Abstract
Very fast amplification of DNA in small volumes can be continuously monitored with a rapid cycler that incorporates fluorimetric detection. Primers were designed to amplify a 157-bp fragment of the rpoB gene spanning codons 526 and 531 and a 209-bp fragment of the katG gene spanning codon 315 of Mycobacterium tuberculosis. Most mutations associated with resistance to rifampin (RMP) and isoniazid (INH) in clinical isolates occur in these codons. Two pairs of hybridization probes were synthesized; one in each pair was 3′ labeled with fluorescein and hybridized upstream of the codon with the mutation; the other two probes were 5′ labeled with LightCycler-Red 640. Each pair of probes recognized adjacent sequences in the amplicon. After DNA amplification was finished by using a LightCycler, the temperature at which the Red 640 probe melted from the product was determined in a 3-min melt program. Twenty M. tuberculosis clinical isolates susceptible to streptomycin, INH, RMP, and ethambutol and 36 antibiotic-resistant clinical M. tuberculosis isolates (16 resistant to RMP, 16 to INH, and 4 to both antimicrobial agents) were amplified, and the presence of mutations was determined using single-strand conformation polymorphism analysis, the LiQor automated sequencer, and the LightCycler system. Concordant results were obtained in all cases. Within 30 min, the LightCycler method correctly genotyped all the strains without the need of any post-PCR sample manipulation. Overall, this pilot study demonstrated that real-time PCR coupled to fluorescence detection is the fastest available method for the detection of RMP and INH resistance-associated mutations in M. tuberculosis clinical isolates.
The emergence of drug-resistant strains of Mycobacterium tuberculosis is an increasing problem for populations and tuberculosis control programs in developed and developing countries alike. Today, rifampin (RMP) and isoniazid (INH) are important components of effective multidrug therapy and prophylaxis for M. tuberculosis infections. However, widespread use of these agents and failure of patients to complete prescribed treatment have led to the emergence of RMP- and INH-resistant strains (5, 7). RMP resistance is well characterized, and 95% of all M. tuberculosis RMP-resistant clinical isolates harbor specific mutations within an 81-bp region of the rpoB gene, which encodes the β subunit of the RNA polymerase (8, 18). In contrast to RMP, genotypic testing for INH resistance is much more complex. Several studies have reported alterations in at least four genes, katG, inhA, ahpC, and kasA (1, 10, 12, 21, 24). Although the importance of katG mutations in INH resistance has been clearly demonstrated, the involvement of the other genes is arguable (6, 10, 11, 15). The highest proportion of mutations is in the katG gene, but only some mutations in this gene have been associated with high-level resistance (e.g., deletions or S315T), whereas others do not appear to confer any resistance to INH (e.g., R463L).
The need to minimize the transmission of drug-resistant strains requires rapid identification procedures. Due to their high sensitivity and specificity, DNA assays have the potential to provide rapid detection of resistance in mycobacterial isolates. Because these procedures do not rely on in vitro growth, the time between diagnosis and the onset of effective therapy is shortened.
In this report we present a single-tube method for detecting mutations associated with resistance to RMP and INH. It combines both rapid-cycle PCR and real-time monitoring of the processing and generation of mutation-specific, fluorescent-probe melting profiles on the LightCycler (Roche Biochemicals). Two fluorescently labeled hybridization probes recognizing adjacent sequences in the amplicon were present in the reaction mixture. The shorter detection probe (sensor [5′-LightCycler-Red 640 labeled]) covers the predicted site of mutation, while the longer probe (anchor [3′ fluorescein labeled]) produces the fluorescent signal. After annealing, the fluorophores are in close proximity and there can be a fluorescence resonance energy transfer between them, providing real-time monitoring of the amplification process. When PCR was completed, fluorescence was monitored as the temperature was increased through the melting temperature (Tm) of the probe-product duplexes, and a characteristic melting profile for each genotype was obtained. We used two different pairs of probes—one designed to detect the two most frequent mutations, Ser531Leu (a change in codon 531 from coding serine to leucine [TCG→TTG]) and His526Asp (a change in codon 526 from coding histidine to aspartic acid [CAC→GAC]) in the rpoB gene related to RMP resistance, and the other designed to detect the most frequent mutation (in codon 315) related to INH resistance in the katG gene. With this method one strain could be genotyped within 30 min without any post-PCR sample manipulation.
MATERIALS AND METHODS
Strains and resistance testing.
Twenty susceptible and 36 resistant clinical isolates of M. tuberculosis (16 resistant to RMP, 16 to INH, and 4 to both), as determined by the radiometric dilution method (23), from 56 different patients were studied. By this method, drug resistance was defined as greater than 1% growth in the presence of 2 μg of streptomycin per ml, 0.2 μg of INH per ml, 2 μg of RMP per ml, or 5 μg of ethambutol per ml. Isolates that were found to be resistant by this method were retested by the proportion method before being reported as resistant (23).
Relatedness of all resistant strains was investigated by restriction fragment length polymorphism of the IS6110 element (16). All the strains had different IS6110 fingerprint patterns. M. tuberculosis H37Rv (susceptible), M. tuberculosis ATCC 35838 (RMP resistant), and M. tuberculosis ATCC 35822 (INH resistant) were used as controls.
Extraction of mycobacterial DNA from the strains.
A rapid DNA extraction procedure for direct testing of M. tuberculosis on Löwenstein-Jensen solid medium was performed as follows. One 10-μl loop of the organisms was suspended in 100 μl of sterile water, and subsequently 100 μl of a 10% suspension of Chelex 100 was added (20). After thorough mixing, the mixture was incubated at 45°C for 45 min, then the suspension was boiled for 5 min, and the bacterial debris was removed by centrifugation (12,000 × g for 5 min). The supernatant was directly used for amplification.
Conventional PCR.
The DNA preparation was amplified with the primers listed in Table 1. PCR primers were designed to amplify the regions between nucleotides 2335 and 2492 of the rpoB gene and nucleotides 2759 and 2967 of the katG gene (19). Conditions for cycling were 95°C for 45 s followed by 35 cycles of 95°C for 15 s, 55°C (annealing temperature for the rpoB gene) and 60°C (annealing temperature for the katG gene) for 15 s, and 72°C for 15 s in a 9600 Perkin Elmer apparatus. PCR products were purified by using the Sephaglas BandPrep kit (Amersham Pharmacia Biotech).
TABLE 1.
Oligonucleotides—primersa and probes—used in the amplification and detection protocol by PCR of the M. tuberculosis strains
| Target | Oligonucleotides | Positionb | Product size (bp) | Annealing temperature (°C) | Tm (°C) |
|---|---|---|---|---|---|
| rpoB | TR8, 5′-GTGCACGTCGCGGACCTCCA | 2492 | 158 | 55 | 68.4 |
| TR9, 5′-TCGCCGCGATCAAGGAGT | 2335 | 61.0 | |||
| rpoB | rpo sensor, 5′-640c-ACCCACAAGCGCCGACTGCTGG-Phosd | 2412 | 70.4 | ||
| rpo anchor, 5′-TTCATGGACCAGAACAACCCGCTGTCGGT-3′Fe | 2379 | 73.8 | |||
| katG | TB86, 5′-GAAACAGCGGCGCTGATCGT | 2759 | 209 | 60 | 64.3 |
| TB87, 5′-GTTGTCCCATTTCGTCGGGG | 2967 | 62.8 | |||
| katG | TB sensor, 5′-640c-TCACCAGCGGCATCGAGGTCGT-Phosd | 2916 | 69.4 | ||
| TB anchor, 5′-CGTATGGCACCGGAACCGGTAAGGACGC-3′Fe | 2886 | 75.3 |
Primer sequences were described by Telenti et al. (19).
The positions of the oligonucleotides correspond to GenBank accession number L27989 for the rpoB gene and accession number X68081 for the katG gene.
640, LightCycler-Red 640 is a fluorophore with maximum absorbance at 622 nm and maximum emission at 638 nm.
The 3′ terminus of the sensor probe was phosphorylated to prevent extension of the probes during PCR.
F, fluorescein.
PCR-single-strand conformation polymorphism analysis (3).
After amplification by the same PCR protocol as above, 50 ng of the purified PCR product was mixed 1:1 with at least 2 μl of loading buffer (99% formamide–5 mM EDTA–0.5% bromophenol blue), heated for 10 min at 95°C, cooled on ice, and loaded in a precast, ready-to-use gel (Genegel Excel 12,5/24 kit; Amersham Pharmacia Biotech). Electrophoresis was run on a GenePhor electrophoresis unit at 600 V, 25 mA, 15 W, and 15°C for about 80 min (until bromophenol blue reached the anode buffer strip). Gels were stained in a Hoefer automated gel stainer with PlusOne DNA silver stain (Amersham Pharmacia Biotech). This method was used as part of the characterization of resistant strains.
DNA sequencing.
Direct sequencing of PCR products of all the strains was performed with a LiQor automated sequencer and corresponding kits from the same manufacturer (Roche Biochemicals).
LightCycler.
All DNA extracts were amplified with the primers used in conventional PCR, with the addition of the fluorescein- and Red 640-labeled probes to the amplification mix. The probes were designed as shown in Fig. 1. For rpoB the 5′-Red 640-labeled probe (rpo sensor) covered the region containing codons 526 and 531. To avoid dimer formation between primer TR8 and the rpo anchor probe, a G→T substitution at position 2407 (GenBank accession number L27989) was introduced in the 3′ terminus of the rpo anchor probe. There was a four-base gap between the rpo sensor and the 3′-fluorescein-labeled probe (rpo anchor). The theoretical Tms were 61 and 68.4°C for TR9 and TR8 primers, respectively, and 70.4 and 73.8°C for the rpo sensor and rpo anchor probes, respectively. For the katG gene, the 5′-Red 640-labeled probe (katG sensor) covered the region containing codon 315. There was a two-base gap between the katG sensor and the 3′-fluorescein-labeled probe (katG anchor). The theoretical Tms were 64.3 and 62.8°C for TB86 and TB87 primers, respectively, and 69.4 and 75.3°C for katG sensor and katG anchor probes, respectively (Table 1). Thus, the Red 640-labeled probes should melt from the PCR product at a lower temperature than the fluorescein-labeled probes so that the fluorescence observed is a measure of the Red 640-labeled probes. All primers and probes were designed and synthesized by TIB MOLBIOL (DNA Synthesis Service; Roche Diagnostics, Berlin, Germany).
FIG. 1.
Design of fluorescent probes for mutation detection in the rpoB (A) and katG (B) genes in the LightCycler. The probes are designed so that the Red 640-labeled probe will detach at a lower temperature than the fluorescein-labeled probe. Primers were those described by Telenti et al. (19).
The components for PCR in a final volume of 20 μl included 2 μl of a commercial ready-to-use reaction mix for PCR (LightCycler-DNA master hybridization probes; Roche Diagnostics) that contains Taq DNA polymerase, reaction buffer, deoxynucleoside triphosphate mix, and 10 mM MgCl2. We added MgCl2 to a final concentration of 4 mM. The primers and probes were added to final concentrations of 0.5 and 0.2 μM, respectively. Finally, we used 2 μl of template DNA. The 20-μl (final volume) reaction mix was placed in glass capillary cuvettes which were filled by pulse centrifugation in a microcentrifuge. Conditions for cycling were 95°C for 45 s, followed by 35 cycles of 95°C for 1 s, 55°C (annealing temperature for the rpoB gene) and 60°C (annealing temperature for the katG gene) for 2 s, and 72°C for 6 s, with monitoring of fluorescence during the annealing phase; this, in turn, was followed by a melting program of 50 to 85°C at 0.1°C/s with continuous monitoring of the fluorescence.
RESULTS
Rapid PCR and continuous monitoring of product.
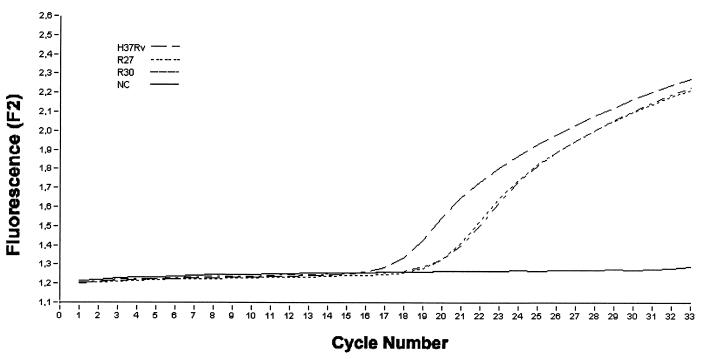
The accumulation of PCR product was monitored by measuring the level of fluorescence. Figure 2 shows the progress of PCR with Chelex 100-extracted DNA as the template. The PCR product was observed to accumulate in an exponential manner, indicating an optimal PCR. It can be seen that the signals started to rise at different times, usually between cycle numbers 15 and 25, because we did not estimate the DNA concentration and started with different amounts of DNA. This did not affect the interpretation of the Tm. We used the same primers for “conventional” PCR and LightCycler, because our target was small enough that this rapid cycling protocol worked well.
FIG. 2.
Accumulation of Red 640 fluorescence during PCR using the katG anchor and sensor probes with three different samples: M. tuberculosis H37Rv strain (susceptible) and R27 and R30 M. tuberculosis strains (INH resistant). NC, negative control. F2 is the channel used by the LightCycler to detect the LightCycler-Red 640 light emission.
Tms of the probes.
The temperatures at which the probes melted from PCR products during the melting program were calculated using the LightCycler software. A summary of the Tms for all of the probes (RMP and INH) together with the changes in Tm for the products derived from resistant and susceptible M. tuberculosis strains is shown in Table 2.
TABLE 2.
Tms for RMP and INH mutation detection probes
| Antimicrobial agent | Mean probe Tm (°C) ± SD for strain or mutationa
|
|||||
|---|---|---|---|---|---|---|
| Wild typeb | 531 (TCG→TTG) | 526 (CAC→GAC) | 315 (AGC→ACC) | 315 (AGC→AAC) | Susceptible strains | |
| RMP | 64.3 ± 0.4 | 66.9 ± 0.5c (18) | 58.1 ± 0.2 (2) | 64.4 ± 0.6 (20) | ||
| INH | 72.8 ± 0.5 | 68.7 ± 0.4c (18) | 67.9 ± 0.07 (2) | 72.4 ± 0.4 (20) | ||
Means were derived from at least three determinations. Numbers in parentheses are numbers of strains tested.
M. tuberculosis H37Rv (susceptible).
Four strains were RMP and INH resistant.
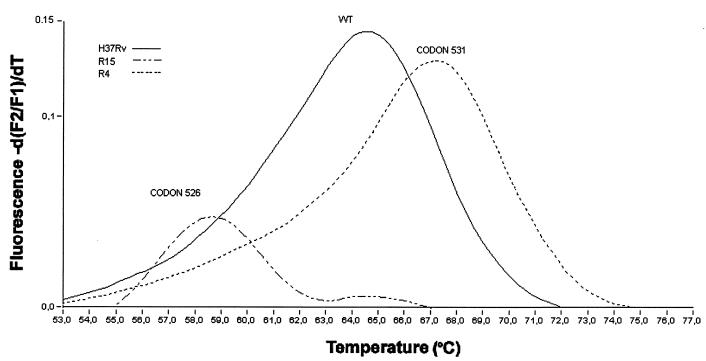
We were able to detect the two most frequent mutations responsible for RMP resistance, using the RMP probe (rpo sensor) (Fig. 3). The Tm for the susceptible strain used as a control (M. tuberculosis H37Rv) was 64.3°C, while the change from wild type to mutant at codon 531 (TCG→TTG) in 18 strains resulted in a >2°C increase in the probe's Tm. In the two strains with mutations at codon 526 (CAC→GAC), we observed a >6°C drop in the probe's Tm.
FIG. 3.
Derivative melting curves (dF/dT) of Red 640-labeled rpo sensor probe with three different samples. This sensor probe was designed to have the sequence CTG at codon 531 and the wild-type sequence (CAC) at codon 526. Strains: H3TRv, M. tuberculosis H37Rv strain (susceptible) with a double mismatch at codon 531 (TCG) resulting in a Tm of 64.4°C; R4, M. tuberculosis R4 strain (RMP resistant) with a 531-codon point mutation (TCG to TTG), which has a higher Tm of 66.9°C showing a single mismatch; R15, M. tuberculosis R15 strain (RMP resistant) with a 526-codon point mutation (CAC to GAC), which has the lower Tm due to the three mismatches, one at codon 526 and two at codon 531. Each trace shows the change in fluorescence ratio with time with respect to temperature, thus allowing calculation of the temperature at which the probe detaches from the PCR product. F2 is the channel used by the LightCycler to detect the LightCycler-Red 640 light emission; F1 is the channel measuring the background.
Figure 4 shows the results with the INH probe (katG sensor). The Tm for the susceptible strain (M. tuberculosis H37Rv) was 72.8°C. The change from wild type (AGC) to ACC at codon 315 in 18 strains resulted in a >3°C drop of Tm of the katG sensor probe. A drop of almost 5°C in the Tm was found in two strains harboring a different mutation at codon 315 (AGC→AAC).
FIG. 4.
Derivative melting curves of Red 640-labeled katG sensor probe with three different samples: M. tuberculosis H37Rv strain (susceptible) with a perfect match to the hybridization probe and a Tm of 72.8°C; M. tuberculosis strain R30 (INH resistant) with the mutation AGC → ACC at codon 315, and a Tm of 68.7°C; and M. tuberculosis strain R27 with the mutation AGC → AAC at codon 315 and a Tm of 67.9°C. Each trace shows the change in fluorescence ratio over time with respect to temperature, thus allowing calculation of the temperature at which the probe detaches from the PCR product. F2 is the channel used by the LightCycler to detect LightCycler-Red 640 light emission; F1 is the channel measuring the background.
All the susceptible M. tuberculosis strains, determined by using either the rpo sensor or the katG sensor probe, showed a Tm that corresponded to the susceptible strain used as control (M. tuberculosis H37Rv). In addition, the sequence inferred from the LightCycler data was always in accordance with the nucleotide sequencing and the single-strand conformational polymorphism pattern.
DISCUSSION
Due to the slow growth of the M. tuberculosis bacillus, unacceptable delays in the diagnosis of drug-resistant tuberculosis can occur when conventional culture-based susceptibility tests are used. Understanding the genetic events that lead to drug resistance in clinical M. tuberculosis isolates is important for elucidation of the action of antimicrobial agents, for design of novel antibiotics that are active against drug-resistant strains, and for development of genetic assays to be adapted to the routine clinical mycobacteriology laboratory. Rapid approaches that use genetic analysis for detection of mutations associated with drug resistance have been proposed (4, 9, 13, 15, 19). However, these methods have restricted applicability in clinical laboratories and are mainly limited to the detection of RMP resistance.
The method described here provides a rapid, accurate, and inexpensive way to detect resistance in M. tuberculosis clinical isolates. Amplification, hybridization, and analysis are all performed simultaneously in sealed capillary tubes. The entire assay, including analysis, can be completed in 30 min. In all cases, the result obtained with the LightCycler corresponded to nucleotide sequence data; therefore, unlike many other nucleic amplification tests (14), the LightCycler amplification assay was 100% specific. This was likely due to the closed-well protocol, which eliminates amplicon carryover. Theoretically, it is possible that natural variability in the sequences where the sensor probes bind could lead to a change in the Tm of the probe that is not associated with resistance mutation. However, previous studies (17) have suggested that the DNA sequences of M. tuberculosis are extraordinarily well conserved and that mutations in the M. tuberculosis genome are almost always associated with drug resistance. This fact makes this method even more useful for resistance detection in M. tuberculosis.
With the RMP probe used in this study, we were able to detect mutations at codon 531 and codon 526. We previously reported (6) that these were the two most frequent residue changes associated with RMP resistance in our study area, and this finding was similar to previous studies (8, 18). Based on our mutation data from strains isolated in the last 6 years (6; unpublished data), if we used an additional probe covering codons 513 and 518, the sensitivity for the detection of RMP resistance would be greater than 95%.
In contrast to RMP resistance testing, genotypic testing for resistance to INH presents many difficulties. Our understanding of the genetic basis of INH resistance is far from complete. Changes in the katG gene, the inhA gene, and the ahpC gene in INH-resistant mycobacterial isolates have been described. The importance of katG mutations in INH resistance is well established, although the extent to which such mutations account for the spectrum of resistance is still debated. Missense mutations at codon 315 of the katG gene have been reported in almost two-thirds of the INH-resistant isolates from different geographic areas including ours (6, 13, 19). The high percentage of mutations at position 315 and the occurrence of different nonsynonymous substitutions demonstrate the importance of this codon for the development of INH resistance among M. tuberculosis strains. Mutations at other positions seem to be rare events. In addition, many of these do not alter enzyme activity, and usually it is suggested that they are involved in the resistance mechanism simply because no other katG gene mutations were observed. For all these reasons, we decided to assess the utility of this new method by using a probe to screen for the presence of mutation in a selected region of the katG gene that includes position 315. The specificity of the test was 100%, and the Tm of the probe allowed the two mutations studied to be distinguished.
We are working on the development of a probe to screen for mutations in the regulatory region of the inhA gene that cause overexpression of this gene. However, in our sample population, we found this alteration (nucleotide substitution at position 209 [C→T]) in only 10% of the strains, and there is still some controversy about the implication of this gene in INH resistance (11).
The presence of mutations in either of these two genes (i.e., katG or inhA) would account for INH resistance, but, according to previous studies on different populations (15), between 20 and 60% of resistant strains have no detectable mutations in those genes. Therefore, more research is needed to elucidate the remaining cases of resistant strains that cannot be attributed to the above-mentioned mutations in katG and inhA genes. Furthermore, in both RMP and INH resistance, the relative frequency of particular (i.e., rare) mutations may reflect local strain populations and/or epidemic strains. Thus, determination of the frequency of mutations underlying drug resistance in isolates from different geographic areas has to be the basis for developing rapid and specific techniques for the detection of drug-resistant strains by molecular genetic techniques.
This new method presents several advantages. First, rapid amplification and analysis allow the test to be completed within 30 min. Because the DNA extraction procedure requires 1 h, the whole detection can be completed in less than 2 h. Second, the LightCycler optical device is capable of measuring fluorescence in two separate channels simultaneously (Red 640 and Red 705 Fluorophore), and it is possible to use Sybr Green I as a generic donor of fluorescence resonance energy transfer (2, 22) (instead of a specific fluorescein probe), thus allowing analysis of different mutations within a single test tube. Third, the assay is run in closed glass capillaries. Postamplification analysis can be performed without opening the capillaries, minimizing the risk of carryover contamination. Fourth, the LightCycler could be used to determine the genetic make-up of some bacterial populations composed of subpopulations with different genotypes. Fifth, the method can be adapted to genotyping resistance information emerging from the identification of additional targets, particularly for INH resistance.
Finally, we emphasize that this is the first time that real-time PCR has been employed to study resistance in M. tuberculosis clinical isolates. Reliable results can be obtained faster than with any other current molecular methods and could be directly applied to clinical samples.
ACKNOWLEDGMENT
This study was financially supported by grant 99/0269 from the Fondo de Investigación Sanitaria, Ministerio de Sanidad y Consumo, Madrid, Spain.
REFERENCES
- 1.Banerjee A, Dubnau E, Quemard A, Balasubramanian S K, Um K, Wilson T, Collins D, De Lisle G, Jacobs W R., Jr InhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 2.Cane P A, Cook P, Ratcliffe D, Mutimer D, Pillay D. Use of real-time PCR and fluorimetry to detect lamivudine resistance-associated mutations in hepatitis B virus. Antimicrob Agents Chemother. 1999;43:1600–1608. doi: 10.1128/aac.43.7.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaubert P, Bautista D, Benhattar J. An improved method for rapid screening of DNA mutations by non-radioactive single-strand conformation polymorphism procedure. BioTechniques. 1993;15:586. [PubMed] [Google Scholar]
- 4.Felmlee T, Liu Q, Whelen C, Williams D, Sommer S S, Pershing P D. Genotyping detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand DNA conformation polymorphism and dideoxy fingerprinting. J Clin Microbiol. 1995;33:1617–1623. doi: 10.1128/jcm.33.6.1617-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frieden T R, Sterling T, Plabos-Mendez A, Kilburn J O, Cauthen G M, Dooley S W. The emergence of drug-resistant tuberculosis in New York City. N Engl J Med. 1993;328:521–526. doi: 10.1056/NEJM199302253280801. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez N, Torres M J, Aznar J, Palomares J C. Molecular analysis of rifampin and isoniazid resistance of Mycobacterium tuberculosis clinical isolates in Seville, Spain. Tuber Lung Dis. 1999;79:187–190. doi: 10.1054/tuld.1998.0195. [DOI] [PubMed] [Google Scholar]
- 7.Heym B, Alzary P, Honoré N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D A, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 8.Kapur V, Li L L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automatic DNA sequencing of mutation in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapur V, Li L L, Hamrick M C, Plicaytis B B, Skinnick T M, Telenti A, Jacobs W R, Banerjee A, Cole S, Yuen K Y, Clarridge III J E, Kreiswirth B N, Musser J M. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automatic DNA sequencing. Arch Pathol Lab Med. 1995;119:131–138. [PubMed] [Google Scholar]
- 10.Kelley C L, Rouse D A, Morris S L. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:2057–2058. doi: 10.1128/aac.41.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khisimuzi M, Sherman D R, Hickey M J, Kreiswirth B N, Morris S, Stover C K, Barry C E., III Biochemical and genetic data suggest that inhA is not the primary target for activated isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 12.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Inhibition of Mycobacterium tuberculosis beta-ketoacyl ACP synthetase by isoniazid. Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 13.Nachamkim I, Kang C, Weinstein M P. Detection of resistance to isoniazid, rifampin, and streptomycin in clinical isolates of Mycobacterium tuberculosis by molecular methods. Clin Infect Dis. 1997;24:894–900. doi: 10.1093/clinids/24.5.894. [DOI] [PubMed] [Google Scholar]
- 14.Noordhoek G T, Van Embden J D A, Kolk A H J. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J Clin Microbiol. 1996;34:2522–2525. doi: 10.1128/jcm.34.10.2522-2525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piateck A S, Telenti A, Murray M G, El-Hajj H, Jacobs W R, Jr, Kramer F R, Alland D. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000;44:103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safi H, Aznar J, Palomares J C. Molecular epidemiology of Mycobacterium tuberculosis strains isolated during a 3-year period (1993–1996) in Seville, Spain. J Clin Microbiol. 1997;35:2472–2476. doi: 10.1128/jcm.35.10.2472-2476.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc Natl Acad Sci USA. 1997;74:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Coltson M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 19.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Takiff H E, Cole S T. Genotyping assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh P S, Metzger D A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:505–513. [PubMed] [Google Scholar]
- 21.Wilson T M, Collins D M. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol Microbiol. 1996;19:1025–1034. doi: 10.1046/j.1365-2958.1996.449980.x. [DOI] [PubMed] [Google Scholar]
- 22.Woo T H S, Patel B K C, Smythe L D, Symonds M L, Norris M A, Dohnt M F. Identification of pathogenic Leptospira genospecies by continuous monitoring of fluorogenic hybridization probes during rapid-cycle PCR. J Clin Microbiol. 1997;35:3140–3146. doi: 10.1128/jcm.35.12.3140-3146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zapata P, Arbeola M, Aznar J. Evaluation of mycobacteria growth indicator tube (MGIT) for drug susceptibility testing of Mycobacterium tuberculosis isolates from clinical specimens. Clin Microbiol Infect. 1999;5:227–230. doi: 10.1111/j.1469-0691.1999.tb00129.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Heym B, Allen B, Young D, Cole S T. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:501–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]