Abstract
Evolutionarily conserved variant histone H2A.Z has been recently shown to regulate gene transcription in Saccharomyces cerevisiae. Here we show that loss of H2A.Z in this organism negatively affects the induction of GAL genes. Importantly, fusion of the H2A.Z C-terminal region to S phase H2A without its corresponding C-terminal region can mediate the variant histone's specialized function in GAL1-10 gene induction, and it restores the slow-growth phenotype of cells with a deletion of HTZ1. Furthermore, we show that the C-terminal region of H2A.Z can interact with some components of the transcriptional apparatus. In cells lacking H2A.Z, recruitment of RNA polymerase II and TATA-binding protein to the GAL1-10 promoters is significantly diminished under inducing conditions. Unexpectedly, we also find that H2A.Z is required to globally maintain chromatin integrity under GAL gene-inducing conditions. We hypothesize that H2A.Z can positively regulate gene transcription, at least in part, by modulating interactions with RNA polymerase II-associated factors at certain genes under specific cell growth conditions.
The eukaryotic genome is packaged into repeated units of a protein-DNA complex called the nucleosome. The nucleosome is composed of four core histones (H2A, H2B, H3, and H4) which form an octamer that wraps 146 bp of DNA. Nucleosomes assemble throughout the genome in a periodical fashion, at every 200 ± 40 bp in metazoans (25) and ≈165 bp in the yeast Saccharomyces cerevisiae (2). Nucleosome depletion experiments in yeast have shown that nucleosomes can have a profound influence on gene expression in eukaryotic cells (8, 10, 43).
Nucleosomal histones are subjected to many types of modifications that can facilitate or inhibit the process of transcription. The chemical modifications include acetylation, phosphorylation, ADP ribosylation, ubiquitination, and methylation (for a review, see reference 30). Nonchemical modification of nucleosomes can be effected by ATP-dependent chromatin-remodeling machines (16, 17). The chromatin-modifying activities are believed to be targeted to promoters either by generally associating with the RNA polymerase II (polII) transcriptional machinery (32, 42) or by being recruited by gene-specific factors (5, 28, 39, 45).
Highly transcribed regions in eukaryotic genomes, especially metazoans, share several characteristic features. These features include increased nuclease sensitivity, hyperacetylated chromatin, and the absence of the linker histone H1 (3). Although various chromatin-remodeling activities and enhancer-specific factors likely contribute to this altered chromatin state, it is also possible that specialized chromatin components such as histone variants have a role in establishing, and perhaps maintaining, a transcriptionally permissive chromatin state. Histone variants have been described for many classes of histones, and perhaps the best studied example is the Z variant of H2A. H2A.Z (also called H2A.F/Z) is evolutionarily conserved from yeast to mammals (14). H2A.Z histones are essential for the viability of Tetrahymena, Drosophila, and mice (4, 22) and constitute roughly 5 to 10% of all cellular H2As (20). Experiments carried out with Drosophila have revealed that the unique feature of the variant histone important for viability resides in the carboxyl-terminal region of H2A.Z (His2AvD) and not in its histone fold (4). Importantly, this variant histone is found to be associated with transcriptionally active chromatin in Tetrahymena (36) and could therefore constitute a form of specialized chromatin that favors gene transcription. Importantly, very recent experiments carried out with Saccharomyces cerevisiae have shown that H2A.Z could regulate transcription and that its function was partially redundant with certain nucleosome-remodeling complexes (35). For example, mutants bearing a deletion in HTZ1, the gene encoding H2A.Z, and a deletion in the SWI2 gene had very dramatic effects on transcription induction of the PHO5 gene, whereas individual mutants have little or no effect on the induction of that gene (35). In addition, Jackson and Gorovsky (15) have also recently shown that htz1Δ yeast cells grew slowly and were particularly sensitive to formamide. Importantly, they also showed that the major H2A genes could not provide the function of H2A.Z and vice versa.
Here we show that loss of yeast variant histone H2A.Z can affect recruitment of the RNA polymerase II transcriptional machinery to the GAL1-10 genes, as well as their transcriptional induction. We also show that the H2A.Z C-terminal region is sufficient to provide the histone variant's unique function in positive regulation of gene transcription, provided that it is appropriately tethered to chromatin. Furthermore, the C-terminal region of H2A.Z is able to interact, directly or indirectly, with components of the transcriptional machinery. Finally, we show that chromatin from htz1Δ cells is globally more sensitive to micrococcal nuclease (MNase) cleavage compared to wild-type cells. Surprisingly, this increase in nuclease sensitivity was observed under GAL gene-inducing conditions but not under GAL-repressing conditions.
MATERIALS AND METHODS
Yeast strains and genetic methods.
W303α, used in our studies, was derived from a germinated spore from a W303 diploid strain (gift from M. A. Osley; MATa/α, ura3-1, leu2-3,112, ade2-1, his3-11,15, trp1-1, can1-100). The htz1Δ strain used in our studies (MAY424) is isogenic to W303α and was made by disrupting one HTZ1 allele from the W303 diploid strain and subsequent sporulation. Cells were typically grown in yeast nitrogen base or yeast extract-peptone-dextrose medium supplemented with the required amino acids and the appropriate carbon source where indicated. Myc-H2A.Z and Myc-H2A strains expressed tagged alleles of each respective histone, which were tagged with nine Myc epitopes by homologous recombination as described by Cosma et al. (5). In the case of Myc-H2A, we observe that the size of the tag decreases from nine Myc to two to three Myc by homologous recombination between the Myc repeats. The strains carrying shorter epitopes are stable and healthy. We therefore used one of these clones for further experiments. The strain used for tagging was W303α. Further details on the tagging procedure can be provided upon request.
Plasmids.
The HTZ1 knockout plasmid contained hisG sequences at each end of the URA3 gene (1) inserted at the BglII-BclI sites of the HTZ1 open reading frame, which deletes most of the coding region. All the H2A and H2A.Z expression plasmids were made by amplifying appropriate PCR products into YIplac211- or YCplac33-derived plasmids, and relevant constructs were sequenced. Glutathione S-transferase (GST)-H2A (amino acids [aa] 96 to 132) and GST-H2A.Z (aa 103 to 134) were made by inserting appropriate H2A and H2A.Z PCR products into the EcoRI-SalI sites of pGEX6P-1 (Amersham-Pharmacia). Further details of plasmid constructions are available upon request.
β-Galactosidase and primer extension analyses.
Yeast β-galactosidase assays were performed essentially as described by Gaudreau et al. (9). For primer extensions, 20 to 30 μg of RNA was used, and primer extension analyses were carried out essentially as described by Ma and Ptashne (26). The oligonucleotide sequences for the primers used were as follows: GAL1, CTCCTTGACGTTAAAGTATAGAGG; GAL7, GGATGGTAACGTCTATGGGAATGGC; GAL10, CCAATGTATCCAGCACCACCTG.
Proteins.
Escherichia coli XA-90 cells were transformed by plasmids expressing GST-H2A (aa 96 to 132), GST-H2A.Z (aa 103 to 134), and GST alone. Typically, cells were grown at an optical density at 600 nm (OD600) of between 0.35 and 0.5 and induced for 2 to 3 h with isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mM. After centrifugation, the cell pellet was resuspended in lysis buffer (20% glycerol, 20 mM HEPES [pH 7.5], 1 mM dithiothreitol [DTT], 150 mM potassium acetate [KoAC], 1 mM EDTA [pH 8]) followed by sonication on ice. Cell lysates were centrifuged at 12,000 rpm for 30 min at 4°C in a Beckman Avanti J30I centrifuge (JA17 rotor), and the supernatant was incubated with 1 ml of glutathione Sepharose 4B (Amersham-Pharmacia) for 2 to 3 h at 4°C. Beads were loaded into a column and washed with buffer A (1 mM EDTA [pH 8.0], 1 mM DTT, 20 mM HEPES [pH 7.5], 20% glycerol, and protease inhibitors) plus 100 mM KOAc.
Interaction assays.
For GST-H2A.Z and GST-H2A interaction experiments, yeast cell extracts (centrifuged at 10,000 rpm) were further centrifuged at 40,000 rpm in a Ti50 rotor for 2 h at 4°C and the pellet was resuspended with buffer A plus 500 mM KOAc on ice (29). After centrifugation as described above, the supernatant was treated with DNase I (10 U/ml) and RNase A (50 μg/ml) for 3 h at 4°C on a rotator followed by dialysis against buffer A plus 50 mM KOAc for 1 h and buffer A plus 100 mM KOAc for 2 h. For pull-down experiments, equal amounts of the different GST fusion proteins were mixed and incubated with 300 μg of chromatin-enriched yeast extract and binding buffer (20 mM HEPES [pH 7.5], 1 mM DTT, 1 mM EDTA [pH 8.0], 150 mM KOAc, 20% glycerol, 0.2% NP-40, and a mixture of protease inhibitors) for 3 h at 4°C on a rotator. Beads were washed four times with the same buffer, and proteins were loaded on a sodium dodecyl sulfate (SDS)–9% polyacrylamide gel electrophoresis (PAGE) gel and subjected to Western blotting with antibodies against Rpb1 (8WG16; BAbCo) and TATA-binding protein (TBP) (18).
Chromatin immunoprecipitations.
Eight hundred milliliters of yeast extract-peptone-dextrose–2% raffinose was inoculated at an OD600 of 0.05 and grown until it reached an OD600 of 0.6. Galactose was then added to a final concentration of 5% to induce GAL genes, and 50-ml samples were collected 0, 10, 20, 40, 60, and 120 min after induction. Formaldehyde was added to the collected samples to a final concentration of 1%, and cells were incubated with agitation for 20 min at room temperature and then overnight at 4°C. Cells were collected by centrifugation, washed three times with 40 ml of ice-cold Tris-buffered saline buffer (20 mM Tris HCl [pH 7.5], 150 mM NaCl), and resuspended in 700 μl of lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 10 μg of aprotinin/μl, 1 μg of leupeptin/μl, and 1 μg of pepstatin/μl). Yeast cells were disrupted by shaking for 2 h in the presence of glass beads (diameter, 0.5 mm) using an Orbital MiniShaker (IKA-Vibrax-VXR). Glass beads were removed, and samples were sonicated four times for 20 s at power 1.5 on a Sonifier II cell disrupter (Branson Ultrasonics) in order to shear chromatin DNA into fragments of 400 bp on average. Samples were centrifuged 5 min at maximum speed in a microcentrifuge, and the supernatant (from now on referred to as whole-cell extract) was used as input material for immunoprecipitation. Two hundred milliliters of whole-cell extract was incubated with either anti-CTD (8WG16; BAbCo), anti-yTBP (18), anti-Gal4 (Santa Cruz Biotechnology), or anti-Myc (9E11; purified from a hybridoma cell line kindly provided by G. Evan) antibody coupled to agarose or magnetic beads (Dynal) overnight at 4°C with agitation. Beads were washed twice with 1 ml of lysis buffer, twice with 1 ml of lysis buffer plus 360 mM NaCl, twice with 1 ml of wash buffer (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, and 1 mM EDTA) and once with 1 ml of Tris-EDTA (TE; 10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Bound material was eluted from beads by resuspending beads in 50 μl of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 1% SDS) and incubating 10 min at 65°C with occasional agitation. Samples were centrifuged briefly and the cross-linking was reversed by incubating 30 μl of supernatant with 120 μl of TE plus 1% SDS overnight at 65°C. Samples were treated with proteinase K, extracted twice with phenol, extracted once with chloroform, precipitated with ethanol, and resuspended in 30 μl of TE. DNA was then treated with RNase A and purified using a PCR purification kit from Qiagen. One microliter of immunoprecipitated DNA and different amounts of input DNA were analyzed by running 20 cycles of 15-μl PCR mixtures including 0.5 μCi of [α-32P]dATP and 250 μM deoxynucleoside triphosphates using 2 μM concentrations of of the following primer pairs: for the GAL1 promoter, TAACCTGGCCCCACAAACCT and CGGCCAATGGTCTTGGTAAT; for the GAL10 promoter, CAGCACCACCTGTAACCAAAAC and GGGGCTCTTTACATTTCCACA; for the GAL1-10 GC-rich region TACGCTTAACTGCTCATTGCT and GCCAATTTTTCCTCTTCATAACC; for the ARN1 promoter, TGCACCCATAAAAGCAGGTGT and GAGAGCTATCGAATGTTTCCTC; for the PHO84 open reading frame, GGTCAATTTGGTTTTGGTACTTT and GAGCAACAGTGGTTTGCAGAAT; for the SSB2 open reading frame, GATTGGTAAGAAGGTTGAAAAGG and GTGCAACAAGGAAACATCGAA; for the ACT1 promoter, TTAAATGGGATGGTGCAAGC and CGCTTACTGCTTTTTTCTTCC; for the YHB1 promoter, CCTTGTACGGAGATCTAAGAGCAA and AAGTCTTTGTGTGGTTTGTTGAA; and for the YJL100W open reading frame, TGCCAAACAGACATGGGAAA and CTGGCTCAAGTGGGTCGTACTTT. PCRs were run on 6% acrylamide–Tris-borate-EDTA gels, dried, and exposed on film. Data were quantified and analyzed by phosphorimaging. The use of increasing amounts of input DNA and immunoprecipitated DNA showed that the PCR amplifications were in the linear range for all the experiments (for an example, see Fig. 3). All experiments were carried out at least three times and gave similar results.
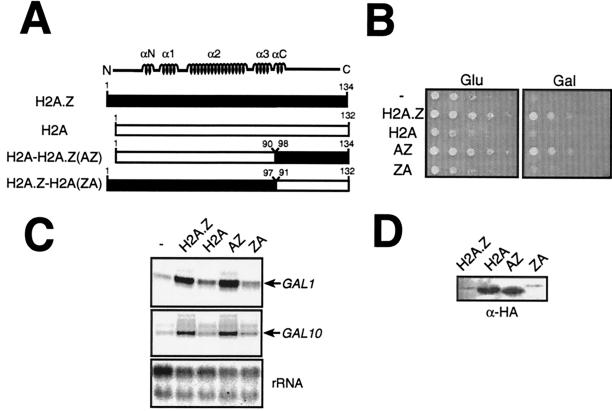
FIG. 3.
The H2A.Z C-terminal region is sufficient to mediate the special function of the variant histone. (A) Drawing of H2A-H2A.Z chimeras used in these experiments. (B) The AZ fusion (H2A [aa 1 to 90]-H2A.Z [aa 98 to 134]) is sufficient to complement the Gal− phenotype of htz1Δ cells. All histone derivatives (H2A.Z, H2A, AZ, and ZA; see text for description) are expressed from the ACT1 (β-actin) promoter on ARS-CEN plasmids and introduced into htz1Δ cells (MAY424). The growth assay was performed as described in the legend to Fig. 1A. (C) The AZ fusion (H2A [aa 1 to 90]-H2A.Z [aa 98 to 134]) is sufficient to fully activate the GAL1-10 genes. The H2A-H2A.Z fusions were assayed by primer extension analyses as for Fig. 2. (D) Histone protein levels were determined by immunoblotting with an anti-HA antibody.
Chromatin analyses.
Preparation of yeast nuclei was as described by Svaren et al. (38) using appropriate carbon sources (as indicated) throughout the procedure in order to avoid altering representative chromatin structures characteristic of particular growth conditions. MNase analysis of chromatin was essentially as described by Svaren et al. (38) for DNase I analysis of chromatin, except that MNase digestion buffer was used in place of the DNase I buffer (15 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1.4 mM CaCl2, 0.2 mM EDTA, 0.2 mM EGTA, 5 mM β-mercaptoethanol). Essentially, nuclei were prepared from approximately 0.5 to 1.0 g of yeast cells and were resuspended in 1.2 ml of digestion buffer. A 0.2-ml aliquot was used for each MNase digestion. Following MNase digestion, DNA was prepared also as described by Svaren et al. (38). Total DNA yields from such digested chromatin ranged from 25 to 50 μg, and 5 to 10 μg was loaded onto a 1.5% agarose gel and stained with Vistra Green (Amersham-Pharmacia) for analysis. Fluorescence was then quantified by fluor imaging. All experiments were carried out at least two to four times and gave similar results.
RESULTS
Deletion of HTZ1 confers slow growth and Gal− phenotypes in yeast and impedes GAL1-10 induction.
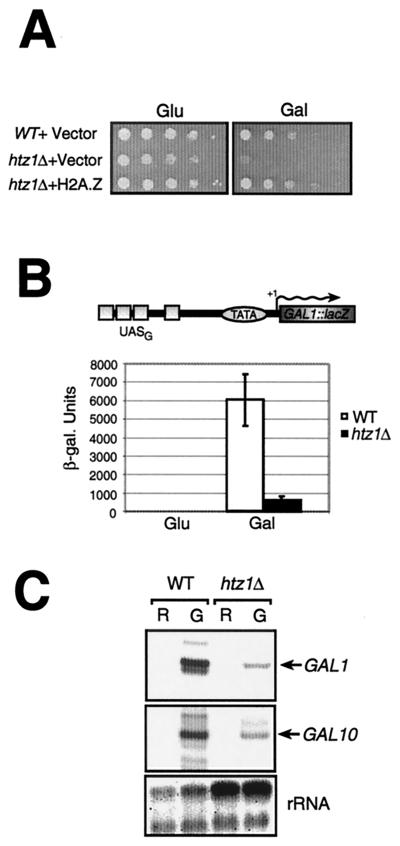
In order to study the role of H2A.Z in gene transcription, we first deleted the gene encoding the histone variant H2A.Z (HTZ1) in the yeast S. cerevisiae. As previously reported by others (15, 35), we observed that htz1Δ spores were able to germinate on selective media, indicating that deletion of this gene does not affect cell viability, although it did produce a slow-growth phenotype. We also observed that htz1Δ cells were sensitive to heat but not to cold (data not shown), a result which is also in accord with those of Santisteban et al. (35). In addition, we observed that htz1Δ cells were unable to grow on minimal media containing galactose as the sole carbon source (Fig. 1A), a result implying that GAL gene transcription was impaired. The results illustrated in Fig. 1A further show that adding back a plasmid expressing wild-type H2A.Z can complement the slow-growth phenotype of htz1Δ cells (see growth on glucose plates) as well as its inability to grow on synthetic complete medium plates containing galactose as the sole carbon source (Gal− phenotype), thus suggesting that the observed phenotypes were due to the HTZ1 deletion. The fact that htz1Δ cells are Gal− suggests that at least some GAL genes are negatively affected by the absence of H2A.Z. In order to directly test this, we first measured transcriptional activation elicited by Gal4—the potent activator of GAL genes—at the GAL1 promoter using a lacZ integrated reporter template. Figure 1B shows that, at that lacZ template, activation is severely impaired (some 10-fold) in the strain with a deletion of HTZ1 compared to an isogenic wild-type strain. The GAL1 and GAL10 genes are divergently transcribed and thus share the same upstream activation sequence (UAS) control elements (UASG). Figure 1C shows primer extension measurements of GAL1 and GAL10 transcript levels. The results show that transcriptional activation of the GAL1 and GAL10 genes is significantly impaired in the strain with a deletion of HTZ1 compared to the isogenic wild-type strain, a result consistent with Fig. 1B and which was recently published by others (for the GAL1 gene) while this report was in preparation (35). Total RNA isolated from wild-type and htz1Δ yeast cultures grown in the presence of either raffinose or raffinose and galactose is shown here (as seen by 18S and 28S rRNAs) as a control. We also tested expression of the GAL7 gene, also induced by Gal4, and saw that it too was crippled for activation in the htz1Δ mutant (data not shown).
FIG. 1.
Deletion of HTZ1 confers slow growth, Gal− phenotypes, and reduced GAL1-10 induction in yeast. (A) htz1Δ cells have a Gal− phenotype. W303α, htz1Δ cells (MAY424), or htz1Δ cells containing a plasmid expressing a wild-type allele of HTZ1 were serially diluted by a factor of 10 on SD media containing either glucose (Glu) or galactose (Gal) as a sole carbon source. Cells were incubated for approximately 2 to 3 days on glucose plates and approximately 4 to 6 days on galactose plates. (B) H2A.Z is required for proper induction of a GAL1::lacZ reporter gene. The strains used in this experiment (W303 and MAY424) contain an integrated reporter template bearing the GAL1 UASG upstream of the GAL1 promoter fused to lacZ. Cells were grown in minimal media with either glucose (Glu) or galactose/raffinose (Gal), and β-galactosidase assays (9) were carried out to measure the extent of gene induction. (C) Ability of htz1Δ cells to induce the GAL1-10 genes. Primer extension analyses were carried out with purified RNA from wild-type (WT) and htz1Δ cells. Yeast cells were grown in raffinose (R), and then galactose (G) to a final concentration of 5% was added to one-half of the culture volume for 6 h in order to induce GAL gene expression.
H2A.Z is not required for Gal4 binding to the UASG.
To directly verify if the deletion of HTZ1 affects cellular levels of Gal4 and its capacity to bind the GAL1-10 UASG, we have performed immunoprecipitations of cross-linked chromatin fragments, followed by quantitative PCR amplification, a method referred to as chromatin immunoprecipitation (ChIP) (11), using an anti-Gal4 antibody (see Materials and Methods for details). The results, shown in Fig. 2, clearly demonstrate that Gal4 binds to the GC-rich region of the GAL1-10 locus to a similar level in both the wild-type and the htz1Δ strains. That GC-rich region contains four Gal4-binding sites known to be responsible for the regulation of GAL1 and GAL10 (44). The result illustrated in Fig. 2 thus indicates that the reduced transcriptional activity of GAL1-10 genes in the htz1Δ mutant is not due to a reduction in Gal4 levels binding to the UASG. We observed no binding of Gal4 in the nearby GAL1 open reading frame, demonstrating that the immunoprecipitation reaction is specific. Interestingly, we saw strong binding of Gal4 to the GC-rich region, even in the absence of galactose, which agrees with previous work by others, showing that the transcriptional activity of Gal4 is not regulated by its DNA-binding activity but rather by the action of Gal80 and Gal3 (for examples, see references 7 and 31). Finally, we tried overexpressing Gal4 from the strong β-actin promoter and assayed for GAL1::lacZ activity in both wild-type and htz1Δ cells. Our results (data not shown) were comparable to those obtained without overexpressing Gal4.
FIG. 2.
ChIP analysis of the binding of Gal4 to the GC-rich region of the GAL1-10 locus. The binding of Gal4 to the GC-rich region and to the GAL1 open reading frame over time after addition of galactose is shown for both the wild-type (WT) and the htz1Δ strains.
The H2A.Z C-terminal region is sufficient to mediate the special function of H2A.Z in GAL gene induction.
Clarkson et al. (4) previously demonstrated that a region encompassing the α3 helix in the C terminus of Drosophila melanogaster H2A.Z conferred the unique function of H2A.Z, relative to H2A, and that region was required for cell viability. Thus, in order to verify if the C-terminal part of H2A.Z was sufficient to provide the special function of H2A.Z in GAL gene induction, compared to S phase H2A, we constructed a chimera (Fig. 3A) bearing the H2A.Z C-terminal region (aa 98 to 134) fused to H2A without its corresponding C-terminal region (aa 1 to 90), a fusion we named AZ. We also fused the H2A C-terminal region (aa 91 to 132) to H2A.Z with its C-terminal region (aa 1 to 97) (Fig. 3A), a fusion we named ZA as a reciprocal control. These fusions were expressed from the strong β-actin promoter to ensure that all the fusions were sufficiently expressed. H2A and H2A.Z used in the experiments illustrated in Fig. 3 were also expressed from the β-actin promoter. The results shown in Fig. 3B show that, as expected, expression of wild-type H2A.Z complements the slow growth and Gal− phenotypes of htz1Δ cells. Figure 3B also shows that overexpression of wild-type H2A does not complement the slow-growth defect of htz1Δ cells—a result also observed by Jackson and Gorovsky (15)—and the Gal− phenotype. Importantly, the AZ fusion was able to complement both phenotypes, while the ZA fusion was not able to. Figure 3C shows that the AZ fusion is also able to fully provide the H2A.Z function in GAL1-10 gene induction but that the ZA fusion cannot. Western blotting experiments reveal that the AZ fusion was expressed to a level comparable to that of H2A and that ZA fusion is expressed to a level comparable to that of H2A.Z (Fig. 3D). We have consistently observed that H2A core derivatives (H2A and AZ) were expressed at much higher levels than H2A.Z core derivatives (H2A.Z and ZA). It is conceivable that H2A.Z cellular levels might be down regulated, compared to H2A, in a way that prevents high concentrations of the molecule in the cell. Alternatively, it is possible that the hemagglutinin (HA) tags on the H2A and H2A.Z N-terminal fragments are not recognized by the anti-HA antibody with comparable efficiency. Our results, consistent with those of the previous section, strongly suggest that the C-terminal region of H2A.Z mediates the special function of the histone variant in GAL1-10 gene induction.
The H2A.Z C-terminal region interacts with components of the transcriptional machinery.
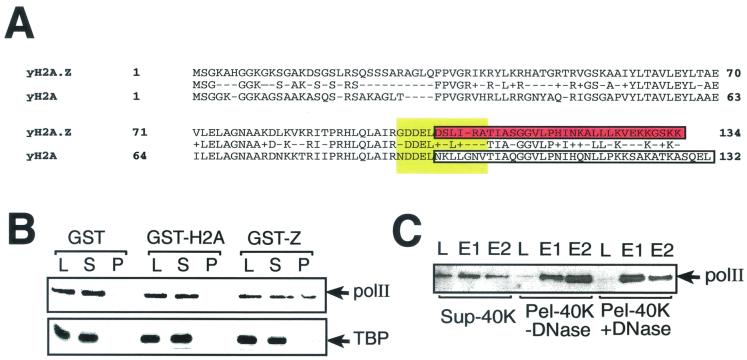
Because the C-terminal part of H2A.Z is thought to be important for its special function in gene transcription, we decided to look for possible interactions between the H2A.Z C-terminal region and components of the transcriptional machinery. Figure 4A shows the aligned amino acid sequences of yeast H2A.Z and H2A. In order to look for possible interactions between the C-terminal region of H2A.Z and some components of the transcriptional machinery, we used recombinant GST-H2A.Z and GST-H2A C-terminal fusions (aa 103 to 134 and 96 to 132, respectively) for interaction assays. A whole-cell extract was prepared as described by Otero et al. (29) except for the ultracentrifugation step (see Materials and Methods). This extract was incubated either with GST alone, with GST-H2A, or with GST-H2A.Z and processed as described in Materials and Methods. Interaction with RNA polII was revealed by Western blotting analyses using an anti-Rpb1 (the large subunit of RNA polII) antibody. The results shown in Fig. 4B show that while GST and GST-H2A do not interact significantly with Rpb1, GST-H2A.Z can significantly interact with the latter (Fig. 4B, upper panel). The same membrane was probed with anti-TBP antibodies, and the result showed (Fig. 4B, lower panel) that TBP could not significantly associate with the GST-H2A.Z fusion. Although we consistently saw weak binding of TBP to H2A.Z under these conditions, the interaction was much weaker than that obtained with RNA polII (compare lanes L and P). We also saw no direct interaction between recombinant TBP and GST-H2A.Z in other experiments (data not shown). The fact that TBP cannot significantly associate with GST-H2A.Z suggests that not all general transcription factors have the ability to interact with the latter, and this provides a specificity control for the RNA polII-H2A.Z association whether it is direct or indirect. Our chromatin-enriched extracts were treated with DNase and RNase to make sure that the interaction we saw was not mediated by an indirect effect of nucleic acids bridging RNA polII components to the histone variant. No significant difference in binding was observed under those conditions (Fig. 4C). Figure 4C further shows that RNA polII is present in greater amounts in the soluble fraction of the extract (Sup-40K) as judged by the band intensities in the load lanes (compare lanes L from Sup-40K, Pel-40K +DNase, and Pel-40K −DNase). We therefore suggest that the putative target of the H2A.Z C-terminal region, as evidenced by the presence of RNA polII, is present at a significantly higher concentration in a chromatin-enriched extract than in a soluble whole-cell extract. Therefore, it is unlikely that the H2A.Z target(s) would directly be RNA polII, but rather some polII-associated factor which predominantly associates with chromatin.
FIG. 4.
The C-terminal region of H2A.Z interacts with components of the transcriptional machinery. (A) Aligned amino acid sequences of yeast H2A.Z and H2A using BLAST (National Center for Biotechnology Information). Boxed areas represent the C-terminal regions that were fused to GST for the experiments illustrated in panels B and C (red and white) and the M6 region in Drosophila H2A.Z required for viability (yellow) (4). (B) GST, GST-H2A (aa 96 to 132), and GST-Z (GST-H2A.Z [aa 103 to 134]) proteins bound to glutathione-Sepharose beads were incubated with a chromatin-enriched yeast extract. L, 2% input of the mixture; S, 2% of the supernatant after pelleting the Sepharose beads; P, washed Sepharose pellet. Samples were analyzed by SDS-PAGE followed by immunoblotting with either an anti-RNA polII antibody or an anti-TBP antibody. (C) The H2A.Z-RNA polII interaction is not mediated by the indirect bridging effect of nucleic acids. The chromatin-enriched extract was treated with or without DNase and RNase and then loaded in a 500-μl glutathione-Sepharose column coupled to GST-H2A.Z (aa 103 to 134). The column was washed and eluted with potassium acetate. L, input of the total reaction; E1 and E2, elutions. Sup-40K is an extract not enriched in chromatin; Pel-40K −DNase is a chromatin-enriched extract not treated with nucleases; Pel-40K +DNase represents the chromatin-enriched extract treated with nucleases. Samples were analyzed as for panel B with an anti-RNA polII antibody.
RNA polymerase II and TBP are not efficiently recruited to the GAL1-10 promoter locus in the absence of H2A.Z.
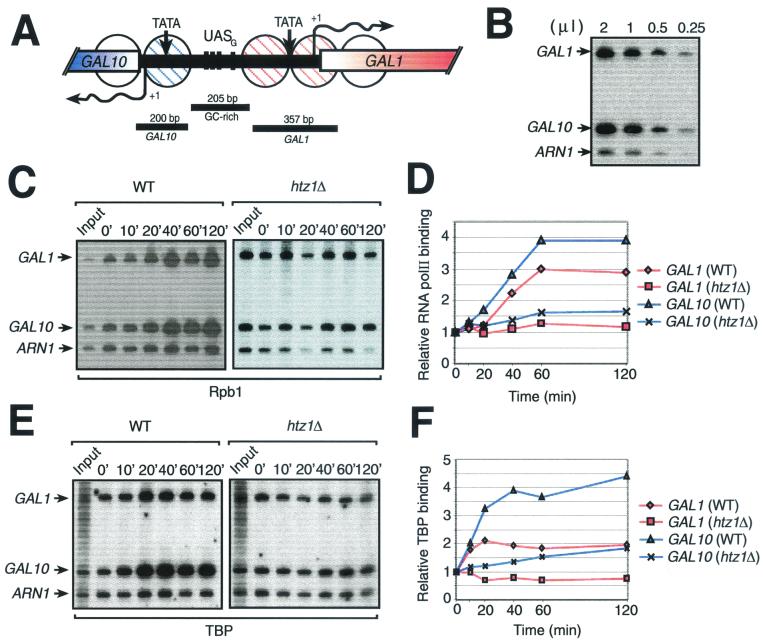
In order to gain some understanding of the mechanism by which H2A.Z affects transcription of RNA polII genes, we examined the in vivo binding of the transcriptional machinery to the GAL1-10 locus in both wild-type (HTZ1) and htz1Δ strains. In order to do so, we performed ChIP experiments. All yeast strains were grown as described for Fig. 2. Under these conditions, wild-type cells showed rapid induction of GAL1 and GAL10 genes whereas the htz1Δ strain shows a slower response, with a substantially lower magnitude (data not shown). Figure 5A shows a representation of the GAL1-10 locus and the regions we PCR amplified in our ChIP assays. Figure 5B shows that our PCRs are within a linear range of amplification. Figure 5C shows that the binding of Rpb1 (the large subunit of RNA pol II) to both the GAL1 and GAL10 promoters increases with time after induction with galactose, reaching its maximum level at 60 min. However, in the htz1Δ strain, the binding of Rpb1 shows no significant increase during the same time course. This shows that efficient recruitment of RNA polII to the GAL1 and GAL10 promoters is dependent on H2A.Z, although it is unclear whether the effect is direct. Moreover, the htz1Δ mutation had a comparable effect on TBP binding (Fig. 5E). Figure 5D and F shows quantifications of RNA polII and TBP binding, respectively, to the GAL1 and GAL10 promoters either in wild-type or htz1Δ cells as shown in Fig. 5C and E.
FIG. 5.
Effect of a htz1Δ mutation on recruitment of the transcriptional machinery to the GAL1-10 locus after galactose induction. (A) Representation of the GAL1-10 locus. GAL1 and GAL10 TATA boxes (TATA), transcriptional initiation sites (arrows with +1), and partial open reading frames are represented. The four Gal4 UASs (UASG) are shown by black crossbars. Circles, positioned nucleosomes covering both GAL1 and GAL10 promoters; stippling, remodeled nucleosomes during galactose induction (24); black bars, regions amplified by PCR in the ChIP experiments shown in panels B, C, and E. (B) Linear PCR amplification of DNA. (C) ChIP analysis of the binding of Rpb1 to the GAL1 and GAL10 promoters. The relative binding of Rpb1 over time after addition of galactose is shown for both wild-type (WT) and htz1Δ strains. ARN1 is used here as an internal control to normalize signals from each lane. (D) Binding of Rpb1 to the GAL1 and GAL10 promoters. Quantification of the experiment illustrated in panel C is shown. (E) ChIP analysis of the binding of TBP to the GAL1 and GAL10 promoters. The procedure was the same as for panel C except that the immunoprecipitation was carried out with an anti-TBP antibody. (F) Binding of TBP to the GAL1 and GAL10 promoters. Quantification of the experiment illustrated in panel E is shown.
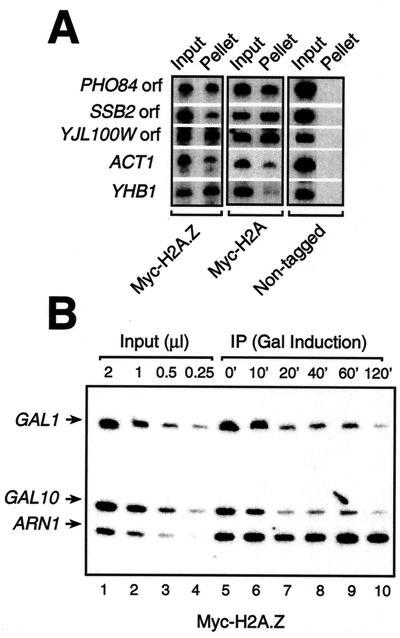
The location patterns of H2A.Z and H2A are similar but not identical.
It has recently been demonstrated that the Drosophila homolog of H2A.Z is spread throughout the genome, but in a nonuniform fashion, as opposed to H2A, which is uniformly distributed across the genome (20). To test whether this was applicable to yeast, we compared the binding of Myc-H2A.Z and Myc-H2A by ChIPs to randomly selected promoters and open reading frames. The data of Fig. 6A show that the binding patterns of both Myc-H2A.Z and Myc-H2A are very similar but not identical to the locus tested. For example, Myc-H2A.Z was efficiently bound to the YHB1 promoter while Myc-H2A was not, and the situation was reversed with the SSB2 open reading frame. These data suggest that H2A.Z is part of nucleosomes, perhaps interspersed with H2A at certain genomic loci. Consistent with this is the recent demonstration of the crystal structure of an H2A.Z-containing nucleosome, a result showing that H2A.Z can be incorporated into nucleosomes, at least in vitro (37). We then looked at Myc-H2A.Z binding to the GAL1 and GAL10 promoters by ChIP using an anti-Myc antibody. Thus, Fig. 6B shows that Myc-H2A.Z is present at both the GAL1 and GAL10 promoters before the addition of galactose (Fig. 6B, lane 5). After addition of galactose, the signal of Myc-H2A.Z at both these promoters decreases with time (Fig. 6B, lanes 6 to 10), a result also observed by Santisteban et al. (35). Figure 6B, lanes 1 to 4, shows that PCR amplifications of these loci are within linear range. This result demonstrates that H2A.Z-containing chromatin is somehow remodeled during the activation of GAL1 and GAL10.
FIG. 6.
DNA binding of H2A.Z in vivo. (A) ChIP analysis of various loci with an anti-Myc antibody on strains expressing either Myc-H2A or Myc-H2A.Z fusion proteins as well as a nontagged strain. Shown are the PHO84, YJL100W, and SSB2 open reading frames, as well as the ACT1 and YHB1 promoters. (B) ChIP analysis of the binding of a Myc-tagged version of H2A.Z to the GAL1-10 promoters after induction by galactose. Lanes 1 to 4, linear PCR amplification of DNA (input DNA); lanes 5 to 10, binding of Myc-H2A.Z to the GAL1-10 promoters and the ARN1 promoter over time after addition of galactose.
Loss of H2A.Z induces a state of increased global chromatin accessibility in yeast cells grown in the presence of galactose.
In an effort to investigate the chromatin state of htz1Δ cells, we digested chromatin from htz1Δ and wild-type cells with MNase. Hence, nuclei purified from htz1Δ and wild-type cells were grown in the presence of either raffinose or raffinose and galactose. Then, the nuclei preparations were treated with 25 U of MNase per ml for increasing periods of time (up to 20 min). Much to our surprise, we saw that chromatin from htz1Δ cells grown in the presence of galactose digested much faster than in wild-type cells, an effect not readily observed with cells grown in the presence of raffinose alone (Fig. 7A; compare right panel with left panel). Figure 7B plots the intensity of bands from the wild-type and htz1Δ digests after 5 and 10 min and, as such, clearly illustrates the faster digestion kinetics of htz1Δ chromatin when cells are grown under GAL-inducing conditions. In order to exclude the possibility that this increase in sensitivity was specific to Gal− cells, we performed similar experiments with gal4Δ cells and found that chromatin from those cells was indistinguishable from that of wild-type cells (data not shown). We next analyzed whether this altered state of chromatin in htz1Δ cells could be reversed by directly adding glucose during nucleus preparation. Interestingly, we found that the simple addition of glucose to these nuclei during their preparation started to reverse this chromatin accessibility defect (Fig. 7C; compare lanes 3 to 7 with lanes 9 to 13). Moreover, Fig. 7 also shows that adding glucose 90 min prior to preparing the nuclei further restores the altered chromatin state (Fig. 7C, lanes 15 to 19). Our results suggest that H2A.Z is required for global chromatin integrity particularly under specific physiological and/or metabolic conditions, and these chromatin alterations can be quickly restored to the original state.
FIG. 7.
Global chromatin analyses of htz1Δ yeast cells. (A) htz1Δ cells have an increased sensitivity to MNase in the presence of galactose and raffinose but not raffinose alone. Yeast nuclei were digested with 25 U of MNase per ml for increasing amounts of time (up to 20 min as indicated). Chromatin DNA was then analyzed by agarose gel electrophoresis. (B) Plot of band intensities (from top to bottom) showing the relative differences in nuclease sensitivity of wild-type (WT) and htz1Δ cells. Bands were scanned from 5- and 10-min digests of WT and htz1Δ cells grown in either raffinose (Raf) or raffinose and galactose (Raf/Gal). (C) Adding glucose to nuclei prepared from galactose- and raffinose-grown htz1Δ cells restores the altered chromatin state. Lane 1, molecular weight marker; lanes 2 to 7, MNase digests (0, 1, 3, 5, 8, and 12 U/ml digested for 20 min) of nuclei prepared from cells grown in raffinose and galactose. In this part of the experiment, raffinose and galactose were added to the nucleus preparation buffers. Lanes 8 to 13, the same MNase digestions from cells also grown in raffinose and galactose but with the addition of glucose to the nuclei at the time of their preparation. Lanes 14 to 19, the same MNase digestions from cells grown in raffinose-galactose but with the addition of glucose 90 min prior to nucleus preparation and throughout their preparation. Samples were analyzed by agarose gel electrophoresis.
DISCUSSION
We have shown that histone variant H2A.Z is required for transcriptional activation of certain genes in S. cerevisiae, a result also recently obtained by others (35). Our results show that GAL gene induction is affected by the HTZ1 deletion, which would account for the Gal− phenotype observed. Importantly, the C-terminal region of H2A.Z, when substituted with the reciprocal C-terminal region in H2A, can complement the GAL transcriptional defect of HTZ1-null cells. On the other hand, replacing the C-terminal region of H2A.Z with that of H2A did not complement these activation defects. Thus, our results clearly show that, by whatever mechanism that may be, the C-terminal region of H2A.Z is important and may be sufficient to mediate the special function of the histone variant in GAL gene induction compared to S phase H2A, provided that it is functionally incorporated into a nucleosome particle. It may well be that the special function of the H2A.Z C-terminal region is actually what prevents this variant histone from complementing HTA1-HTA2 deletions (the genes encoding S phase H2A) and vice versa (15). Although our results and those of Santisteban et al. (35) suggest a positive role for H2A.Z in gene regulation at certain genes, it is conceivable that it might also be a negative regulator at other genes. Accordingly, a recent report has shown that H2A.Z was important for silencing at HMR (6). Positive and negative modulations of transcription have also been observed with histone H4, where preventing its expression in S. cerevisiae affects the activity of genes in either a positive or a negative fashion (43). Furthermore, disruption of the Swi/Snf chromatin-remodeling machine in S. cerevisiae, as well as the histone acetyltransferase Gcn5, also seems to affect genes in either a negative or a positive fashion (13). In light of these observations, it is interesting that different histone mutations have different effects on transcription. For example, a class of H2A N-terminal tail mutants show specific transcriptional defects of some, but not all, Swi/Snf-dependent genes (12). H3 N-terminal tail mutants increase GAL1 activation while H4 N-terminal mutations decrease GAL1 activation (40).
We have shown that H2A.Z was important for proper recruitment of RNA polII components to certain promoters. Hence, chromatin immunoprecipitation experiments using RNA polII antibodies have demonstrated that RNA polII was not efficiently recruited to the GAL1-10 promoters under inducing conditions in the absence of the variant histone, a condition which supports our transcription measurements at the GAL promoters. Interestingly, TBP recruitment was also affected in the htz1Δ mutant to an extent similar to that observed with RNA polII recruitment. Since TFIID and the RNA polymerase II holoenzyme are known to bind to the GAL1-10 promoters cooperatively (19, 21), it is hard to assess if the effect seen on the binding of TBP is actually a consequence of a strong defect in RNA polII holoenzyme recruitment or vice versa. Under those conditions, we were able to show that Gal4 could be bound as efficiently in wild-type cells as in htz1Δ cells, a result which suggests that the effect of deleting HTZ1 on the binding of RNA polII to the GAL promoters is not a consequence of a defect in Gal4 binding to the UASG. Interestingly, we have also shown that the H2A.Z C-terminal region could interact with some component(s) of the transcriptional machinery in vitro, as judged by the presence of RNA polII in our protein-interaction assays. This interaction was most obvious when using a chromatin-enriched extract, thereby suggesting that the interaction between H2A.Z and RNA polII would be mediated by a factor predominantly associated with chromatin components. We therefore suggest that H2A.Z is a cofactor of certain genes that acts specifically by facilitating the recruitment of the RNA polII transcriptional machinery to some promoters, as exemplified here for the GAL genes. This interaction could be important for H2A.Z to mediate the recruitment of the RNA polII holoenzyme—or chromatin-remodeling machines which transiently associate with RNA polII—to GAL genes and presumably other genes. Recent studies (37) involving the crystallization of a nucleosome core particle containing the mouse variant histone H2A.Z revealed an altered surface, compared to H2A, in the C-terminal region of the molecule which binds a metal ion (His112). The authors of those studies even propose that this altered surface may serve to recruit protein factors, some of which may be involved in chromatin assembly and remodeling. Interestingly, His112 is conserved in the yeast molecule (His118).
Surprisingly, H2A.Z was found to be present at most genomic locations tested, including the GAL1-10 UASG, a result also observed by others (35). Moreover, the location pattern of the variant histone was similar, but not identical to that of H2A, suggesting that H2A.Z's function in gene transcription may not be that of creating chromosomal domains where H2A is replaced by H2A.Z. However, this type of replacement could still happen at the level of single nucleosomes. In agreement with our observations is a recent report by Leach et al. (20) showing that Drosophila H2A.Z is widely distributed in the genome. However, the study showed that the variant's distribution was not uniform and that the banded pattern of H2A.Z on polytene chromosomes was complex and did not parallel the concentration of DNA as was the case for H2A. It is thus conceivable that certain regions of the genome would preferentially be occupied by variant histones and perhaps interspersed by regular histones. In fact, we propose that the H2A-to-H2A.Z ratio at a given gene might be detrimental to an appropriate regulation of its expression.
Intriguingly, loss of H2A.Z creates a global increase in chromatin accessibility under GAL gene induction conditions. Moreover, this increase in nuclease sensitivity was not observed when cells were grown in the presence of raffinose or glucose. Such increases in global chromatin accessibility have been observed also with certain yeast mutants, for example, with Sin− versions of histone H4 (41) and yeast cells with a deletion of Sin4, an RNA polII holoenzyme component believed to modulate chromatin organization (27). We have also shown that the simple Gal− phenotype of htz1Δ cells cannot account for the nuclease hypersensitivity observed, as gal4Δ cells did not show such increased chromatin accessibility under the same conditions. Moreover, the fact that simply adding galactose to raffinose-grown cells is sufficient to induce this nuclease hypersensitivity suggests that GAL-inducing conditions trigger some special function of the variant histone. It is conceivable that loss of H2A.Z, under GAL-inducing conditions, specifically affects the expression of protein factors required to maintain global chromatin integrity. Alternatively, the requirement for H2A.Z might be more detrimental under certain growth conditions to the fine-tuning of global gene expression. In accord with this possibility is the recent finding that changes in global gene expression do occur when cells are grown in galactose versus glucose (33, 34). Indeed, the expression of approximately 10% of the genome is affected by a factor of 2, either positively or negatively (33), and some GAL genes are known to be induced more than 1,000-fold (23). These changes in global transcription patterns could thus create specific requirements for the specific function of H2A.Z.
ACKNOWLEDGMENTS
This work was supported by grants from the CIHR and NSERC of Canada and the FCAR of Québec to L.G. F.R. holds a fellowship from the NCI of Canada; L.G. is a research scholar of the CIHR/CRS Inc. of Canada.
We are grateful to Mary Ann Osley for gifts of yeast strains and Gerard Evan for the 9E11 hybridoma cell line. We thank Martin Gorovsky, Jocelyn Beaucher, and Karine Lemieux for discussions and comments on the manuscript. We also thank Nancy Hannett for the Myc tagging of H2A and H2A.Z and Daniel Paradis for technical help. We are especially thankful to Richard Young for all of his support during the course of this study.
M.A., F.R., and M.L. contributed equally to this work.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multicopy disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bash R, Lohr D. Yeast chromatin structure and regulation of GAL gene expression. Prog Nucleic Acid Res Mol Biol. 2000;65:197–259. doi: 10.1016/s0079-6603(00)65006-7. [DOI] [PubMed] [Google Scholar]
- 3.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 4.Clarkson M J, Wells J R E, Gibson F, Saint R, Tremethick D J. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. doi: 10.1038/21436. [DOI] [PubMed] [Google Scholar]
- 5.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 6.Dhillon N, Kamakaka R T. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 7.Dudley A M, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durrin L K, Mann R K, Grunstein M. Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1621–1629. doi: 10.1128/mcb.12.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudreau L, Keaveney M, Nevado J, Bryant G O, Struhl K, Ptashne M. Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 11.Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 12.Hirschhorn J N, Bortvin A L, Ricupero-Hovasse S L, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eucaryotic genome. Cell. 1998;98:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 14.Jackson J D, Falciano V T, Gorovsky M A. A likely histone H2A.F/Z variant in Saccharomyces cerevisiae. Trends Biochem Sci. 1996;21:466–467. doi: 10.1016/s0968-0004(96)20028-3. [DOI] [PubMed] [Google Scholar]
- 15.Jackson J D, Gorovsky M A. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 2000;19:3811–3816. doi: 10.1093/nar/28.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 17.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 18.Koleske A J, Buratowski S, Nonet M, Young R A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- 19.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires PolII holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 20.Leach T J, Mazzeo M, Chotkowski H L, Madigan J P, Wotring M G, Glaser R L. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J Biol Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- 21.Li X-Y, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Li B, Gorovsky M A. Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol Cell Biol. 1996;16:4305–4311. doi: 10.1128/mcb.16.8.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 24.Lohr D. Nucleosome transactions on the promoters of the yeast GAL and PHO genes. J Biol Chem. 1997;272:26795–26798. doi: 10.1074/jbc.272.43.26795. [DOI] [PubMed] [Google Scholar]
- 25.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 27.Macatee T, Jiang Y W, Stillman D J, Roth S Y. Global alterations in chromatin accessibility associated with loss of SIN4 function. Nucleic Acids Res. 1997;6:1240–1247. doi: 10.1093/nar/25.6.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neely K E, Hassan A H, Wallberg A E, Steger D J, Cairns B R, Wright A P H, Workman J L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 29.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M G, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Martin J. Chromatin and transcription in Saccharomyces cerevisiae. FEMS Microbiol Rev. 1999;23:503–523. doi: 10.1111/j.1574-6976.1999.tb00410.x. [DOI] [PubMed] [Google Scholar]
- 31.Platt A, Reece R J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 33.Ren B, Robert F, Wyrick J J, Aparicio O, Jennings E G, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert T L, Wilson C J, Bell S P, Young R A. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 34.Roth F P, Hugues J D, Estep P W, Church G M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 35.Santisteban M S, Kalashnikova T, Smith M M. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 36.Stargell L, Bowen J, Dadd C A, Dedon P C, Davis M, Cook R G, Allis C D, Gorovsky M A. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993;7:2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- 37.Suto R K, Clarkson M J, Tremethick D J, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;12:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 38.Svaren J, Venter U, Hörz W. In vivo analysis of nucleosome structure and transcription factor binding in Saccharomyces cerevisiae. Methods Mol Genet. 1995;6:153–167. [Google Scholar]
- 39.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 40.Wan J S, Mann R K, Grunstein M. Yeast histone H3 and H4 N-termini function through different GAL1 regulatory elements to repress and activate transcription. Proc Natl Acad Sci USA. 1995;92:5664–5668. doi: 10.1073/pnas.92.12.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wechser M A, Kladde M P, Alfieri J A, Peterson C L. Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 43.Wyrick J J, Holstege F C P, Jennings E G, Causton H C, Shore D, Grunstein M, Lander E S, Young R A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 44.Yocum R R, Hanley S, West R, Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yudkovski N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]