P overnight at 4°C at 35 mV constant voltage. The blots were blocked overnight at 4°C in BLOTTO (5% nonfat dry milk in 20 mM Tris, 137 mM NaCl, 0.25% Tween, pH 7.6). After six 10-min washes in TBST (20 mM Tris, 137 mM NaCl, 0.05% Tween, pH 7.6), the blots were incubated in primary antibodies for 3 h. Primary antibodies used were cyclin E HE-12 and cyclin D1 monoclonal antibodies (Santa Cruz Biochemicals, Santa Cruz, Calif.) at 1 μg/ml, anti-FLAG polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) at 0.25 μg/ml, monoclonal antibodies to p27, CDK2, and CDK4 (Transduction Laboratories, Lexington, Ky.) and monoclonal antibody to p21 (Oncogene Research Products/Calbiochem, San Diego, Calif.), all at 0.1 μg/ml in BLOTTO, and actin monoclonal antibody (Roche Molecular Biochemicals) at 0.63 μg/ml in BLOTTO. Following primary antibody incubation, the blots were washed and incubated with goat anti-mouse immunoglobulin-horseradish peroxidase conjugate at a dilution of 1:5,000 in BLOTTO for 1 h and finally washed and developed with the Renaissance chemiluminesence system as directed by the manufacturer (NEN Life Sciences Products, Boston, Mass.).
For immunoprecipitations followed by Western blot analysis, 300 μg of cell extracts was used per immunoprecipitation with polyclonal antibody to FLAG (Santa Cruz Biochemicals), as previously described (9). The immunoprecipitates were then electrophoresed in 13% gels, transferred to Immobolin P, blocked, and incubated with the indicated antibodies at dilutions described above. For the histone H1 and GST-Rb kinase assays, the immunoprecipitates were incubated with kinase assay buffer containing 60 μM cold ATP, 5 μCi of [32P]ATP, and 5 μg of histone H1 (Roche Molecular Biochemicals) or 1 μg of GST-Rb (Santa Cruz Biochemicals) in a final volume of 30 μl at 37°C for 30 min. The products of the reactions were then analyzed on either a 13% (histone H1) or 10% (GST-Rb) SDS-PAGE gel. The gels were then stained, destained, dried, and exposed to X-ray film. For quantitation, the protein bands corresponding to histone H1 and GST-Rb were excised and the radioactivity of each band was measured by Cerenkov counting.
In vitro transcription and translation assays.
To transcribe and translate the cyclin EL-FLAG constructs cloned in the pcDNA3.1 vector, we used the TNT Coupled Reticulocyte Lysate system (Promega, Madison, Wis.). Briefly, 1 μg of pCDNA3.1 plasmid containing either the cyclin EL-FLAG inserts or no insert was added to rabbit reticulocyte lysate in the presence of T7 RNA polymerase and 1 mM complete amino acid mixture in a total volume of 50 μl and incubated at 30°C for 90 min. One-microliter volumes of each of the translation products, containing both the rabbit reticulocyte lysate and the synthesized cyclin E protein, were then separated on SDS-PAGE gels and subjected to Western blot analysis using either a monoclonal antibody to cyclin E (clone HE-12) or a polyclonal antibody to FLAG (both from Santa Cruz Biochemicals).
Plasmid construction.
Block deletions (e.g., from aa 67 through 69, creating DEL6769) and amino acid substitutions (e.g., methionine substituted by alanine at position 16, creating M16A) were prepared using quick-change site-directed mutagenesis (5) or PCR cloning. The plasmid pCDNA 3.1, containing the EL form of cyclin E and a FLAG epitope (boldface) 5′-GCAAGCTTTTCACTTGTCATCGTCGTCCTTGTAGTCCGCCATTTCCGGCCCGCT-3′ at the carboxy terminus (pCDNA 3.1 EL-FLAG, [23]) was used as the starting template for all of the mutations. The following oligonucleotides were synthesized along with their antiparallel version to form a double-stranded DNA pair containing the desired mutation centered in the sequence: M16A, AAGGAGCGGGACACCGCGAAGGAGGACGGCGG; M46A, CCAGATGAAGAAGCGGCCAAAATCGAC; V67D, GACAATAATGCAGACTGTGCAGACCCC; C68D, AATAATGCAGTCGATGCAGACCCCTGC; A69D, AATGCAGTCTGTGATGACCCCTGCTCC; D70P, GCAGTCTGTGCACCCCCCTGCTCCCTG; D70A, GCAGTCTGTGCAGCCCCCTGCTCCCTG; DEL6769, TGGGACAATAATGCAGACCCCTGCTCCCTG; DEL6469, AGCCAGCCTTGGGACGACCCCTGCTCCCTG; DEL7075, CAATAATGCAGTCTGTGCACCCACACCTGACAAAG; DEL1621, TGCGAAGGAGCGGGACACCGCGGAGTTCTCGGCT; DEL2227, AAGGAGGACGGCGGCTCCAGGAAGAGGAAG; DEL2833, GAGTTCTCGGCTCGCAACGTGACCGTTTTT; DEL3439, AGGAAGAGGAAGGCACAGGATCCAGATGAA; DEL4045, GTGACCGTTTTTTTGATGGCCAAAATCGAC. PCRs were performed with 1 μg of template (pCDNA 3.1-EL-FLAG) using Pfu polymerase (Stratagene) in the buffer provided and 125 ng of each oligonucleotide pair for 16 cycles of 95°C for 30 s, 55°C for 60 s, and 68°C for 12 min. The methylated template plasmid (from growth in Escherichia coli host DH10B) was digested with DpnI restriction enzyme at 37°C for 1 h. The DpnI-resistant, unmethylated PCR product containing the mutation was desalted on a G-50 Sephadex spin column and electroporated into the E. coli host DH10B. The E. coli cells were plated on ampicillin plates, and colonies were picked and checked for successful mutation by restriction analysis and sequencing. The following mutations were made with a different strategy using oligonucleotides to PCR amplify the plasmid pLXSN containing the cyclin EL (42) cDNA as template. The DEL7681 forward primer (GTCTGTGCAGACCCCTGCTCCCTGATCGATGATGACCGG) was used with the reverse primer CE1247 (AGCAAACGCACGCCTCCGCTGCAACA). The DEL8287 (ACACCTGACAAAGAACCAAACTCAACGTGCAAG) and DEL8893 (ACACCTGACAAAGAAGATGATGACCGGGTTTACCCTCGGATTATTGCA) forward primers were combined with the bridge forward primer CE196 (GCAGTCTGTGCAGACCCCTGCTCCCTGATCCCCACACCTGACAAAGAA) and used with the reverse primer CE1247. The PCRs were performed with 10 ng of template (pLXSN-cyclin E EL) using Taq polymerase in the buffer provided for 18 cycles of 95°C for 30 s, 65°C for 20 s, and 72°C for 30 s. The PCR products were ligated into the T/A cloning vector pCRII and transformed into DH10B, and colonies were selected as above. Plasmids containing the correct mutation were digested with the restriction enzymes BsgI and AgeI, and the fragment was ligated into the same site of pCDNA 3.1 EL-FLAG. Transformation and colony selection were as described above.
Expression of the LMW forms of cyclin E in insect cells.
Plasmids pVLCDK2, pVLcycE, pVLcycET1, and pVLcycET2 were constructed by subcloning the full-length cDNA fragments for CDK2, the EL form of cyclin E, and its truncated forms (i.e., T1 and T2), respectively, into the multiple cloning sites downstream of the polyhedrin promoter of baculovirus transfer vector pVL1392 (PharMingen, San Diego, Calif.) to generate the high-expression vector systems for these proteins. Recombinant viruses were produced by cotransfection of the above purified plasmids, separately, with linearized BaculoGold virus DNA (PharMingen) into Sf9 insect cells. For the resultant recombinant viruses, B.G.CDK2, B.G.cycE, B.G.cycET1, and B.G.cycET2, titers were determined by end-point dilution assay and amplifications were performed by infecting the Sf9 insect cells at a low multiplicity of infection (MOI) (0.7), maintaining the high percentage (99%) of the recombinant viruses in the large stock and therefore assuring the high expression of the desired proteins. The supernatants of viruses amplified were harvested around 60 h postinfection (p.i.). Protein extracts were subjected to Western blotting, immunoprecipitation, and H1 kinase analysis, as described above.
Coinfection of the viruses B.G.CDK2 plus B.G.cycE, B.G.CDK2 plus B.G.cycET1, and B.G.CDK2 plus B.G.cycET2 was carried out by mixing the two viruses at a 1:1 ratio and coinfecting the Sf9 insect cells at an MOI of 1.4 (0.7 of each virus). As controls, the Sf9 cells were also infected with the four different viruses separately, each at an MOI of 1.4, at the same time. After around 60 h p.i., infected cells were harvested and protein extracts from these cells were subjected to Western blotting, immunoprecipitation, and H1 kinase analysis, as described before.
Porcine pancreatic elastase digestion.
Plasmids containing the full-length cyclin EL, block deletions A to E, A′, and point mutations of cyclin E FLAG-tagged constructs as well as the vector-alone construct were translated in vitro using a TNT kit (Promega) as described above. A total of 10−4 U of the porcine pancreatic elastase (Sigma Biochemicals) was mixed with 0.25 μl of the TNT reaction mixture (i.e., 5 μl of a 1:20 dilution) in the elastase reaction buffer (50 mM Tris [pH 8.5], 250 mM NaCl) and incubated at 30°C for 5 min. At the end of incubation the entire reaction mixture (total volume, 20 μl) was subjected to SDS-PAGE and Western blot analysis with anti-FLAG antibody.
Cyclin E purification and MS.
Cyclin E was purified from 500 mg of MDA-MB-157 cell extract prepared by sonication and centrifugation as described previously (45). The extract was precleared with three successive incubations with 1 ml of Sepharose CL6B rotated at 4°C for 2 h. After preclearing, the extract was incubated overnight with 1 ml of the monoclonal antibody HE-111 cross-linked to Sepharose (Santa Cruz). The antibody resin was washed with Buffer A (50 mM Tris [pH 8.0], 1 mM EDTA, 0.5 M NaCl, 0.5% Nonidet P-40), Buffer B (50 mM Tris [pH 8.0], 1 mM EDTA, 0.15 M NaCl, 0.1% SDS, 0.5% Nonidet P-40), and Buffer C (50 mM Tris [pH 8.0], 1 mM EDTA, 0.15 M NaCl, 0.5% Nonidet P-40) and then eluted by boiling in SDS-polyacrylamide, reducing, sample buffer. The eluted sample was loaded onto a 10% polyacrylamide gel, electrophoresed, and silver stained (omitting the glutaraldehyde step [38]). A gel slice was excised and a 1-mm fragment was electrophoresed again and evaluated by Western blotting to confirm the presence of cyclin E. The gel slice was digested using modified trypsin from Boehringer Mannheim. Reactive cysteines were reduced with 10 mM dithiothreitol (DTT) and blocked with iodoacetamide (aminocarboxy methylation). The gel slice was dehydrated in acetonitrile for 30 min and then dried in a vacuum. The slice was rehydrated in 10 mM DTT, 100 mM ammonium bicarbonate at 55°C for 1 h. This solution was replaced with 55 mM iodoacetamide, 100 mM ammonium bicarbonate in the dark for 45 min at room temperature with vortexing. The slice was then washed with 100 mM ammonium bicarbonate for 10 min, dehydrated in acetonitrile for 30 min, and dried in a vacuum. One more round of hydration-dehydration was performed, and the gel slice was swollen in digestion buffer (50 mM ammonium bicarbonate, 5 mM CaCl2, 12.5 ng of trypsin/μl) and digested at 37°C for 18 h. The peptides were extracted and analyzed by matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) as follows. The gel slice was extracted twice with 0.1% trifluoroacetic acid (TFA) and 70% acetonitrile in a small volume for 45 min. The sample was concentrated using a Speed-Vac. The combined extracts were loaded onto a C18 Zip Tip (Millipore), washed with 0.1% TFA, and eluted with 10 to 70% acetonitrile. One microliter of the eluted sample was spotted on target along with 1 μl of matrix (10 mg of alpha-cyano-4-hydroxycinnamic acid/ml) solubilized in 70% acetonitrile and analyzed on a Bruker Reflex delayed-extraction MALDI-TOF apparatus.
RESULTS
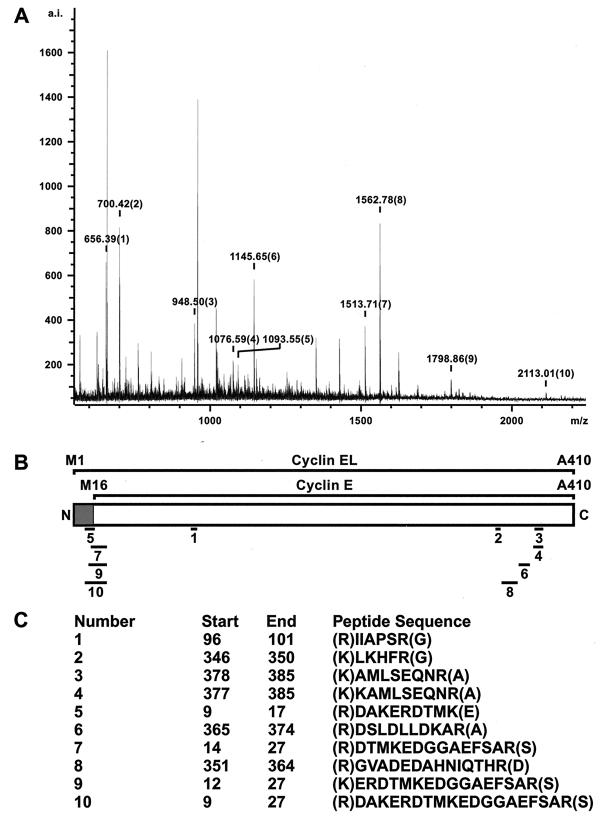
MS identifies the predominant cyclin E as the EL form.
The characteristic pattern of cyclin E processing found in tumor cells and tumor tissues includes a number of LMW forms ranging in size from 33 to 45 kDa, in addition to the 50-kDa full-length form which is found in both normal and tumor cells (31). We used MS to identify the full-length 50-kDa cyclin E in tumor cells by determining the N terminus directly from tryptic peptides of the purified cyclin E protein. To this end, we purified cyclin E from 2 × 1010 MDA-MB-157 cells (500 mg of total cellular extract) with a cyclin E monoclonal antibody (clone HE-111) affinity column. The purified cyclin E was eluted from the column and resolved by SDS-PAGE. After silver staining the gel, the band corresponding to the full-length (i.e., 50 kDa) cyclin E was excised, reduced, alkylated, and digested with trypsin according to established procedures (52). The purified cyclin E tryptic peptides were extracted from the gel slice and analyzed by MALDI-TOF MS, which yielded 10 peptides with internally calibrated masses matching predicted cyclin E peptides from both the N- and C-terminal domains of cyclin E (Fig. 1). The mass spectrum of the purified cyclin E tryptic digests as analyzed by MALDI-TOF is depicted in Fig. 1A. The alignments of the 10 peptides to cyclin E are shown underneath the schematic representation of cyclin EL (Fig. 1B). The shaded region defines the unique cyclin EL sequences not found in the 45-kDa cyclin E. Cyclin EL is the 15-aa elongated variant of cyclin E identified in 1995 (42), while the 45-kDa cyclin E corresponds to the gene identified in the yeast complementation studies first identifying cyclin E (33, 34). Four of these tryptic peptides (numbers 5, 7, 9, and 10) contain sequences from the N terminus of the EL form of cyclin E which are not present in the 45-kDa form of cyclin E (Fig. 1C). All four peptides contain an internal methionyl residue from position 16 (M16) of the intact EL form of cyclin E, which could only derive from the EL cyclin E and not the 45-kDa cyclin E, with a predicted translation start at M16. This MS analysis of cyclin E purified from breast cancer cells confirms that the 50-kDa full-length cyclin E found in these cells is the cyclin EL. Therefore, we selected the EL form of cyclin E as the backbone for all subsequent expression vectors.
FIG. 1.
MS identified cyclin EL as the 50-kDa cyclin found in tumor cells. (A) MALDI-TOF analysis of the tryptic digests of the purified cyclin E band excised from a silver-stained PAGE gel. The major identified peaks all correspond to cyclin E when compared to the available databases through Protein Prospector. Cyclin E peaks are designated with a mass and a number. Unlabeled peaks correspond to trypsin self-digestion products or were not identified. (B) Schematic representation of cyclin EL and cyclin E. Solid bars show alignments of peptides to different regions of cyclin EL. The shaded region is unique to cyclin EL. (C) Peptide sequences obtained from MALDI-TOF analysis of cyclin E tryptic digests. The numbering corresponds to the peaks identified in panel A. The internally calibrated masses submitted to Protein Prospector are within 0.06 Da of the peptides matched.
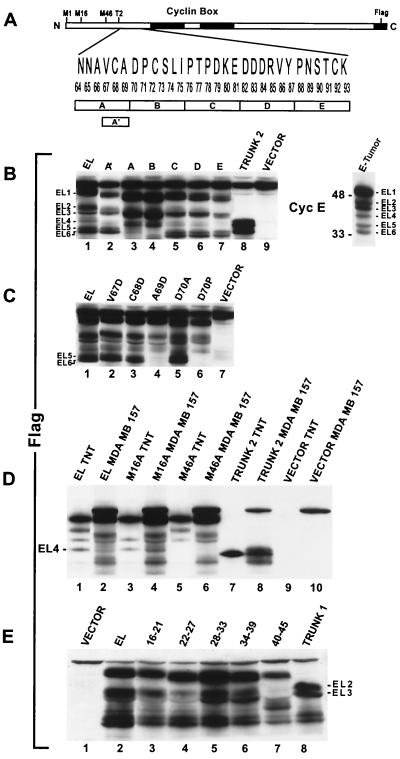
Block deletions and amino-acyl substitutions define cyclin E proteolysis in tumor cells.
Previously, we showed that the processing of cyclin E required to generate the LMW forms detected only in tumor cells occurs posttranslationally (23). To identify the exact position where cyclin E is proteolytically processed, we transiently transfected the full-length and selectively deleted and/or mutated forms of C-terminus epitope-tagged, cyclin EL-FLAG into tumor cells and analyzed their expression by Western blotting using a FLAG antibody (Fig. 2B). The results revealed that in the MDA-MB-157 tumor cell line the exogenously expressed FLAG-tagged cyclin EL (Fig. 2B, lane EL) is processed very similarly to the endogenous cyclin E (Fig. 2B, lane E-Tumor), generating the same Western blotting pattern of bands. We used a numbering system to facilitate the analysis of the cyclin E LMW forms. The full-length 50-kDa form of cyclin E is EL1. Below EL1 is a doublet migrating at 45 and 44 kDa and designated as EL2 and EL3. A single band migrating at 40 kDa is EL4, and another doublet below this at 35 and 33 kDa is designated EL5 and EL6. The Western blotting patterns of cyclin EL-FLAG isoforms EL1 to -6 were comparable in other transfected breast cancer cell lines (see below).
FIG. 2.
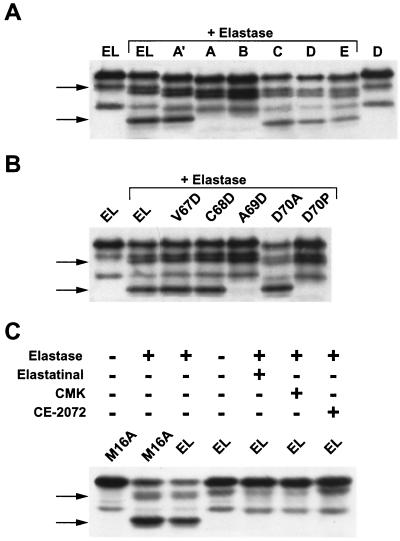
Deletional analysis of cyclin E identifies sequences responsible for generating the LMW forms. (A) Schematic representation of cyclin EL, highlighting the sequences used for deletional analysis. Five block deletions of 6 aa each were made in a 30-aa region (aa 64 to 93) designated A-E. The block deletion A′ contains a 3-aa deletion spanning aa 67 to 69. T2 represents the start of cyclin EL-Trunk2. All constructs were made with an 8-aa FLAG peptide fused at the carboxy terminus. (B) The block deletions A to E and A′ as well as the cyclin EL, Trunk 2, and vector alone were transiently transfected into MDA-MB-157 cells. Protein was extracted 24 h posttransfection and subjected to Western blot analysis with an anti-FLAG antibody. Note that the block deletions A to E are each 6 aa shorter than EL and migrate faster on the gel. The band above cyclin EL1 is protein that is cross-reactive with the FLAG antibody and can be seen in all lanes including the vector-alone lane. On the left of the Western blot, the relative mobilities of the LMW bands are labeled EL1 to -6. Shown on the right is the cyclin E Western blot of the total cell extract from the tumor cell line MDA-MB-157 (untransfected), with the LMW bands (EL1 to -6) labeled. (C) Transient transfection of cyclin EL-FLAG constructs harboring the indicated point mutations in aa 67 to 70, followed by Western blotting with FLAG antibody. (D) The EL, M16A, M46A, and Trunk 2 plasmid constructs with the FLAG tag were transiently transfected into MDA-MB-157 cells. Protein was extracted 24 h posttransfection and subjected to Western blot analysis with an anti-FLAG antibody. The same plasmids were also transcribed or translated in vitro (TNT assay) and subjected to SDS-PAGE and Western blot analysis in the indicated lanes. The LMW band labeled EL4 is not translated in vitro or in transient transfection of MDA-MB-157 cells when the methionyl at position 46 is mutated to an alanyl residue (lanes 5 and 6). Vector TNT and vector MDA-MB-157 represent in vitro-translated and -transfected empty vectors, respectively. (E) Transient transfection of cyclin EL-FLAG constructs harboring the indicated block deletions in aa 16 to 45, followed by Western blotting with FLAG antibody.
The region of cyclin E where N-terminal proteolysis occurs was previously refined with a series of truncated versions of cyclin E (23) to a 30-aa region from residues 64 to 93 (Fig. 2A). To define the exact amino-acyl residues involved in the proteolysis, a series of five successive 6-aa block deletions of cyclin EL as well as conservative and nonconservative substitutions were made in amino-acyl residues 64 to 93. The mutated cyclin E plasmids were then transiently expressed in tumor cells and evaluated by Western blot analysis with FLAG antibody (Fig. 2B).
These results show that two 6-aa block deletions, designated A (residues 64 to 69) and B (residues 70 to 75) (see Fig. 2A for schematic representation of these block deletions), were able to prevent the proteolysis of cyclin E into bands EL5 and EL6 when transfected into the cell line MDA-MB-157 (Fig. 2B, lanes A and B). None of the other bands appeared to be affected except for the reduction in mass due to the 6-amino-acyl deletion (Fig. 2B, lanes C to E). A smaller (three amino-acyl) block deletion, named A′, slightly reduced but did not eliminate processing (Fig. 2B, lane A′). Any other large block deletion further toward the C terminus had no effect on processing (Fig. 2B, lanes C to E).
We suspected that the A deletion contained the protease recognition motif; therefore, this domain was targeted for site-specific amino-acyl substitution. The B deletion may not contain a specific protease recognition sequence, but the deletion brings P76 adjacent to A69, disrupting the ability of the protease to cleave at this site. To test these possibilities, we constructed a series of conserved and nonconserved amino-acyl substitutions from aa 67 to 75 (Fig. 2C). These substitutions confirmed that small hydrophobic amino acids at positions 67, 68, and 71 to 75 are not essential to generate cyclin EL5 and EL6, as shown by the presence of these two LMW forms following individual substitutions of any of these residues with alanine or aspartate (Fig. 2C, lanes 2 and 3, and data not shown). However, the amino acids at positions 69 and 70 are required for the generation of both cyclin forms EL5 and EL6. When the alanine at position 69 (A69) is substituted with aspartate (A69D) or when aspartate at position 70 (D70) is substituted by proline (D70P), cyclin E is not cleaved into bands EL5 and EL6 (Fig. 2C). Additionally, when alanine is substituted at D70 (D70A), not only is cyclin E cleaved into the EL5 and EL6 bands but also the levels of these products are induced (Fig. 2C, lane 6), suggesting that the D70A substitution may be a superior substrate for the cyclin E protease compared to the wild type (Fig. 2C, compare D70A to EL). There is precedent for this finding, as proline substitution classically converts a substrate into an inhibitor when placed into the P1′ position of the scissile bond of the proteolytic cleavage site (3).
Lastly, transfection of an N-terminal-truncated cyclin EL, termed Trunk 2 (i.e., a 64-aa N-terminal truncation with translation initiating at a methionyl residue replacing an asparaginyl residue at position 65; Fig. 2A) served two purposes. First, it helped pinpoint the region of proteolysis, since its transfection into MDA-MB-157 resulted in further processing of this construct into an LMW form comigrating with EL5 and -6 (Fig. 2B, lane 8). Additionally, processing so close to the N-terminal deletion suggests that the protease only requires a domain extremely local to the point of proteolysis. We have identified this domain as a sequence targeted by the elastase class of proteases (see below).
The cyclin E 45-kDa form is not expressed in tumor cells.
We suspected that bands EL2 and -3 represented protein translated from the M16 start site originally thought to be the primary form of cyclin E (33, 34). However, we were surprised when an M16A substitution did not affect the expression of bands EL2 and -3 (Fig. 2D). For these analyses, we compared the in vitro-translated products of cyclin EL1 and of cyclin EL1 mutated at M16 or M46 to an alanyl residue, with the proteolytically processed protein products from transfected tumor cells (Fig. 2D). When the protein from EL1-FLAG-transfected MDA-MB-157 was run side by side with the in vitro-translated cyclin E products from the same vector, the bands EL2 and EL3 clearly migrated faster than the in vitro translation products from the M16 translation start site (Fig. 2D, compare lanes 1 and 2). This difference in migration between the M16 in vitro-translated band and EL2 and -3 in the tumor cells was a hint that M16 may not be required to generate EL2. EL2 is the LMW band we suspected was the 45-kDa cyclin E wild type (33, 34). We then replaced the methionyl residues at positions 16 and 46 with an alanyl residue, knocking out any alternative translation initiation from these sites (Fig. 2D). The M16A and M46A substitutions clearly eliminated translation starts from these sites when translated in vitro (lanes 3 and 5). However, only the M46A substitution eliminated EL4 from transfected MDA-MB-157 cells (lane 6), while the M16A substitution had no effect on the generation of EL2 (lane 4). Even though EL2 and -3 migrate closely to the cyclin E translated from M16, these bands are most likely created by proteolysis rather than by alternate translation at M16. The in vitro mutagenesis of cyclin E shown here and the MS data (Fig. 1) collectively provide strong evidence that the high-molecular-weight, full-length form of cyclin E found in tumor cells is translated from the cyclin EL mRNA (42) and not the cyclin E wild-type form initially cloned as this G1 cyclin (33, 34). Furthermore, while the EL5 and -6 and EL2 and -3 LMW forms are due to proteolytic processing, EL4 can be accounted for by an alternate translation start site, as shown by the elimination of this band after the mutation of the methionyl at residue 46 into alanyl, both in transfected cells and when the protein is translated (i.e., TNT assay) in vitro (Fig. 2D, lanes 5 and 6).
Lastly, an examination of the proteolytic process generating EL2 and EL3 was performed similarly to the process used to define EL5 and EL6. Successive 6-aa block deletions in cyclin E were created in the EL-FLAG vector used to transfect the tumor cell line MDA-MB-157. These block deletions span the region from M16 to M46 (Fig. 2E). The results from FLAG Western blot analysis of cells transfected with these block deletions reveal that the processing of cyclin E into EL2 and EL3 is not eliminated by block deletions spanning aa 16 to 39. However, the block deletion of aa 40 to 45 results in the prevention of cyclin E proteolysis into bands EL2 and EL3 when transfected into MDA-MB-157 cells (Fig. 2D). These results suggest that the amino acids targeted by proteolysis to generate cyclin EL2 and EL3 are within residues 40 to 45 of the cyclin EL protein.
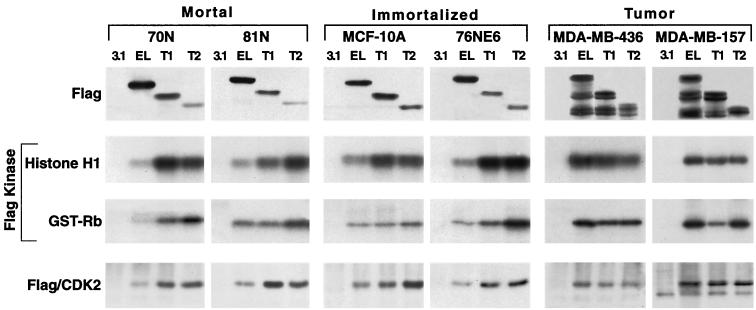
The LMW forms of cyclin E are hyperactive.
To examine the biochemical activity of the LMW forms of cyclin E compared to that of the full-length form, we used three different constructs of cyclin E that were FLAG tagged at the C terminus in transfection assays using normal and tumor cells. The three constructs are cyclin EL-FLAG, Trunk 1, and Trunk 2, as described previously (23). Trunks 1 and 2 bracket only the LMW isoforms of cyclin E and not the full-length form, while cyclin EL represents the full-length form. Trunk 1 initiates at aa 40 and brackets EL2–6 (Fig. 2E, lane 8), and Trunk 2 initiates at aa 65 and brackets EL5 and EL6 (Fig. 2D, lane 7). We have examined the biochemical activities associated with the protein products of cyclin EL, Trunk 1, and Trunk 2 constructs in several normal and tumor cell lines (Fig. 3).
FIG. 3.
The LMW forms of cyclin E are hyperactive. Cyclin EL-FLAG and cyclin E-FLAG constructs Trunk 1 and Trunk 2 were transfected into two mortal cell strains (81N and 76N), two immortalized cell lines (MCF-10A and 76NE6), and two tumor cell lines (MDA-MB-157 and MDA-MB-436), harvested 16 h posttransfection, and subjected toWestern blot analysis with anti-FLAG antibody, histone H1 and GST-Rb kinase analysis, or immune complex formation with CDK2. For Western blot analysis, 50 μg of protein extract from each condition was analyzed with polyclonal antibody to FLAG. For kinase activity and immune complex formation, equal amounts of protein (250 μg) from cell lysates were prepared from each condition and immunoprecipitated with anti-FLAG antibody (polyclonal) coupled to protein A beads. Histone H1 and GST-Rb were used as substrates in the kinase reaction. For each condition, the resulting autoradiogram of the histone H1 and GST-Rb SDS-PAGE are shown. Immune complex formation with CDK2 was assessed by subjecting the anti-FLAG immunoprecipitates to Western blot analysis using a monoclonal antibody to CDK2.
Transfection of the two normal mortal cell strains (70N and 81N) and the two immortalized cell lines (MCF-10A and 76NE6) with each of the three cyclin E-FLAG constructs results predominantly in the expression of the full-length form of each construct, as shown in the Western blot analysis using FLAG antibody (Fig. 3). Following transient transfection of each cell line with the cyclin E-FLAG constructs, the activities of the resultant protein products were assessed by histone H1 and GST-Rb phosphorylation and immune complex formation with CDK2 (Fig. 3). Histone H1 and GST-Rb were used as substrates for active cyclin E-CDK2 complexes in immunoprecipitates prepared with an antibody to FLAG. This analysis revealed that the truncated forms of cyclin E activate a greater amount of kinase activity than the EL form in the four normal cell lines (mortal or immortalized). In all four normal cell types, Trunk 1 and Trunk 2 phosphorylated histone H1 and GST-Rb between two- and sixfold more effectively than the full-length cyclin E. (The bands corresponding to histone H1 or GST-Rb were excised and quantitated by scintillation counting [data not shown].) The expression levels of the truncated forms are generally less than that of the EL form (Fig. 3, top panel, lanes EL, T1, and T2). In spite of this lower protein expression, the kinase activity is significantly (i.e., two- to sixfold) higher and this effect is more noticeable in the non-tumor cell lines. The same non-tumor cell lines also show an increased amount of CDK2 immunoprecipitating with the truncated cyclin E. Hence, the kinase activity associated with each construct was proportional to the amount of complex formation between cyclin E-FLAG constructs and CDK2 in each of the normal cell lines (Fig. 3, compare Flag kinase panels with the Flag/CDK2 panel). Although this could represent an increased affinity of the truncated forms of cyclin E for CDK2, it is more likely it involves an upregulation of CDK2 protein levels, since the CDK2 levels increase after transfection as shown by Western blotting (Fig. 4A). Collectively, these results suggest that in normal cells the forced expression of the LMW forms of cyclin E has the ability to bind to and activate CDK2 in order to phosphorylate substrates such as histone H1 and GST-Rb much more effectively than the full-length form.
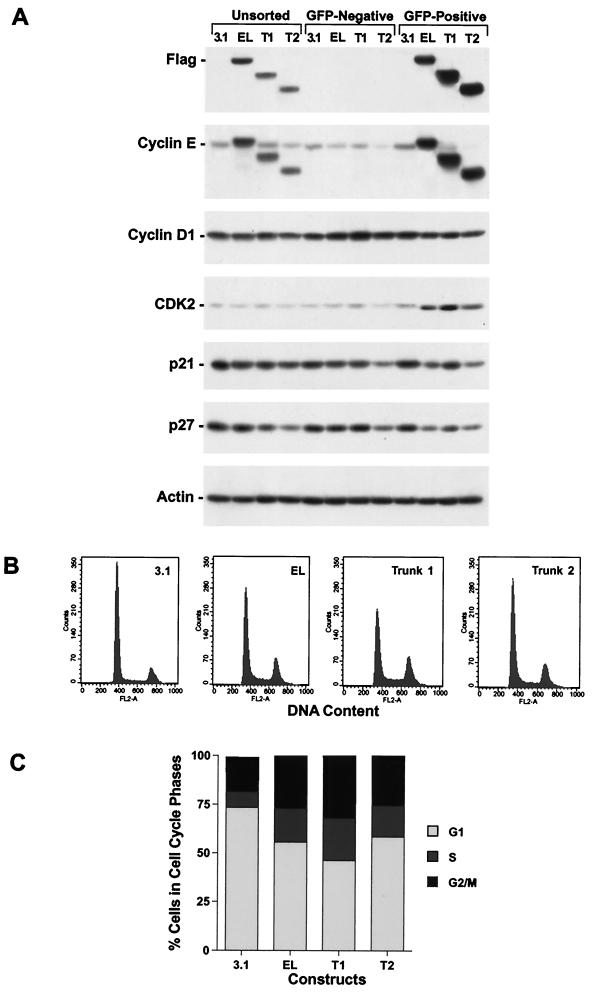
FIG. 4.
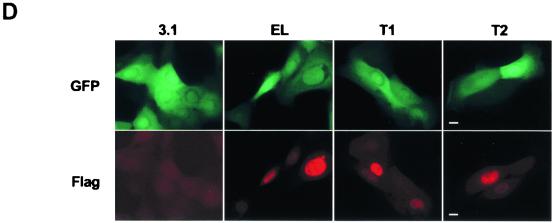
The LMW forms of cyclin E are biologically active. 81N mortal human mammary epithelial cells were cotransfected with either vector alone backbone (i.e., 3.1), cyclin EL-FLAG, cyclin E-Trunk 1-FLAG, or cyclin E-Trunk 2-FLAG and a GFP-containing vector at a 3:1 ratio. (A) Sixteen hours posttransfection, cells were harvested, GFP-containing cells were sorted, and the sorted and unsorted cells were then subjected to Western blot analysis with the indicated antibodies. (B and C) The GFP-positive cells were then stained with PI and subjected to flow cytometry analysis. Panel B shows the FACS scan raw data of the GFP-sorted cells, and panel C graphically shows the distribution of cells in each phase of the cell cycle. (D) Sorted cells were also plated on microscope slides, allowed to attach for 16 h at 37°C, and stained with FLAG antibody. Following antibody staining, samples were mounted and examined with a Nikon Optiphot microscope equipped with an oil immersion 60×, 1.4 numerical aperture, differential interference contrast objective. Digital images were collected with a SPOT camera (Diagnostics Instruments Inc.) using filters for fluorescein and Texas red, and for each channel, the exposure times were kept constant. Bar, 10 μm.
Tumor cells behave differently than normal cells in processing and activating these constructs. For example, transfection of the two tumor cell lines, MDA-MB-157 and MDA-MB-436, with the three constructs resulted in the processing of several LMW forms as well as the full-length form of each construct (Fig. 3). Additionally, the kinase activity associated with the full-length construct was higher than that for Trunk 1, which was in turn higher than that for Trunk 2 in tumor cells. Lastly, immune complex formation of the three cyclin E-FLAG constructs to CDK2 in tumor cells does not show a significant difference in binding of the full-length cyclin E-FLAG construct to CDK2 than either Trunk 1-FLAG or Trunk 2-FLAG constructs. Since tumor cells have the machinery to process the full-length cyclin E into its LMW forms, the activity associated with the cyclin EL-FLAG is a combination of the full-length form and all the LMW forms it is processed to. However, unlike tumor cells, normal cells do not process cyclin EL-FLAG into its LMW forms. Hence, the forced expression of the LMW forms of cyclin E in normal cells provides us with an ideal model system to assess the activities of the individual protein products of Trunk 1 and Trunk 2 constructs on cell cycle progression compared to activities of the full-length form.
The LMW forms of cyclin E facilitate S-phase entry.
Next, we examined the biological activity of the LMW forms of cyclin E by addressing if their overexpression in normal cells would have a mitogenic effect on the cell cycle and increase progression of cells through S phase. For these studies, we transfected the three cyclin E constructs (i.e., cyclin EL-FLAG, Trunk 1-Flag, and Trunk 2-Flag) into a normal mammary epithelial 81N cell strain. These constructs were cotransfected with a plasmid containing enhanced GFP (EGFP). Sixteen hours following cotransfection, cells were harvested and subjected to FACS analysis to isolate the GFP-positive cells from GFP-negative cells. Following sorting of cells, the EGFP-positive cells were divided into three samples. The first group of EGFP-positive cells was extracted and prepared for Western blot analysis along with the EGFP-negative sorted and unsorted cells (Fig. 4A). The second group of EGFP-positive cells was fixed and prepared for cell cycle analysis (DNA content with PI) using flow cytometry (Fig. 4B and C). The third sample was plated onto coverslips for analysis by anti-Flag immunofluorescence (Fig. 4D).
Sorting cells based on EGFP cotransfection proved effective as a means of selecting Flag-tagged cyclin E-expressing cells (Fig. 4A, top panels). The unsorted cells in the first four lanes showed a typical pattern of expression of the Flag-tagged cyclin E constructs in normal cells (Fig. 4A, Unsorted). The next four lanes represent the EGFP-negative cells, dramatically demonstrating the efficiency of FACS. There is no detectable Flag signal present in these EGFP-negative sorted cells. The final four lanes show the Western blot signal from the EGFP-positive cells. Complementing the findings shown in the previous four lanes, these lanes show a strong Flag signal. Compared to the unsorted cells, the EGFP-positive cells have a much stronger Flag signal, reflecting the lack of dilution from the untransfected cells. These results indicate that most, if not all, of the GFP transfected cells also expressed the protein products of the cyclin E-FLAG constructs, while none of the GFP-negative cells expressed the cyclin E-FLAG protein products and hence were not cotransfected with the FLAG constructs.
If cyclin E-FLAG has mitogenic activity on the cell cycle, it may also affect the expression of other cell cycle regulators. To test this hypothesis, we examined the expression of CDK2, cyclin E (endogenous), cyclin D1, p21, and p27 in the GFP-expressing cells (Fig. 4A). Our results revealed that CDK2 levels increased substantially in cells transfected with the cyclin E-FLAG constructs compared to vector-alone controls. Ectopic expression of Trunk 1, one of the LMW forms of cyclin E, had the most profound effect on CDK2 expression. However, the full-length cyclin E and Trunk 2 also increased CDK2 expression over vector-alone transfectants. Furthermore, the levels of p27 and p21 were decreased (slightly, however) in the FLAG transfectants. Levels of cyclin D1 were unaffected. Lastly, the mere overexpression of the exogenous cyclin E-FLAG constructs, up to 10-fold over that of endogenous cyclin E, did not result in proteolytic processing of cyclin E in these normal cells (Fig. 4A, Cyclin E panel). Collectively, these results suggest that the LMW forms of cyclin E, and to a lesser degree the full-length form, are involved in a positive feedback loop, where CDK2 levels, and presumably its activity, are increased and p27 and p21 levels are decreased. Such a positive feedback loop would be predictive of facilitated progression through G1 and into S phase.
We next examined the cell cycle progression of EGFP-positive, FLAG-transfected cells by staining the sorted cells with PI and analyzing them by flow cytometry for DNA content (Fig. 4B and C). These results revealed that transfection of full-length cyclin E (i.e., EL), Trunk 1, or Trunk 2 resulted in a significant progression of the cells into S and G2/M. Trunk 1 was the most mitogenic form of cyclin E, decreasing the G1 phase from 70 to 45% and increasing the S-plus-G2/M phases from 30 to 55%. However, full-length cyclin E and Trunk 2, the smallest of the LMW forms of cyclin E, were also quite effective in facilitating G1-to-S-phase progression. On average, the G1 phase in cells harboring EL, Trunk 1, or Trunk 2 dropped between 20 and 37% while the S-plus-G2/M content of cells increased by 147 to 265%, i.e., over twofold in Trunk 1-transfected cells (Fig. 4C). These results suggest that overexpression of at least two of the LMW forms of cyclin E in normal cells has a profound mitogenic effect, stimulating the cells to progress through the cell cycle. The effect on G2 accumulation by Trunk 1 and Trunk 2 is different, with Trunk 1 having a more profound G2 accumulation than Trunk 2. These observations suggest that there may be two different mechanisms at work promoting tumor survival, one involving Trunk 1 (EL2/3) and another involving Trunk 2 (EL5/6).
We also examined the intracellular localization of the LMW forms of cyclin E compared to that of the full length. It has been well established that the endogenous cyclin E full-length form is predominantly nuclear (42). To determine the subcellular localization of the LMW forms of cyclin E, the GFP-positive sorted cells were stained with affinity-purified anti-FLAG antibody (Fig. 4D). The fluorescence microscopy images in Fig. 4D show transfected, EGFP-positive sorted cells in the upper panels. In the same cells, the immunofluorescence localization of the cotransfected Flag-tagged cyclin E is shown in the lower panels. In each case, Flag-tagged cyclin E localized to the nucleus. The expression levels of the Flag-tagged cyclin E isoforms varied from cell to cell, yet even the highest expression levels showed no cytoplasmic staining above background. This shows that the nuclear localization of cyclin E can occur even when a significant amount of the N terminus has been deleted (Fig. 4D, T2 panels).
Hyperactive LMW forms of cyclin E have increased affinity for CDK2.
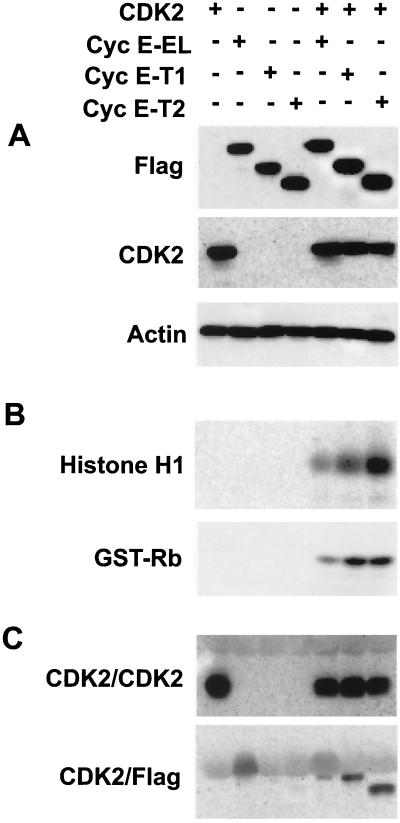
The results from our transfection experiments (Fig. 3) revealed that the tumor-specific LMW forms of cyclin E promote more kinase activity than the full-length protein, explaining their selection in tumor cells. Furthermore, these LMW forms of cyclin E can also induce the expression of more total CDK2 protein in transfected human cells (Fig. 4A). These data raise the question of whether the LMW forms are hyperactive due to their preferentially inducing the expression of more CDK2 or whether these forms are hyperactive regardless of how much CDK2 is present in the cell. To directly address this question quantitatively, we overexpressed full-length cyclin E and its LMW forms and CDK2 in insect cells using a baculovirus expression system (Fig. 5). Insect cells were coinfected with the recombinant baculovirus containing CDK2 and either full-length cyclin E, cyclin E-T1, or cyclin E-T2 cDNAs (Fig. 5). As a control for expression of individual proteins, insect cells were also infected with only one baculovirus, representing each of the aforementioned cDNAs. Sixty hours p.i., cells were harvested and subjected to Western blotting (Fig. 5A) and histone H1 and GST-Rb kinase (Fig. 5B) and immunoprecipitation (Fig. 5C) analyses. Western blot analysis with FLAG or CDK2 antibodies showed that there were similar levels of expression of the three cyclin E forms and of CDK2 in the infected insect cells. Furthermore, whether CDK2 was infected alone or in combination with the three cyclin E forms, equal levels of the protein were expressed in these cells. CDK2 kinase assays using either histone H1 or GST-Rb as substrate revealed that the truncated forms of cyclin E activate a greater amount of kinase activity than the EL form (Fig. 5B) under conditions where equal amounts of CDK2 were immunoprecipitated from each sample (Fig. 5C). In the coinfected insect cells, Trunk 1 and Trunk 2 phosphorylated histone H1 and GST-Rb between two- and sixfold more effectively than the full-length cyclin E. (The bands corresponding to histone H1 and GST-Rb were excised and quantitated by scintillation counting [data not shown].) Interestingly, the fold increase in the hyperactivity of the LMW forms of cyclin E over the full length was very similar in the insect cell baculovirus expression system and the transfection experiments in normal mammary epithelial cells (Fig. 3). Immunoprecipitation analysis revealed that the hyperactivity of the LMW forms of cyclin E was not due to different levels of CDK2 in the cell, since the amount of CDK2 immunoprecipitated was identical in all the coinfected samples (Fig. 5C). What is different among the three forms of cyclin E is their ability to bind to CDK2. The LMW forms of cyclin E bind more effectively to CDK2 than the full-length form, as was apparent in CDK2 immunoprecipitates blotted with FLAG (Fig. 5C) (cyclin E immunoblots showed identical results [data not shown]). These experiments suggest that even though the same amounts of cyclin E and CDK2 are present in the cell and equal amounts of CDK2 are immunoprecipitated from each sample, the LMW forms of cyclin E can bind to CDK2 more effectively than the full-length form, resulting in their hyperactivity. The results from the insect cell baculovirus expression system (Fig. 5) clearly recapitulated the results from our transient-transfection assays (Fig. 3 and 4) and collectively suggest that the LMW forms of cyclin E are indeed hyperactive.
FIG. 5.
The LMW forms of cyclin E bind more effectively to CDK2 than does the full-length form of the protein. Cell lysates were prepared from insect cells coinfected with baculovirus containing the indicated cyclin E constructs and CDK2. (A) At 60 h p.i., equal amounts (50 μg) of protein were added to each lane; the gel was then subjected to Western blot analysis with the indicated antibodies. (B) Histone H1 and GST-Rb kinase assays were also performed on the same cell extracts by immunoprecipitating equal amounts of cell lysate with polyclonal antibody to CDK2 coupled to protein A beads, using histone H1 or GST-Rb as substrates. The autoradiograms of the histone H1 and GST-Rb SDS-PAGE are depicted. (C) Immune complex formation with CDK2 was assessed for the same samples by subjecting the anti-CDK2 immunoprecipitates to Western blot analysis using monoclonal antibodies to CDK2 and FLAG.
Identification of an elastase class of enzymes as the protease generating the LMW forms of cyclin E in vitro.
Since the LMW isoforms of cyclin E have a significant biochemical and biological function in inducing G1-to-S-phase progression, we next set out to identify the putative protease responsible for the aberrant cleavage of cyclin E in tumor cells. The results shown in Fig. 2 suggest a novel type of proteolysis cleaving cyclin E into EL5 and EL6. We refer to the cyclin E protease domain stretching from A66 to P71 as AVCADP. This is a short hydrophobic stretch of amino acids followed by aspartate-proline and embedded in a random coil (GOR4-predicted) secondary structure (termed loop 2) bearing a striking similarity to the scissile domains of the serine proteinase inhibitors plasminostrepsin and streptomyces subtilisin inhibitor, making the AVCADP sequence accessible to the protease (3). A second cleavage point nearer to the N terminus in the domain from K32 to K48 (anticipated to generate EL2/3) has the sequence VFLQDP, similar to AVCADP, appears to be in a similar random coil structure (GOR4), and is termed loop 1.
The AVCADP and VFLQDP sequences in cyclin E, whose cleavage generates EL5/6 and EL2/3, respectively, are consensus sequences for the elastase class of serine proteases (3). In order to demonstrate the elastase quality of cyclin E proteolysis, cyclin E was synthesized through in vitro translation using rabbit reticulocyte lysate and then partially digested with porcine pancreatic elastase (Fig. 6). Each of the mutations described in Fig. 2B and C were also used in this in vitro study. These analyses clearly showed that the proteolysis of cyclin E in cells is very similar to proteolysis of cyclin E in vitro using commercial elastase (Fig. 6A and B). In vitro-translated (reticulocyte lysate) wild-type cyclin E and similar proteins translated from constructs containing mutations previously used for transfection all digest in vitro with a porcine pancreatic elastase, generating a Western blot pattern similar to the in vivo (i.e., transfected) pattern (compare Fig. 6A with Fig. 2B). Incubation of the purified elastase enzyme with in vitro-translated cyclin EL products resulted in generation of EL3 and EL6 (Fig. 6A; compare lanes 1 and 2). Block deletions A and B knocked out EL6 production, while block deletions A′, C, D, and E were incapable of knocking out the EL6, similar to tumor cell transfection of FLAG-tagged cyclin EL mutant constructs (i.e., Fig. 2B). Incubation of the point mutations of cyclin EL constructs by elastase also generated or knocked out the LMW forms of cyclin E as seen with the tumor cell transfection of the same constructs (compare Fig. 6B with Fig. 2C). For example, incubation of the in vitro-translated mutant constructs V67D and C68D generated EL6 and EL3, while mutant constructs A69D and D70P eliminated EL6 but not EL3. Additionally, elastase enzyme also induced the cleavage of cyclin E (presumably at the loop 1 domain) to generate EL3 in all EL and mutant vectors containing V67D, C68D, and A69D point mutations (Fig. 6B). This result clearly demonstrates that the cleavage site for generation of the EL2/3 doublet is distinct from the site that gives rise to EL5/6. Hence, elastase cleaves preferentially at two sites, one defined by the mutations shown in Fig. 2B and C and Fig. 6A and B as well as a site downstream of the M16 position (Fig. 2E). This is seen as the appearance of a second band below the M16 start site (Fig. 6C, lane 2). This band is generated by elastase cleavage, as clearly seen in Fig. 6C, lanes 1 and 2, where the M16A mutation lacking a band at the M16 position is still cleaved by elastase. Furthermore, as was the case with EL5/6, we believe the generation of the EL2/3 doublet in cell transfection is the result of posttranslational modification (e.g., phosphorylation) of EL3, which cannot be generated in vitro.
FIG. 6.
Elastase digestion of in vitro-translated cyclin E. (A and B) Plasmids containing full-length cyclin EL and its block deletions and point mutations were used to synthesize cyclin E using the TNT reticulocyte lysate system. After digestion with porcine pancreatic elastase, the proteins were separated by SDS-PAGE and analyzed by Western blotting with a FLAG antibody. (C) M16A and EL TNT protein products were incubated in the presence of no elastase, elastase alone, or elastase plus the indicated elastase inhibitors. CMK, methoxysuccinyl-Ala-Ala-Pro-Val-chloromethylketone; the arrows point to the elastase-mediated cleavage sites at EL3 (top arrow) and EL6 (bottom arrow).
The cleavage of cyclin E in vitro by elastase is entirely due to the elastase activity and not to a minor contamination, as shown by the inhibition of elastase activity against cyclin E with three specific elastase inhibitors used at low micromolar concentrations (Fig. 6C). The three different inhibitors of elastase used in this study were elastatinol, methoxysuccinyl-Ala-Ala-Pro-Val-chloromethylketone (MPCMK), and an α-ketooxadiazole (CE-2072). Elastatinol and MPCMK are potent elastase-specific substrate-based inhibitors of elastases (37, 56). CE-2072 is a specific and potent reversible inhibitor of human neutrophil elastase (i.e., with a Ki of 0.025 nM) (12, 59). We found that with all three inhibitors, the elastase-mediated cleavages of cyclin EL into EL3 and EL6 were inhibited (Fig. 6C).
Cyclin E processing takes place intracellularly.
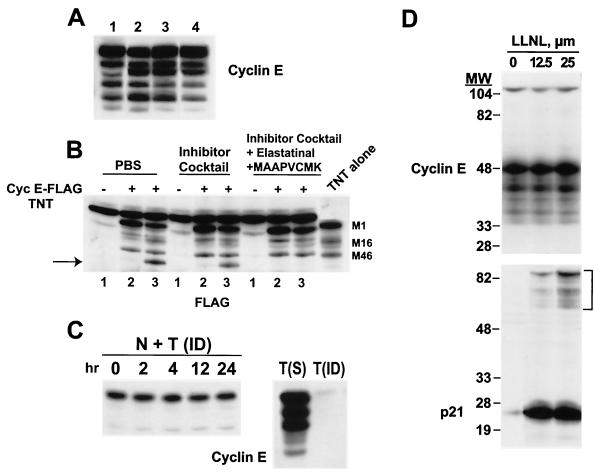
The data presented in Fig. 6 suggest that the LMW forms of cyclin E are enzymatically produced, very likely by the elastase class of proteases. Since elastase is a secreted enzyme, it may be possible that upon breaking open the cells, the secreted elastase can come into contact with the intracellular cyclin E and induce the cleavage in lysed cells. We have addressed this issue and found by using three different approaches that the fragmentation pattern observed with cyclin E in tumor cells occurs intracellularly prior to homogenization of cells (Fig. 7A to C). Initially (Fig. 7A), we blocked the membrane-bound proteases from contacting cyclin E during the lysis procedures and found that the LMW forms of cyclin E in tumor cells preexist prior to lysis in intact cells. Our usual procedure for preparing tumor and normal cell extracts is to sonicate the cells in PBS containing a protease inhibitor cocktail (Fig. 7). Under these conditions, we detected the full complement of the LMW forms of cyclin E (Fig. 7A, lane 3). Additionally, in order to prevent active extracellular proteases from contacting cyclin E during lysis, sample buffer (containing β-mercaptoethanol and 10% SDS) was added directly to cells, immediately boiled for 10 min, and then subjected to SDS-PAGE and Western blot analysis with cyclin E antibody (lane 1). We also prepared cell extracts by sonicating them in only PBS (lane 2). This experiment revealed that the fragmentation pattern was the same regardless of the buffers used. Next, to address directly whether any extracellular elastase activity could potentially cleave cyclin E during the lysis procedure, resulting in the generation of LMW forms, we added a cocktail of elastase inhibitors during lysis (lane 4). The pattern of cyclin E LMW forms remained unchanged by the elastase inhibitors (compare lanes 1 and 4). These results indicate that the LMW forms of cyclin E in tumor cells are likely to preexist in the cells prior to extraction.
FIG. 7.
Cyclin E proteolysis occurs in the cell, not during protein extraction. (A) Cells from the human breast tumor cell line MDA-MB-157 were harvested, and the cell pellets were suspended in either SDS-PAGE sample buffer and boiled immediately (lane 1) or PBS (lane 2), protease inhibitor cocktail (25 μg of leupeptin/ml, 25 μg of aprotinin/ml, 10 μg of pepstatin/ml, 1 mM benzamidine, 10 μg of soybean trypsin inhibitor/ml, 0.5 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 0.5 mM vanadate) (lane 3), or protease inhibitor cocktail plus 25 μM concentrations of the elastase inhibitors elastatinal and MPCMK (lane 4). Following resuspension, cells in lanes 2 to 4 were sonicated in a circulating ice water bath, centrifuged for 45 min at 100,000 × g, and subjected to Western blot analysis with cyclin E antibody. (B) Cyclin E-FLAG recombinant protein (4 μl of TNT reticulocyte lysate reaction mixture) was added either to tumor cell extracts following sonication (lanes 2) or directly to cells prior to sonication (lanes 3). Following sonication in the indicated buffers, the cyclin E-FLAG–tumor cell extract mixture was subjected to Western blot analysis with an anti-FLAG antibody. Lane 1 of each buffer set depicts the addition of reticulocyte lysate reaction mixture with empty vector to the tumor cell extract. The “TNT alone” lane is cyclin E-FLAG TNT reticulocyte lysate reaction mixture without mixing of the tumor cell extracts. The arrow points to the cleaved cyclin E. (C) Tumor cells were immune-depleted of cyclin E by passing tumor cell extracts on a cyclin E affinity column for three rounds. The immune-depleted cyclin E tumor cell extracts were then mixed (1:1 ratio) with normal cell extracts, incubated for the indicated times at 37°C, and subjected to Western blot analysis with anti-cyclin E antibody (lanes 1 to 5). Lanes 6 and 7 depict cyclin E levels in tumor cell extracts prior to (lane 6) and following (lane 7) immune depletion. N, normal cells; T, tumor cells; ID, immune-depleted cells; S, starting material. (D) MDA-MB-157 cells were incubated with the indicated concentrations of LLnL for 36 h and subjected (50 μg/lane) to Western blot analysis with antibodies to cyclin E and p21. The bracket in the p21 panel indicates the high-molecular-weight laddering of p21, which is diagnostic of polyubiquitination.
To determine whether tumor cell extracts contain a protease activity that is capable of digesting recombinant cyclin E in vitro and that can be specifically inactivated, we next mixed cell extracts prepared from tumor cells with recombinant cyclin E protein (Fig. 7B). Tumor cell extracts were prepared by sonication in either PBS, protease inhibitors, or protease-plus-elastase inhibitors, mixed with in vitro-transcribed or -translated (i.e., TNT assay) cyclin E-FLAG recombinant protein, and subjected to Western blot analysis with a FLAG antibody (Fig. 7B). In Fig. 7B, the lane “TNT alone” shows the pattern of in vitro-transcribed/translated recombinant cyclin E (unmixed) with translational start sites at the M1, M16, and M46 positions. When the recombinant cyclin E was mixed with tumor cell extracts prepared in different buffers, no additional processing of this protein was detected in any of the mixed extracts, as a banding pattern similar to that for the unmixed sample was revealed. (compare lane 2 of buffer set with the TNT alone lane). These results suggest that once the tumor cells are sonicated, they no longer retain elastase activity capable of cleaving recombinant cyclin E into its LMW forms. To determine whether prior to sonication the cells have retained any extracellular (i.e., secreted) elastase activity, the recombinant cyclin E was first added directly to the intact cells and then the mixture of tumor cells and recombinant cyclin E was sonicated in the three different buffer conditions as before. Addition of recombinant cyclin E to cells prior to sonication resulted in the processing of the recombinant cyclin E into its LMW forms (Fig. 7B, compare lanes 2 and 3). Furthermore, such activity is completely inhibited by the addition of elastase inhibitors (lane 3 of the elastatinal set). These results suggest that tumor cells contain an extracellular protease that is present in intact cells, prior to sonication, which can cleave cyclin E into its LMW forms. Furthermore, this protease is of the elastase class, as addition of elastase inhibitors can inhibit the processing of recombinant cyclin E. However, sonication of cells neutralizes this activity, since addition of the recombinant cyclin E to cell extracts (i.e., after sonication) resulted in no in vitro cleavage of cyclin E (Fig. 7B, lanes 2).
Next, we directly addressed whether sonicated tumor cell extracts contain a protease which could process cyclin E into its LMW forms (Fig. 7C). Cyclin E was first immune-depleted from tumor cell extracts by using a cyclin E affinity column. Cyclin E immune-depleted tumor extracts were mixed with extracts from normal cells, incubated for up to 24 h at 37°C, and subjected to Western blot analysis with cyclin E antibody (Fig. 7C). Normal cells predominantly expressed the 50-kDa form and none of the LMW forms of cyclin E (Fig. 7C, left panel) Under these conditions, no changes in the mobility of cyclin E in normal cells were detected and no LMW forms of cyclin E were generated following incubation of cyclin E immune-depleted tumor cell extracts with normal cell extracts for up to 24 h. These results indicate that the tumor cell extracts, once sonicated, are devoid of any protease activity capable of processing cyclin E from normal cells into LMW forms.
The turnover of cyclin E is thought to be through the ubiquitin-proteasome pathway and is regulated by CDK2 binding and by its site-specific autophosphorylation at threonine 386 in the PEST domain at the C terminus of cyclin E (11, 61). To examine the contribution of the proteasome pathway in the generation of the LMW forms of cyclin E in breast cancer, we treated MDA-MB-157 cells with N-acetyl-l-leucinyl-l-leucinal-l-norleucinal (LLnL), a potent inhibitor of this enzyme. LLnL resulted in dramatic induction of p21 in a dose- (Fig. 7D) and time- (data not shown) dependent fashion. Also shown in Fig. 7D is the high-molecular-weight laddering of p21, which is diagnostic for polyubiquitination of p21. However, LLnL was completely ineffective in modulating the levels of either the full-length or any of the LMW forms of cyclin E. Furthermore, no high-molecular-weight laddering of cyclin E was noted. In addition to LLnL, we also used several other inhibitors of the proteasome, such as lactacystin, MG-132, and proteasome inhibitor I. None of these proteasome inhibitors altered the pattern of the LMW forms of cyclin E, although proteins such as p21 and p27 were highly induced (data not shown). These studies suggest that the proteolytic processing of cyclin E into LMW forms occurs intracellularly and is distinct from the destruction of cyclin E that is mediated through the ubiquitination-proteasome pathway in tumor cells.
DISCUSSION
In this report we describe a novel proteolysis of cyclin E, occurring only in tumor cell lines and tissues, which generates the LMW forms of cyclin E in tumor cells. A total of six cyclin E proteins were detected in tumor cells ranging in molecular mass from 50 to 33 kDa. We have termed the 50-kDa cyclin E as EL1 and the five cyclin E protein products from 45 to 33 kDa as the LMW isoforms cyclin EL2 to EL6. All of the detectable LMW forms of cyclin E found in tumor cells can be explained as the result of three mechanisms at work. First, alternate translation at M46, but not at M16, accounts for the band we call EL4 (Fig. 2D). M16 alternate translation does not occur in cells, even though it can readily occur in reticulocyte lysate (Fig. 2D and 6A and B). Second, proteolysis of cyclin E occurs around the carboxyl side of A69, generating bands at EL5 and -6 (Fig. 2B and C). These bands are a doublet resulting from the third process, a posttranslation modification such as phosphorylation or deacetylation. Finally, bands EL2 and -3 arise from a similar proteolysis and posttranslational modification as EL5 and -6 at aa 40 to 45. We have also identified the full-length cyclin E as the EL form (Fig. 1 and 2D) described previously as the 15-aa alternatively spliced elongated form of cyclin E (42).
The proteolytic processing of cyclin E is unique to tumor cell lines and tissues. This tumor-specific pattern is not the result of overexpression or constitutive expression since normal cells transfected with cyclin E under a strong constitutive promoter do not further process cyclin E into its LMW forms (23; Fig. 3). The proteolysis we have described also differs from the proteasome-dependent proteolysis of cyclin E described by two other laboratories (11, 62). In these studies, the ubiquitination of cyclin E was dependent upon a specific phosphorylation at threonine 380. Although the destruction of cyclin E may be mediated through the ubiquitination-proteasome pathway in tumor cells, the generation of the LMW forms observed under steady state conditions in tumor cells and tissues is not likely to be generated by the proteasome pathway. Additionally, the LMW forms of cyclin E are present constitutively in the tumor cells and are not subject to cell cycle regulation (29). Furthermore, the LMW forms of cyclin E do not seem to represent the intermediate proteolytic products of degradative machinery (Fig. 7D). We conclude that these LMW forms of cyclin E are more likely due to the action of a different protease.
The way cyclin E is processed into LMW isoforms is also different than the proteolysis of other cyclins in the cell. For example, cyclin D1 is degraded by a calpain protease (10). Cyclin A is cleaved in vitro by a p27-dependent protease which removes the destruction box and allows evasion from proteasomal degradation (4). The proteolysis we describe here for cyclin E at A69 to D70 does not appear to occur to other cyclins. This site is homologous to elastase protease substrate sequences containing essential small hydrophobic residues (3). When A69 is replaced with an aspartyl residue, the proteolysis is nearly completely blocked (Fig. 2 and 6). A D70P substitution completely blocks degradation, presumably due to the unique imino-peptide bond afforded by the proline residue within the scissile bond. The effect of proline at position 70 is not surprising since this is commonly used in the design of peptide inhibitors of proteases (46). The involvement of an aspartyl residue in the cleavage site raises the possibility that a caspase protease might be involved, yet the typical DEVD motif is not present. In addition, the cyclin E protease appears to cleave on the N-terminal side of D70, contrary to a caspase cleavage on the C-terminal side of aspartyl residues. For example, a caspase activity which cleaves cyclin A during apoptosis in Xenopus embryos cleaves downstream of a critical aspartyl residue (54). In the case of cyclin E, the aspartyl residue is not essential since the D70A mutation does not block proteolysis of cyclin E and in fact may encourage it (Fig. 2).
The sequence analysis of the specific regions in cyclin E cleaved to produce EL5/6 (loop 2, A66 to P71; the AVCADP motif) and EL2/3 (loop 1, K32 to K48; the VFLQDP motif) suggested that the protease cleaving these regions is a serine protease of the elastase class (3). These regions within cyclin E form a hairpin loop type of structure making them accessible to the protease. We showed that in vitro-translated wild-type and mutant cyclin E proteins all digest in vitro with a porcine pancreatic elastase, generating a Western blot pattern similar to the in vivo pattern (compare Fig. 2 and 6). The striking similarity of in vivo and in vitro cleavage is supporting evidence that the elastase class of enzymes may actively cleave cyclin E in the cell. There are two possible ways to interpret how cyclin E is cleaved into its LMW forms by the elastase class of enzymes in tumor cells but not normal cells: (i) there is more elastase activity in tumor cells than in normal cells; or (ii) normal cells express high levels of elafin, an inhibitor of elastase. There is precedent for both these possibilities. Several studies have shown that the elastases are overexpressed in human cancer (20, 63, 64). Additionally, recent studies have reported that elafin, a potent elastase inhibitor for human leukocyte elastase and porcine pancreatic elastase (60), is differentially expressed in normal versus tumor-derived mammary epithelial cells, with very high levels in normal cells and undetectable levels in breast cancer cells (65). Collectively, these studies suggest that normal cells may be protected from the proteolytic effects of elastase by overexpressing elafin, while in tumor cells elastase expression may promote increased cyclin E activation and subsequent accelerated S-phase entry.
The specific cyclin E proteolysis at the AVCADP and VFLQDP motifs may separate a substrate-targeting domain (28) from the active cyclin E-CDK2 complex or affect the binding of an inhibitor protein. In addition, since the efficiency of proteasome proteolysis does not leave partial peptides, we presume the proteolysis of cyclin E generating bands EL2 through EL6 occurs prior to the reported proteasome degradation (11, 61). Hence, although the proteasome pathway is involved in degrading cyclin E, the proteolysis of cyclin E described here is a newly discovered level of cyclin regulation distinct from the ubiquitin-proteasome pathway. The elastase proteolysis of cyclin E may have a specialized role in the regulation of cyclin E-CDK2 activity, substrate selection, or subcellular localization rather than a role regulating the timely destruction of cyclin E.
The LMW forms of cyclin E seem to have a regulatory role distinct from that of the full-length protein. Our results clearly show that the LMW forms are biochemically hyperactive, as they can phosphorylate substrates such as histone H1 and GST-Rb much more efficiently than the full-length form. We used two different systems to examine the biochemical activity of the LMW forms of cyclin E compared to the full-length form. The two systems used, namely the transient-transfection assay in normal human mammary epithelial cells and the insect cell baculovirus expression system, both revealed that the LMW forms of cyclin E are two- to sixfold more active than the full-length form. Furthermore, the insect cell system, which is more quantitative, revealed that the hyperactivity of the LMW forms of cyclin E is in part due to an increased affinity of these forms for CDK2. Cyclin kinase inhibitors may also play a role in the cyclin E LMW form hyperactivity (data not shown). This high level of kinase activity associated with the LMW forms of cyclin E may be a significant contributor to the tumor phenotype directly through the action of the overexpressed kinase. The effect of the LMW forms (i.e., T1 and T2 transient expression) on normal cell growth also reflects this stimulated CDK2 kinase activity. When normal cells, which are devoid of any LMW forms, are forced to express the LMW forms of cyclin E, the cells progress through S phase very effectively. This fits the pattern seen in tumor cells, which already overexpress the LMW forms of cyclin E. Apparently, the tumor cells have adapted to this state since they are less responsive to the T1 and T2 transient expression (Fig. 3 and data not shown). Lastly, we showed that the LMW forms of cyclin E appear to retain the ability to localize to the nucleus even with a significant loss of the N terminus. Obviously, the nuclear targeting or retention signal is not found in the N terminus of cyclin E (19). The conclusion from these studies is that the LMW forms of cyclin E are not only functional but also hyperactive compared to the full-length form, providing the tumor cells with an added growth advantage. The LMW forms of cyclin E can phosphorylate substrates more effectively than normal cells, which only express the full-length form, and as a result tumor cells can progress through G1 and into S phase, bypassing the restriction point. The oncogenic potential of these LMW forms in vivo is currently under study and should elucidate how extensive a role these LMW forms have in the transformation phenotype.
Lastly, the LMW forms of cyclin E could provide a novel target for rational drug design for the treatment of metastatic breast cancer without harming normal proliferating cells in the body. Our studies have implicated a protease that is induced during metastatic progression (20, 63, 64) in the proteolytic regulation of cell cycle progression. By identifying the specific protease (i.e., of the elastase class) which cleaves cyclin E into its LMW forms that is found exclusively in tumor cells and tissues, we may be able to design cyclin E-specific protease inhibitors. These inhibitors would then help control the progression through the cell cycle in invasive cells, thus limiting the ability of these cells to populate distant metastatic sites.
ACKNOWLEDGMENTS
We thank Andrew Koff for the cDNA to cyclin EL, John Cheronis for CE-2072, and David Johnson, Lei Li, Sharon Roth, Philip Tofilon, and Matthew Callister (all at M. D. Anderson Cancer Center) for critical reading of the manuscript. We also gratefully acknowledge the use of the Wadsworth Center's mass spectrometry (Charles Hauer), tissue culture, molecular biology, and photography/graphics core facilities.
R.H. was supported by a fellowship (BC980981) from the U.S. Army Medical Research Acquisition Activity (USAMRAA). This research was supported in part by grant no. DAMD-17-94-J-4081 from the USAMRAA and by grant no. R29-CA666062 from the National Cancer Institute (both to K.K.).
REFERENCES
- 1.Band V, DeCaprio J A, Delmolino L, Kulesa V, Sager R. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells J. Virol. 1991;65:6671–6676. doi: 10.1128/jvi.65.12.6671-6676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements Proc. Natl Acad Sci USA. 1990;87:463–467. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett A J, Salvesen G. Proteinase inhibitors. Vol. 12. Cambridge, Mass: Elsevier; 1986. [Google Scholar]
- 4.Bastians H, Townsley F M, Ruderman J V. The cyclin-dependent kinase inhibitor p27(Kip1) induces N-terminal proteolytic cleavage of cyclin A. Proc Natl Acad Sci USA. 1998;95:15374–15381. doi: 10.1073/pnas.95.26.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boel E, Hjorth A L, Moller K B, Moller P H. A short synthetic adaptor as second-strand primer in the construction of cDNA libraries by the vector-primer method. BioTechniques. 1991;11:26–28. [PubMed] [Google Scholar]
- 6.Bortner D M, Rosenberg M P. Induction of mammary gland hyperplasia and carcinomas in trangenic mice expressing human cyclin E. Mol Cell Biol. 1996;17:453–459. doi: 10.1128/mcb.17.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckley M F, Sweeney K J E, Hamilton J A, Sini R L, Manning D L, Nicholson R I, deFazio A, Watts C K W, Musgrove E A, Sutherland R L. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- 9.Chen X, Lowe M, Keyomarsi K. UCN-01 mediated G1 arrest in normal but not tumor cells is pRb dependent and p53 independent. Oncogene. 1999;18:5691–5702. [PubMed] [Google Scholar]
- 10.Choi Y H, Lee S J, Nguyen P, Jang J S, Lee J, Wu M L, Takano E, Maki M, Henkart P A, Trepel J B. Regulation of cyclin D1 by calpain protease. J Biol Chem. 1997;272:28479–28484. doi: 10.1074/jbc.272.45.28479. [DOI] [PubMed] [Google Scholar]
- 11.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Turnover of cyclin E by the ubiquitin-proteosome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 12.Coeshott C, Ohnemus C, Pilyavskaya A, Ross S, Wieczorek M, Kroona H, Leimer A H, Cheronis J. Converting enzyme-independent release of tumor necrosis factor-α and IL-1β from a stimulated human monocytic cell line in the presence of activated neutrophils or purified proteinase 3. Proc Natl Acad Sci USA. 1999;96:6261–6266. doi: 10.1073/pnas.96.11.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou Q-P, Levin A H, Zhao S, Pardee A B. Cyclin E and cyclin A as candidates for the restriction point protein. Cancer Res. 1993;53:1493–1497. [PubMed] [Google Scholar]
- 14.Dulic V, Drullinger L, Lees E, Reed S, Stein G. Altered regulation of G1 cyclins in senescent human diploid fibroblasts: accumulation of inactive cyclin E-cdk2 and cyclin D1-cdk2 complexes. Proc Natl Acad Sci USA. 1993;90:11034–11038. doi: 10.1073/pnas.90.23.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 16.Ewen M E, Sluss H K, Sherr C J, Natsushime H, Kato J-Y, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cylins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 17.Fisher R P, Morgan D O. A novel form of cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 18.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 19.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 20.Grant A J, Lerro K A, Wu C-W. Cell associated elastase activites of rat mammary tumour cells. Biochem Int. 1990;22:1077–1084. [PubMed] [Google Scholar]
- 21.Gray-Bablin J, Zalvide J, Fox M P, Knickerbocker C J, DeCaprio J A, Keyomarsi K. Cyclin E, a redundant cyclin in breast cancer. Proc Natl Acad Sci USA. 1996;93:15215–15220. doi: 10.1073/pnas.93.26.15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper J W, Elledge S J. The role of cdk7 in CAK function, a retro-retrospective. Genes Dev. 1998;12:285–289. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 23.Harwell R M, Porter D C, Danes C, Keyomarsi K. Processing of cyclin E differs between normal and tumor breast cells. Cancer Res. 2000;60:481–489. [PubMed] [Google Scholar]
- 24.Hessling J J, Miller S E, Levy N L. A direct comparison of procedures for the detection of mycoplasma in tissue culture. J Immunol Methods. 1980;38:315–324. doi: 10.1016/0022-1759(80)90280-x. [DOI] [PubMed] [Google Scholar]
- 25.Hunt T. Cell cycle gets more cyclins. Nature. 1991;350:462–463. doi: 10.1038/350462a0. [DOI] [PubMed] [Google Scholar]
- 26.Hunter T, Pines J. Cyclins and cancer. Cell. 1991;66:1071–1074. doi: 10.1016/0092-8674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- 27.Hunter T, Pines J. Cyclins and cancer II: cyclin D and cdk inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 28.Kelly B L, Wolfe K G, Roberts J M. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci USA. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keyomarsi K, Conte D, Toyofuku W, Fox M P. Deregulation of cyclin E in breast cancer. Oncogene. 1995;11:941–950. [PubMed] [Google Scholar]
- 30.Keyomarsi K, Herliczek T. The role of cyclin E in cell proliferation, development and cancer. In: Meijer L, Guidet S, Philippe M, editors. Progress in cell cycle research. Vol. 3. New York, N.Y: Plenum Press; 1997. pp. 171–191. [DOI] [PubMed] [Google Scholar]
- 31.Keyomarsi K, O'Leary N, Molnar G, Lees E, Fingert H J, Pardee A B. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 32.Keyomarsi K, Pardee A B. Redundant cyclin overexpression and gene amplification in breast cancer cells. Proc Natl Acad Sci USA. 1993;90:1112–1116. doi: 10.1073/pnas.90.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, Philippe M, Roberts J M. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991;66:1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- 34.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 35.Makela T P, Tassan J P, Nigg E A, Frutiger S, Hughes G J, Weinberg R A. A cyclin associates with the CDK-activating kinase MO15. Nature. 1994;371:254–257. doi: 10.1038/371254a0. [DOI] [PubMed] [Google Scholar]
- 36.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molla A, Hellen C V, Wimmer E. Inhibition of proteolytic activity of poliovirus and rhinovirus 2A proteinases by elastase-specific inhibitors. J Virol. 1993;67:4688–4695. doi: 10.1128/jvi.67.8.4688-4695.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrissey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen N H, Arnerlov C, Emdin S O, Landberg G. Cyclin E overexpression, a negative prognostic factor in breast cancer with strong correlation to oestrogen receptor status. Br J Cancer. 1996;74:874–880. doi: 10.1038/bjc.1996.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtsubo M, Roberts J M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter D C, Keyomarsi K. Novel splice variants of cyclin E with altered substrate specificity. Nucleic Acids Res. 2000;28:E101. doi: 10.1093/nar/28.23.e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter P L, Malone K E, Heagerty P J, Alexander G M, Gatti L A, Firpo E J, Daling J R, Roberts J M. Expression of cell-cycle regulators p27 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 45.Rao S, Lowe M, Herliczek T, Keyomarsi K. Lovastatin mediated G1 arrest in normal and tumor breast cells is through inhibition of CDK2 activity and redistribution of p21 and p27, independent of p53. Oncogene. 1998;17:2393–2402. doi: 10.1038/sj.onc.1202322. [DOI] [PubMed] [Google Scholar]
- 46.Read R J, James M N G. Introduction to the protein inhibitors: X-ray crystallography. Amsterdam, The Netherlands: Elsevier Science Publishers BV; 1986. [Google Scholar]
- 47.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson H, O'Keefe L V, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- 49.Scott K, Walker R. Lack of cyclin E immunoreactivity in non-malignant breast and association with proliferation in breast cancer. Br J Cancer. 1997;76:1288–1292. doi: 10.1038/bjc.1997.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sgambato A, Doki Y, Schieren I, Weinstein I B. Effects of cyclin E overexpression on cell growth and response to transforming growth factor β depend on cell context and p27kip1 expression. Cell Growth Diff. 1997;8:393–405. [PubMed] [Google Scholar]
- 51.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 52.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 53.Spruck C H, Won K-A, Reed S I. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 54.Stack J H, Newport J W. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin A degradation during interphase of the cell cycle. Development. 1997;1997:3185–3195. doi: 10.1242/dev.124.16.3185. [DOI] [PubMed] [Google Scholar]
- 55.Steeg P S, Zhou Q. Cyclins and breast cancer. Breast Cancer Res Treat. 1998;52:17–28. doi: 10.1023/a:1006102916060. [DOI] [PubMed] [Google Scholar]
- 56.Stein R L, Trainor D A. Mechanism of inactivation of human leukocyte elastase by a chloromethyl ketone: kinetic and solvent effect studies. Biochemistry. 1986;25:5414–5419. doi: 10.1021/bi00367a011. [DOI] [PubMed] [Google Scholar]
- 57.Tsai L-H, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- 58.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Wieczorek M, Gyorkos A, Spruce L W, Ettinger A, Ross S E, Kroona H S, Burgos-Lepley C E, Bratton L D, Drennan T S, Garnert D L, Von Burg G, Pilkington C G, Cheronis J C. Biochemical characterization of α-ketooxidiazole inhibitors of elastases. Arch Biochem Biophys. 1999;367:193–201. doi: 10.1006/abbi.1999.1249. [DOI] [PubMed] [Google Scholar]
- 60.Wiedow O, Schroder J-M, Gregory S H, Young J A, Christophers E. Elafin: an elastase-specific inhibitor of human skin. Purification, characterization, and complete amino acid sequence. J Biol Chem. 1990;265:14791–14795. [PubMed] [Google Scholar]
- 61.Won K-A, Reed S I. Activation of cyclin E/cdk2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 62.Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 63.Yamashita J-I, Ogawa M, Ikei S, Omachi H, Yamashita S-I, Saishoji T, Momura K, Sato H. Production of immunoreactive polymorphonuclear leucocyte elastase in human breast cancer cells: possible role of polymorphonuclear leucocyte elastase in the progression of human breast cancer. Br J Cancer. 1994;69:72–76. doi: 10.1038/bjc.1994.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeydel M, Nakagawa S, Biempica L, Takahashi S. Collagenase and elastase production by mouse mammary adenocarcinoma primary cultures and cloned cells. Cancer Res. 1986;46:6438–6445. [PubMed] [Google Scholar]
- 65.Zhang M, Zou Z, Maass N, Sager R. Differential expression of elafin in human normal mammary epithelial cells and carcinomas is regulated at the transcriptional level. Cancer Res. 1995;55:2537–2541. [PubMed] [Google Scholar]