Abstract
The aim of our study was to compare the performances of two new hepatitis B virus (HBV) DNA assays, a cross-linking assay (NAXCOR) and a hybrid-capture amplification assay (Digene), versus the widely used branched-DNA (bDNA) assay (Chiron) in the monitoring of HBV DNA levels during antiviral treatment. Serial serum samples from 12 chronically HBV infected patients undergoing a phase II trial of an antiviral drug, 2′,3′-dideoxy-5-fluoro-3′-thiacytidine (FTC), were studied. A total of 96 serum samples were tested for HBV DNA using the cross-linking, hybrid-capture amplification, and bDNA assays. In the comparison of the cross-linking and bDNA assays, concordant results were found in 77 (80.3%) samples, no significant difference was found between the median log10 HBV DNA levels (6.66 versus 7.17 meq/ml), and the results of the two assays were closely correlated (r = 0.95). In the comparison of the hybrid-capture amplification and bDNA assays, concordant results were found in 79 (82.3%) samples, no significant difference was found between the median log10 HBV DNA levels (6.98 versus 6.99 meq/ml), and the results of the two assays were closely correlated (r = 0.99). Six (6.3%) samples by the cross-linking assay and 10 (10.4%) samples by the bDNA assay required retesting because of unacceptably high within-run coefficients of variance. No sample required retesting in the hybrid-capture amplification assay according to the internal validation. In conclusion, the cross-linking and hybrid-capture amplification assays were as sensitive as the bDNA assay for HBV DNA detection and can be recommended for monitoring of HBV DNA levels during antiviral treatment.
Diagnosis of chronic hepatitis B virus (HBV) infection has long been based on HBV serology and measurement of liver enzymes. With the development of therapies for chronic HBV infection, including alpha interferon and nucleotide analogues, serology and liver enzymes are no longer sufficient to monitor antiviral response and the success of therapy. Seroconversion of HBeAg does not always imply cessation of viral replication or disease remission, as certain viral mutants do not produce HBeAg but remain replicative (2–4, 17). Assays of HBV DNA, the most reliable parameter for monitoring viral replication, have been increasing used as the most important marker of therapeutic efficacy in most treatment trials (5, 10, 15, 19, 22).
Early HBV DNA assays, including semiquantitative dot blot hybridization assays, a column-based solution hybridization assay (Abbott Laboratories, Chicago, Ill.), and a microplate-based hybridization assay (Digene Hybrid Capture; Murex Diagnostic Ltd., Dartford, United Kingdom), have been proven insensitive in detecting low HBV DNA titers (14, 18, 24). In a recent study, the majority of patients with undetectable HBV DNA levels by a solution hybridization assay (Abbott) during lamivudine therapy experienced relapse and resumption of viral replication after cessation of medication (15). The study was criticized for missing low-grade viral activity during treatment due to the relative insensitivity of the assay. More-sensitive assays are needed for monitoring of treatment response and prediction of responders to antiviral therapy. The recently developed hybridization assay with branched-DNA (bDNA) molecules and signal amplification (Chiron Diagnostic, Emeryville, Calif.) detects HBV DNA down to 7 × 105 viral copies/ml (1, 12). The bDNA assay has been used as the standard for HBV DNA quantification in more recent antiviral treatment trials (8, 11).
A new HBV DNA assay based on nucleic acid cross-linking has been developed with a lower limit of HBV DNA detection of 5 × 105 viral equivalents/ml (NAXCOR XLnt; NAXCOR, Menlo Park, Calif.). This assay has been shown to yield sensitive and reproducible results comparable to those of the bDNA assay (Chiron) (16). Digene Diagnostics Inc. (Silver Spring, Md.) has also produced a second-generation HBV DNA assay (Hybrid Capture II) based on hybrid-capture amplification technology, and this assay has also been found to be able to detect low levels of HBV DNA (6). Both new assays do not require overnight incubation, unlike the bDNA assay, and can be completed within a few hours. A sensitive assay that gives reliable results at very low levels of HBV DNA is very much in need for monitoring the therapeutic effects of antiviral agents. However, there has to date been no study to evaluate the use of cross-linking and hybrid-capture amplification assays in the monitoring of serial HBV DNA levels during treatment. In this study, we aimed to evaluate the performance of cross-linking and hybrid-capture amplification assays versus the bDNA assay in the detection and quantification of HBV DNA in serial serum samples of patients undergoing antiviral treatment.
MATERIALS AND METHODS
Patients and samples.
Serial serum samples from 12 chronic hepatitis B patients who underwent a multicenter, open-labeled phase II study of a new antiviral drug, 2′,3′-dideoxy-5-fluoro-3′-thiacytidine (FTC) (Triangle Pharmaceuticals, Durham, N.C.), were studied (9). All patients were HBV DNA positive by the bDNA assay at baseline and were treated with FTC for 42 days. Serum samples were taken at baseline (day 0) and at 1, 7, 14, 28, 42, 56, and 84 days after the commencement of therapy, aliquoted, and stored at −70°C. In total, 96 serum samples were tested for HBV DNA using the cross-linking (NAXCOR), hybrid-capture amplification (Digene), and bDNA (Chiron) assays. Since the bDNA assay has been the most widely used test among the three, the cross-linking and hybrid-capture amplification assays were compared against the bDNA assay for their performance.
Cross-linking assay (NAXCOR).
Oligonucleotides complementary to sequences in the entire 3.2-kb HBV genome were synthesized as described previously (23). Two types of DNA probes were synthesized: fluorescein-containing reporter probes and biotin-containing capture probes. Both types of probes were modified with light-activated cross-linking agents. The assay was run according to the manufacturer's instructions. In brief, 300 μl of the serum sample was mixed with 30 μl of a lysis reagent (proteinase K, sodium dodecyl sulfate) at 65°C for 30 min. A 6.3-μl volume of denaturation reagent (sodium hydroxide) was then added and boiled at 100°C for 15 min. After cooling, 125 μl of the sample was transferred into 2 wells of a 96-well microplate. Three positive standards, containing HBV DNA at levels of 0.5, 60, and 6,000 meq/ml, and a negative standard were placed in two wells each. DNA probes and a neutralization solution were then added into the sample and control wells for hybridization at 45°C for 20 min, followed by irradiation with UV light for cross-linking at 45°C for 30 min. Streptavidin-coated magnetic-bead reagents were then added to capture the cross-linked probe-target hybrid at 37°C for 30 min. After two washes, the beads were incubated with an anti-fluorescein antibody-alkaline phosphate (AP) conjugate at 37°C for 20 min. After four further washes, Attophos (JBL Scientific, San Luis Obispo, Calif.) was added and incubated at 37°C for 60 min to react with AP. The final fluorescent product was detected by measuring the fluorescent signal with a fluorometer, and the HBV DNA concentration was calculated by comparison to a standard calibration curve obtained from the assaying of the positive standards. Samples with coefficients of variation (CV) higher than 15% between the results of the duplicate tests were retested. The range of HBV DNA quantification was 0.5 to 6,000 meq/ml.
Hybrid-capture amplification assay (Digene).
The hybrid-capture amplification assay was run according to the manufacturer's instructions. In brief, 30 μl of each serum sample, each of five calibrators (numbered 1 to 5, with HBV DNA at 0, 0.142, 28.3, 566, and 1,700 meq/ml, respectively), and each of two positive controls of the entire HBV genome were mixed with 30 μl of denaturation reagent in the wells of a microplate and centrifuged at 1,100 rpm for 1 min. The microplate was then incubated at 65°C for 30 min. An HBV probe mix was prepared by mixing HBV RNA probes specific to HBV strains ad and ay with probe diluent at a 1:25 dilution. A 30-μl portion of HBV probe mix was added into each microwell, and the mixtures were centrifuged at 1,100 rpm for 1 min and then incubated at 65°C for 60 min for hybridization. A 75-μl portion of the RNA-DNA hybrid was transferred to a capture microplate coated with anti-RNA-DNA hybrid antibodies and centrifuged at 1,100 rpm for 60 min at 20 to 25°C. The capture mixture was then aspirated out and microplate blotted. AP-conjugated antibodies to RNA-DNA hybrids were added to the wells of the capture microplate and incubated at 20 to 25°C for 30 min for hybrid detection. Several AP molecules were conjugated to each antibody, and multiple conjugated antibodies bound to each captured hybrid, resulting in substantial amplification. After decanting and blotting, a chemiluminescent substrate was added and incubated at 20 to 25°C for 15 min. Light emitted from cleavage of the chemiluminescent substrate by AP was detected by reading the microplate on the DML 2000 luminometer (Digene). HBV DNA results were internally validated based on the variance of the calibrators that were run with each plate of samples, and no duplicates were required, according to the manufacturer's recommendation. HBV DNA levels were then calculated by comparing the sample results to a standard curve obtained from the results of the calibrators. The range of HBV DNA quantification was 0.142 to 1,700 meq/ml.
bDNA assay (Chiron).
The bDNA-based assay was performed according to the manufacturer's instructions as described in detail previously (12). Briefly, HBV viral particles released from the serum sample (in duplicate), HBV standards, and controls (positive and negative) were hybridized overnight at 63°C with capture probes that were bound to the wall of the microwell plate as well as target probes that bound with different sequences of the HBV genome. The wells were then washed, and bDNA molecules were hybridized to extender probes. Subsequently, oligonucleotide-linked AP molecules were added to attach to the bDNA polymer. The captured DNA was detected by the chemiluminescence produced after reaction of the bound AP with a dioxetane substrate and was quantified by comparing the signal with a standard curve obtained from the assay of the HBV standards. Samples with results greater than the lower quantification limit but with CV of >20%, as well as samples below the quantification limit with CV of >25% between the two replicates, were retested, as recommended by the manufacturer. The range of HBV DNA quantification was 0.7 to 5,700 Meq/ml.
Statistics.
A two-tailed Fisher exact test was used to compare categorical data. A two-tailed Mann-Whitney U test was used to compare medians of results from different assays. Regression analysis was used to define the relationship between the results of different assays on the same serum samples. P values of <0.05 indicated statistical significance.
RESULTS
Overall, HBV DNA levels were measurable in 41 samples and undetectable in 27 samples by all three assays. Pretreatment samples from all patients had measurable HBV DNA levels by the three assays, while all samples with undetectable HBV DNA levels by the three assays were either on-treatment or post-treatment samples. Discordant results, with HBV DNA detectable by one or two assays only, were obtained for the remaining 28 samples. Three samples had HBV DNA levels higher than 1,700 meq/ml (the upper limit of detection by the hybrid-capture amplification assay), but none had HBV DNA levels over the upper limit of detection by the cross-linking and bDNA assays.
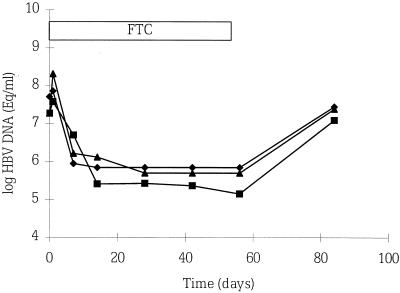
The serial HBV DNA levels of a patient typical of the 12 patients studied by the cross-linking, hybrid-capture amplification, and bDNA assays are shown in Fig. 1. A dramatic reduction in HBV DNA levels was documented by all three assays on day 7. After discontinuation of FTC on day 42, a resurgence of HBV DNA was detected by all three assays on day 84. This pattern of HBV changes was reported by Gish et al. in the FTC phase II study (9). HBV DNA remained undetected by all three assays on day 84 in only 1 of the 12 cases.
FIG. 1.
Serial HBV DNA levels of a patient as measured by the bDNA (⧫), hybrid-capture amplification (■), and cross-linking (▴) assays. HBV DNA levels decreased to below the limit of detectability during FTC treatment, but rose again after the cessation of treatment on day 42.
Precision of the assays.
Within-run reproducibility was determined for the cross-linking and bDNA assays as recommended by the manufacturers, while the validation of results of the hybrid-capture amplification assay was based on the variation of the internal calibrators. With samples in both tests run in duplicate, the mean within-run CV for the cross-linking and bDNA assays were 7.3 and 12.3%, respectively. Six (6.3%) and 10 (10.4%) samples required repeat testing in the cross-linking and bDNA assays, respectively, as their CV exceeded the limit set by the manufacturers (P = 0.44). No sample required repeat testing in the hybrid-capture amplification assay, as the variance of calibrators was within normal limits in all runs.
Performance of the cross-linking versus the bDNA assay.
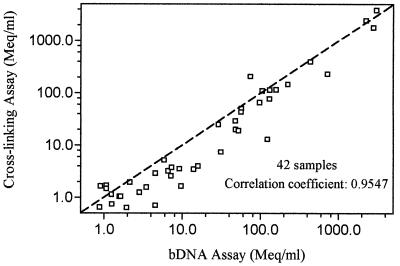
For the cross-linking and bDNA assays, concordant results were obtained for 77 (80.3%) samples; HBV DNA was detectable in 42 samples and undetectable in 35 samples by both assays. HBV DNA was detectable by the cross-linking assay alone in 15 samples (median HBV DNA level, 0.80 meq/ml; range, 0.60 to 150.8 meq/ml) and by the bDNA assay alone in 4 samples (median HBV DNA level, 0.94 meq/ml; range, 0.74 to 1.69 meq/ml). There was no significant difference between the median log10 HBV DNA levels in samples detected by the cross-linking and bDNA assays, which were 6.66 (range, 5.81 to 9.58) and 7.17 (range, 5.99 to 9.48) meq/ml, respectively (P = 0.25), and the results of the two assays were closely correlated (r = 0.95; P < 0.001; slope = 1.007) (Fig. 2). Accordingly, the formula derived for the interassay conversion of results is log(HBV DNA level by cross-linking assay) = 1.007 × log(HBV DNA level by bDNA assay) − 0.233, simplified as (HBV DNA level by cross-linking assay) = 0.595 × (HBV DNA level by bDNA assay)1.007, or (HBV DNA level by bDNA assay) = 1.702 × (HBV DNA level by cross-linking assay)0.993.
FIG. 2.
Comparison of HBV DNA levels among 42 serum samples determined by the cross-linking and bDNA assays. The line passing through the data with a slope equal to 1 is the hypothetical line that all data points would fall on if the two assays yielded results in complete agreement with each other.
Performance of the hybrid-capture amplification versus the bDNA assay.
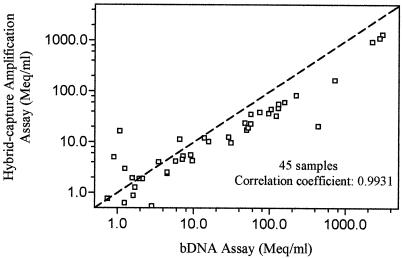
For the hybrid-capture amplification and bDNA assays, concordant results were found in 79 (82.3%) samples; HBV DNA was detectable in 45 samples and undetectable in 34 by both assays. HBV DNA was detectable by the hybrid-capture amplification assay alone in 16 samples (median HBV DNA level, 0.27 meq/ml; range, 0.16 to 56.03 meq/ml) and by the bDNA assay alone in 1 sample (HBV DNA level, 0.85 meq/ml). No significant difference was found between the median log10 HBV DNA levels in the hybrid-capture amplification and bDNA assays, which were 6.98 (range, 5.87 to 9.48) and 6.99 (range, 5.44 to 9.12) meq/ml, respectively (P = 0.30), and the results of the two assays were closely correlated (r = 0.99; P < 0.001; slope = 0.876) (Fig. 3). Accordingly, the formula derived for the interassay conversion of results is log(HBV DNA level by hybrid-capture assay) = 0.876 × log(HBV DNA level by bDNA assay) − 0.0970, simplified as (HBV DNA level by hybrid-capture assay) = 0.800 × (HBV DNA level by bDNA assay]0.876, or (HBV DNA level by bDNA assay) = 1.291 × (HBV DNA level by hybrid-capture assay)1.412.
FIG. 3.
Comparison of HBV DNA levels among 45 serum samples determined by the hybrid-capture amplification and bDNA assays. The line passing through the data with a slope equal to 1 is the hypothetical line that all data points would fall on if the two assays yielded results in complete agreement with each other.
DISCUSSION
The results of our study have shown a very good correlation among the cross-linking, hybrid-capture amplification, and bDNA assays in the detection and quantification of HBV DNA over a wide range of HBV DNA levels. The cross-linking and hybrid-capture amplification assays tend to detect HBV DNA in more samples than the bDNA assay, particularly over the low-level viremia range. This potentially increases the sensitivity of identification of persistent low-grade viremia in patients receiving antiviral treatments, but the clinical significance of this finding requires further investigation. Discordant results do occur within the quoted detection range of the three assays. The reasons for the discrepancy are not entirely clear. It could be related to assay-specific inhibition, a difference in the target probe sequence, or nonspecific hybridization.
The cross-linking and hybrid-capture amplification assays have the advantage over the bDNA assay that they require less time to complete. The cross-linking and hybrid-capture amplification assays take approximately 5 and 3 h, respectively, while the bDNA assay requires overnight incubation. The higher mean within-run CV in both the cross-linking and bDNA assays in our study compared with that reported previously might be related to the smaller number of replicates performed in our study (16). Although no samples required retesting in the hybrid-capture amplification assay, we are still concerned about the reliability of validation by the use of calibrators alone, as recommended by the manufacturer. It has been shown that the within-run CV of the hybrid-capture amplification assay ranged from 5 to 21% among three different centers (7).
Three of the 96 (3%) samples in our study had HBV DNA levels exceeding the upper detection limit of the hybrid-capture amplification assay, which was about 1/2 log unit lower than that of the cross-linking and bDNA assays. The failure to quantify HBV DNA in the very high level viremic patients may potentially limit the use of the hybrid-capture amplification assay in predicting high-risk patients who may need higher doses or a longer duration of antiviral treatment.
HBV DNA remained undetectable in 28% of serum samples by all three of the assays in our study. Whether this indicates very low levels of virus or complete clearance of HBV DNA is not clear. Our previous study has shown that approximately 50% of patients who had undetectable HBV DNA levels by the bDNA assay were in fact PCR positive by our in-house PCR assay (3). Although PCR is highly sensitive and can detect HBV DNA levels as low as 100 copies/ml, this very low level of viremia is of doubtful clinical significance. Moreover, PCR assays are prone to contamination, giving rise to false-positive results (20). A quantitative PCR assay with a sensitivity of 103 viral copies/ml has recently been introduced (13, 21). Whether it is useful and reliable in the monitoring of response to antiviral treatments requires further evaluation.
In summary, we found that the cross-linking assay and the hybrid-capture amplification assay are at least as sensitive as the bDNA assay for the detection and quantification of HBV DNA. These two new assays required shorter procedure times than the bDNA assay. Based on these results, the cross-linking and hybrid-capture amplification assays can be recommended for monitoring HBV DNA levels in antiviral therapies.
ACKNOWLEDGMENTS
This study was supported by the Cheng Suen Man Shook Foundation Centre for Hepatitis Studies.
We acknowledge Michael Wood and Minna Sung of NAXCOR and Mark van Asten of Digene Diagnostic Technology, Pymble, New South Wales, Australia, for providing the HBV DNA assays for this study; and Triangle Pharmaceuticals for allowing us to use FTC trial data in this study.
REFERENCES
- 1.Butterworth L A, Prior S L, Buda P J, Faoagali J L, Cooksley W G E. Comparison of four methods for quantitative measurement of hepatitis B viral DNA. J Hepatol. 1996;24:686–691. doi: 10.1016/s0168-8278(96)80264-9. [DOI] [PubMed] [Google Scholar]
- 2.Carman W F, Jacyna M R, Hadziyannis S, Karayiannis P, McGarvey M J, Makris A, Thomas H C. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;ii:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 3.Chan H L Y, Leung N W Y, Hussain M, Wong M L, Lok A S F. Are pre-core HBV mutants becoming more prevalent? Hepatology. 1998;28:484A. [Google Scholar]
- 4.Davis G L, Hoofnagle J H, Waggoner J G. Spontaneous reactivation of chronic hepatitis B virus infection. Gastroenterology. 1984;86:230–235. [PubMed] [Google Scholar]
- 5.Dienstag J, Perrillo R, Schiff E, Bartholomew M, Vicary C, Rubin M. A preliminary trial of lamivudine for chronic hepatitis B infection. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Meijide M, Brill J, Eddington N, Zhu J, Makar A, Larincz A T. Performance characteristics of the Digene Hybrid Capture II hepatitis B DNA test. Hepatology. 1998;28:486A. [Google Scholar]
- 7.Garcia-Meijide M, Eddington N, Brill J, Niesters H G M, Fries E, Krajden M, et al. Multi-center evaluation of the second-generation Digene Hybrid Capture II HBV test. Hepatology. 1998;28:579A. [Google Scholar]
- 8.Gilson R J C, Murray-Lyon I M, Nelson M R, Rice S J, Tedder R S, Murray A, et al. Extended treatment with adefovir dipivoxil in patients with chronic hepatitis B virus infection. Hepatology. 1998;28:491A. doi: 10.1046/j.1365-2893.1999.00182.x. [DOI] [PubMed] [Google Scholar]
- 9.Gish R G, Leung N W Y, Wright T L, Trinh H, Robertson A T, Harris J J, et al. Anti-hepatitis B virus (HBV) activity and pharmacokinetics of FTC in a 2-month trial in HBV-infected patients. Gastroenterology. 1999;116:A1216. [Google Scholar]
- 10.Grellier L, Mutimer D, Ahmed M, Brown D, Burroughs A K, Rolles K, McMaster P, Beranek P, Kennedy F, Kibbler H, McPhillips P, Elias E, Dusheiko G. Lamivudine prophylaxis against reinfection in liver transplantation for hepatitis B cirrhosis. Lancet. 1996;348:1212–1215. doi: 10.1016/s0140-6736(96)04444-3. [DOI] [PubMed] [Google Scholar]
- 11.Heathcote J, Chan R, McHutchison J, Lee W M, Sherman M, Rutkiewicz V, et al. A phase II multi-center study of oral lobucavir for treatment of chronic hepatitis B. Hepatology. 1998;28:318A. [Google Scholar]
- 12.Hendricks D A, Stowe B J, Hoo B S, Kolberg J, Irvine B D, Neuwald P D, Urdea M S, Perillo R P. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am J Clin Pathol. 1995;104:537–546. doi: 10.1093/ajcp/104.5.537. [DOI] [PubMed] [Google Scholar]
- 13.Kessler H H, Pierer K, Dragon E, Lackner H, Santner B, Stunzner D, Stelzl E, Waitzl B, Marth E. Evaluation of a new assay for HBV DNA quantitation in patients with chronic hepatitis B. Clin Diagn Virol. 1998;9:37–43. doi: 10.1016/s0928-0197(97)10008-3. [DOI] [PubMed] [Google Scholar]
- 14.Krajden M, Minor J, Cork L, Comanor L. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J Viral Hepatol. 1998;5:415–422. doi: 10.1046/j.1365-2893.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- 15.Lai C L, Chein R N, Leung N W Y, Chang T T, Guan R, Tai D I, Ng K Y, Wu P C, Dent J C, Barber J, Stephenson S L, Gray D F. A one-year trial of lamivudine for chronic hepatitis B. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 16.Lai V C H, Guan R, Wood M L, Lo S K, Yuen M F, Lai C L. Nucleic acid-based cross-linking assay for detection and quantification of hepatitis B virus DNA. J Clin Microbiol. 1999;37:161–164. doi: 10.1128/jcm.37.1.161-164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lok A S, Lai C L, Wu P C, Leung E K, Lam T S. Spontaneous hepatitis B e antigen to antibody seroconversion and reversion in Chinese patients with chronic hepatitis B virus infection. Gastroenterology. 1987;92:1839–1843. doi: 10.1016/0016-5085(87)90613-5. [DOI] [PubMed] [Google Scholar]
- 18.Pawlotsky J, Bastie A, Lonjon I, Remire J, Darthuy F, Soussy C, Dhumeaux D. What technique should be used for routine detection and quantification of HBV DNA in clinical samples? J Virol Methods. 1997;65:245–253. doi: 10.1016/s0166-0934(97)02196-4. [DOI] [PubMed] [Google Scholar]
- 19.Perillo R P, Schiff E R, Davis G L, Bodenheimer H C, Jr, Lindsay K, Payne J, Dienstag J L, O'Brien C, Tamburro C, Jacobson I M, et al. A randomized, controlled trial of interferon alfa-2b alone and after prednisone withdrawal for the treatment of chronic hepatitis B. N Engl J Med. 1990;323:295–301. doi: 10.1056/NEJM199008023230503. [DOI] [PubMed] [Google Scholar]
- 20.Quint W G V, Heijtink R A, Schirm J, Gerlich W H, Niesters H G M. Reliability of methods for hepatitis B virus detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranki M, Schatzl H M, Zachoval R, Uusi-Oukari M, Lehtovaara P. Quantification of hepatitis B virus DNA over a wide range from serum for studying viral replicative activity in response to treatment and in recurrent infection. Hepatology. 1995;21:1492–1499. [PubMed] [Google Scholar]
- 22.Wong D K H, Cheung A M, O'Rourke K, Naylor C D, Detsky A S, Heathcote J. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann Intern Med. 1993;119:312–323. doi: 10.7326/0003-4819-119-4-199308150-00011. [DOI] [PubMed] [Google Scholar]
- 23.Wood M, Albagli D, Cheng P, Huan B, Van Atta R. Nucleic acid cross-linking probes for DNA/RNA diagnostics. Clin Chem. 1996;42:S196. [Google Scholar]
- 24.Zaaijer H L, ter Borg F, Cuypers H T M, Hermus M C A H, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]