Abstract
Bacteroides spp. are opportunist pathogens that cause blood and soft tissue infections and are often resistant to antimicrobial agents. We have developed a combined PCR-restriction fragment length polymorphism (RFLP) technique to characterize the 16S rRNA gene for identification purposes and the nitroimidazole resistance (nim) gene for detection of resistance to the major antimicrobial agent used to treat Bacteroides infections: metronidazole (MTZ). PCR-RFLP analysis of 16S ribosomal (rDNA) with HpaII and TaqI produced profiles that enabled discrimination of type strains and identification of 70 test strains to the species level. The 16S rDNA PCR-RFLP identification results agreed with routine phenotypic testing for 62 of the strains. The discrepancies between phenotypic and PCR-RFLP methods for eight strains were resolved by 16S rDNA sequencing in three cases, but five strains remain unidentified. The presence of nim genes was indicated by PCR in 25 of 28 strains that exhibited reduced sensitivity to MTZ. PCR-RFLP of the nim gene products identified the four reported genes (nimA, -B, -C, and -D) and indicated the presence of a previously unreported nim gene in 5 strains. This novel nim gene exhibited 75% DNA sequence similarity with nimB. These rapid, accurate, and inexpensive methods should enable improved identification of Bacteroides spp. and the detection of MTZ resistance determinants.
The genus Bacteroides sensu stricto has undergone dynamic taxonomic changes in recent years and, at present, contains 12 species of anaerobic, non-spore-forming, gram-negative bacilli formerly termed the Bacteroides fragilis group (20). Bacteroides spp. form part of the endogenous human gastrointestinal microflora and are opportunist pathogens that cause infection in a range of sites (3, 7). They are the most common anaerobic isolates from blood, intra-abdominal infections, perirectal abscesses, and soft tissue infections (8). Conventional methods that have been used to identify Bacteroides spp. are based upon phenotypic characteristics, including carbohydrate fermentation and other biochemical tests (6, 12, 20). However, phenotypic methods have an inherent risk of misidentification because of (i) the variable nature of some biochemical reactions and their dependence upon environmental conditions, (ii) the need for subjective interpretation of results, (iii) the fact that some related species differ by only one phenotypic reaction, and (iv) the occurrence of “intermediate” organisms with characteristics inconsistent with recognized species.
In the United Kingdom, the 5-nitroimidazole drug, metronidazole (MTZ), has been the antibiotic of choice for prophylaxis and therapy of infections caused by Bacteroides spp. and other anaerobes for nearly three decades. Reports of resistance to MTZ (1, 2, 15, 22) are of potential importance because Bacteroides spp. are often resistant to other clinically relevant antibiotics, including tetracycline, clindamycin, and β-lactams (9). They are also worrisome because many laboratories in the United Kingdom still rely upon MTZ sensitivity to identify the presence of anaerobes in primary cultures from clinical specimens (4), resulting in the possibility that MTZ resistance in anaerobes has been under-reported.
Despite the global use of MTZ, studies on resistance have been limited, and most research has been done in France (1, 2, 5, 10, 17, 18, 22). Although a number of resistance mechanisms have been suggested (for a review, see reference (17), some MTZ-resistant strains have been shown to possess specific nitroimidazole resistance genes (nimA to nimD) (10, 17, 18). Carlier et al. (5) have suggested that these genes encode a nitroimidazole reductase that converts 4- or 5-nitroimidazole to 4- or 5-aminoimidazole, thus avoiding the formation of toxic nitroso radicals that are essential for antimicrobial activity. The four reported nim genes are carried on plasmids (nimA, nimC, and nimD) or the chromosome (nimB), but despite the transferable nature of the plasmids (1), epidemiological studies have found that 75% of resistant isolates possess chromosomally encoded resistance mechanisms (2).
PCR amplification, followed by restriction digest analysis, is a simple technique that has been applied to species identification and could be used to analyze nim genes. Restriction fragment length polymorphism analysis of amplified small subunit rRNA gene (16S rDNA PCR-RFLP) has been shown to be a rapid, accurate, and effective method for the identification of clinically important anaerobes, including clostridia (23) and actinomycetes (11). The aim of the present study was to develop a PCR-RFLP technique for the reliable identification of Bacteroides spp. and for the characterization of nim genes present in MTZ-resistant isolates.
MATERIALS AND METHODS
Bacterial culture and identification of test strains.
Bacteroides strains were cultured on Fastidious Anaerobe Agar (FAA; Lab M, Bury, United Kingdom) supplemented with 5% (vol/vol) horse blood in an anaerobic atmosphere (10% [vol/vol] CO2 and 10% [vol/vol] H2 in N2) at 36°C. A total of 74 well-characterised strains, including the type strain of each species, were analyzed: Bacteroides caccae (3 strains), Bacteroides distasonis (8 strains), Bacteroides fragilis (29 strains), Bacteroides merdae (2 strains), Bacteroides ovatus (3 strains), Bacteroides splanchnicus (2 strains), Bacteroides stercoris (2 strains), Bacteroides uniformis (5 strains), Bacteroides variabilis (1 strain), Bacteroides vulgatus (3 strains), Bacteroides eggerthii (1 strain), and Bacteroides thetaiotaomicron (15 strains). A further eight clinical strains that gave ambiguous results in phenotypic tests have been analyzed. Strains were obtained from the National Collection of Type Cultures (NCTC), the American Type Culture Collection (ATCC), and the collection of the Anaerobe Reference Unit (ARU), University Hospital of Wales, Cardiff. All strains were identified by conventional biochemical analysis according to methods documented previously (12, 16).
Of the 82 strains analyzed, 28 exhibited no zone, or a reduced zone, of susceptibility to a 5-μg MTZ disk. Quantitative estimates of MTZ resistance for these 28 strains and 9 fully sensitive strains were determined with E-test strips according to the manufacturer's instructions (AB Biodisk, Solna, Sweden) on FAA.
16S rRNA and nim gene PCR.
Crude template nucleic acid was prepared with GeneReleaser (Cambio, Cambridge, United Kingdom). Briefly, a single colony harvested after culture for 18 h on FAA was resuspended in 20 μl of TE buffer (10 mM Tris; 1 mM EDTA, pH 8.0). GeneReleaser (20 μl) was added, vortex mixed for 20 s, and treated for 6 min at full power in a microwave oven (750 W) before centrifugation (12,000 × g for 2 min). Template nucleic acid in the supernatant was stored at −20°C.
16S rRNA genes were amplified with the universal primers (13), pA (5′-AGAGTTTGATCCTGGCTCAG; positions 8 to 27, Escherichia coli numbering) and pH (5′-AAGGAGGTGATCCAGCCGCA; positions 1540 to 1520). Template nucleic acid (5 μl) was included in a 50-μl PCR reaction mixture (10 mM Tris-HCl, pH 8.3; 1.5 mM MgCl2; 50 mM KCl; gelatin, 0.01%; 200 μM concentrations of each deoxynucleoside triphosphate; 0.2 μM concentrations of each primer; and 1U of Taq DNA polymerase [Promega, Madison, Wis.]). Reaction mixtures were denatured for 3 min at 95°C and subjected to 30 cycles of denaturation at 95°C for 45 s, annealing at 55°C for 1 min, and polymerization at 72°C for 90 s.
The presence of nim genes in 28 strains that exhibited reduced sensitivity or resistance to MTZ and in 9 MTZ-sensitive strains was assessed by PCR with primers NIM-3 and NIM-5 according to methods validated previously (22). Positive control strains containing nim genes included B. fragilis BF8 (nimB), B. fragilis 638R containing plasmid pIP417 (nimA), B. fragilis 638R containing plasmid pIP419 (nimC), and B. fragilis 638R containing plasmid pIP421 (nimD) (9, 17, 20). B. fragilis NCTC 11295 was included as a nim gene-negative control that is resistant to MTZ.
PCR products were resolved by agarose (1.5%) gel electrophoresis with a molecular weight standard (100 bp; Advanced Biotechnologies, Epsom, United Kingdom), stained with ethidium bromide (0.5 μg ml−1), and visualized with UV light.
RFLP analysis.
Amplification products from 16S rDNA PCR (n = 82) and nim gene PCR (n = 25) were treated with the restriction endonucleases HpaII and TaqI, according to the manufacturer's instructions (Promega). Digestion products were resolved in Metaphor agarose (3% [wt/vol]; FMC Bioproducts) at 100 V in TAE (40 mM Tris-acetate buffer; 1 mM EDTA, pH 8.0) for 2.5 h and visualized with UV light after staining for 20 min with ethidium bromide (0.5 μg ml−1). To enable normalization of RFLP patterns a molecular weight standard (100 bp; Advanced Biotechnologies) was run at five-lane intervals. Restriction profiles were analyzed with GelCompar (Applied Maths, Kortrijk, Belgium), and dendrograms were produced with the hierarchic cluster comparison algorithm UPGMA (unweighted-pair-group method using arithmetic averages) with fine alignment (21).
Discrepancies in RFLP profiles were investigated by sequencing the 16S rRNA or nim gene PCR products. Briefly, amplification products were cleaned with QIAquick spin PCR clean-up columns (Qiagen, Ltd., Crawley, West Sussex, United Kingdom) and sequenced using the ABI-PRISM Dye Terminator Cycle Sequencing kit (Perkin-Elmer, Warrington, United Kingdom). Sequences were compared to those in the EMBL database with BLAST N and analyzed further with DNASIS (Hitachi Software, Yokohama, Japan).
Nucleotide accession numbers.
The novel nim gene DNA sequence from B. fragilis ARU 6881 has been assigned the EMBL accession no. AJ244018.
RESULTS
Routine phenotypic identification.
All strains were nonmotile, anaerobic, gram-negative bacilli. They were also strongly saccharolytic and bile resistant and produced succinic acid as a major end product of fermentation. Tests that were useful in the differentiation of type strains and clinical strains are shown in Table 1. The type strains of B. merdae and B. distasonis could not be distinguished reliably by routine phenotypic methods because larch arabinogalactan is no longer available (12, 16). Of the 70 clinical strains analyzed, 42 (60%) were identified unequivocally, 20 (28%) gave weak reactions for critical tests and were only identified on repeat testing, and 8 (11%; ARU 9523, 12484, 12592, 11946, 11441, 11476, 12719, and 13188) could not be assigned reliably to a recognized species.
TABLE 1.
Distinguishing phenotypic characteristics of Bacteroides spp.a
| Species | No. of strains tested | Resultb with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Indole | α-Fucosidase | Arabinose | Glycogen | Melibiose | Rhamnose | Salicin | Sucrose | Trehalose | Xylan | ||
| B. caccae | 3 | − | + | + | + | + | v | − | + | + | − |
| B. distasonis | 8 | − | − | v | + | + | + | + | + | + | − |
| B. eggerthii | 1 | + | − | + | + | − | + | − | − | − | + |
| B. fragilis | 29 | − | + | − | + | + | − | − | + | − | − |
| B. merdae | 2 | − | − | − | + | + | + | + | + | + | − |
| B. ovatus | 3 | + | + | + | + | + | + | + | + | + | + |
| B. stercoris | 2 | + | + | − | + | − | + | − | + | − | − |
| B. splanchnicus | 2 | + | + | v | − | v | − | − | − | − | − |
| B. thetaiotaomicron | 15 | + | + | + | + | + | + | v | + | + | − |
| B. uniformis | 5 | + | + | + | + | + | v | + | + | − | − |
| B. variabilis | 1 | + | + | + | + | + | + | + | + | − | + |
| B. vulgatus | 3 | − | + | + | + | + | + | − | + | − | v |
16S rDNA PCR-RFLP identification.
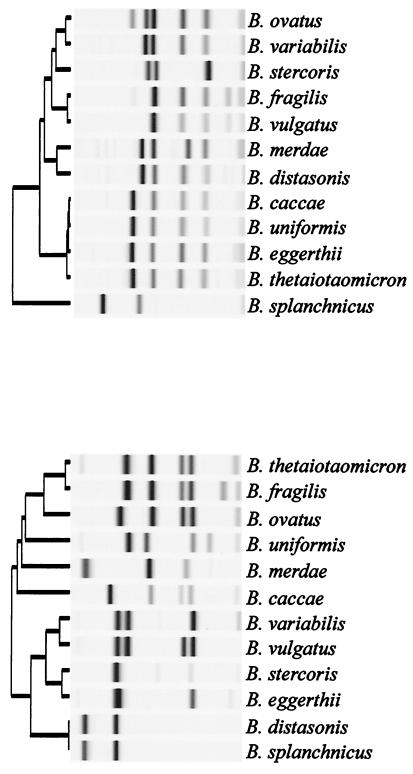
UPGMA dendrograms of the 16S rDNA PCR restriction profiles obtained for type strains of Bacteroides spp. with HpaII and TaqI are shown in Fig. 1 (top and bottom panels, respectively). HpaII profiles enabled the 12 type strains to be differentiated into 8 groups, while TaqI differentiated them into 11 groups. All type strains could be separated using the two-enzyme strategy, and comparison with the reference profiles permitted unequivocal identification of 65 (93%) of the 70 clinical strains. Three strains which were not identified phenotypically (ARU 9523, 12484, and 12592) were identified as B. ovatus by 16S rDNA PCR-RFLP; 16S rDNA sequencing also indicated 100% similarity with the 16S rDNA gene of the B. ovatus type strain. The five strains unidentified by 16S rDNA PCR-RFLP or phenotypic methods (ARU 11946, 11441, 11476, 12719, and 13188) yielded identical RFLP profiles. These strains produce profiles identical to those of B. thetaiotaomicron strains with HpaII but also identical to those of B. ovatus strains with TaqI. Comparison of 16S rDNA sequences from strains ARU 11476 and 11946 with sequences for the type strains and the data in EMBL resulted in no definitive match. The closest similarity (97%) was to the 16S rRNA gene of B. ovatus NCTC 11153T. These five strains may represent a previously unrecognized species.
FIG. 1.
UPGMA dendrograms of HpaII (top) and TaqI (bottom) 16S rDNA PCR-RFLP profiles obtained with the 12 types strains of Bacteroides spp.
The total unit consumable cost of PCR-RFLP identification has been estimated to be £2.18 ($3.50 based on batch analysis of 10 strains). This does not take into account any capital equipment costs, however.
Nitroimidazole resistance.
The control strains containing the four nim genes (10, 18, 22) all gave visible PCR product (Table 2; Fig. 2), whereas strain NCTC 11295 did not. Of the 37 clinical strains analyzed, 28 gave an MTZ MIC of ≥3 mg/liter, and 25 of these produced a PCR product with primers NIM-3 and NIM-5 (Table 2). No PCR product was found in nine MTZ-sensitive strains with MIC of ≤0.38 mg/liter. Three MTZ-resistant strains also failed to yield a nim gene PCR product.
TABLE 2.
MIC of MTZ and type of nim gene present for 42 strains of Bacteroides
| Species | Strain | MTZ MIC (mg/liter) | nim gene |
|---|---|---|---|
| B. fragilis | NCTC 11295a | >32 | |
| B. fragilis | Bf-8a | >32 | B |
| B. fragilis | 638R(pIP417)a | 24 | A |
| B. fragilis | 638R(pIP419)a | 6 | C |
| B. fragilis | 638R(pIP421)a | 16 | D |
| B. fragilis | ARU 1970 | >32 | A |
| B. fragilis | ARU 2592 | 6 | |
| B. distasonis | ARU 3448 | 0.25 | |
| B. distasonis | ARU 3523 | >32 | A |
| B. thetaiotaomicron | ARU 3690 | >32 | E |
| B. fragilis | ARU 4096 | >32 | D |
| B. fragilis | ARU 4408 | 8 | C |
| B. fragilis | ARU 5589 | 24 | |
| B. fragilis | ARU 6381 | >32 | A |
| B. fragilis | ARU 6881 | >32 | E |
| B. fragilis | ARU 7420 | 24 | B |
| B. distasonis | ARU 8259 | >32 | C |
| B. fragilis | ARU 9276 | >32 | A |
| B. fragilis | ARU 9750 | >32 | B |
| B. fragilis | ARU 10177 | 24 | A |
| B. thetaiotaomicron | ARU 10769 | >32 | E |
| B. fragilis | ARU 10887 | 8 | A |
| B. fragilis | ARU 11477 | 6 | C |
| B. fragilis | ARU 11562 | >32 | B |
| B. fragilis | ARU 11563 | >32 | E |
| B. ovatus | ARU 11564 | >32 | E |
| B. fragilis | ARU 11696 | >32 | A |
| B. fragilis | ARU 11721 | >32 | A |
| B. distasonis | ARU 11849 | 6 | A |
| B. thetaiotaomicron | ARU 11850 | 3 | A |
| B. thetaiotaomicron | ARU 12371 | >32 | B |
| B. vulgatus | ARU 12487 | 0.19 | |
| B. thetaiotaomicron | ARU 12496 | 0.38 | |
| B. distasonis | ARU 12593 | >32 | C |
| B. fragilis | ARU 12605 | >32 | |
| B. distasonis | ARU 12641 | 0.25 | |
| B. thetaiotaomicron | ARU 12719 | 0.38 | |
| B. fragilis | ARU 12902 | 0.19 | |
| B. vulgatus | ARU 12912 | 0.19 | |
| B. fragilis | ARU 12947 | 0.25 | |
| B. fragilis | ARU 12963 | >32 | B |
| B. fragilis | ARU 12984 | 0.25 |
Control strains (22).
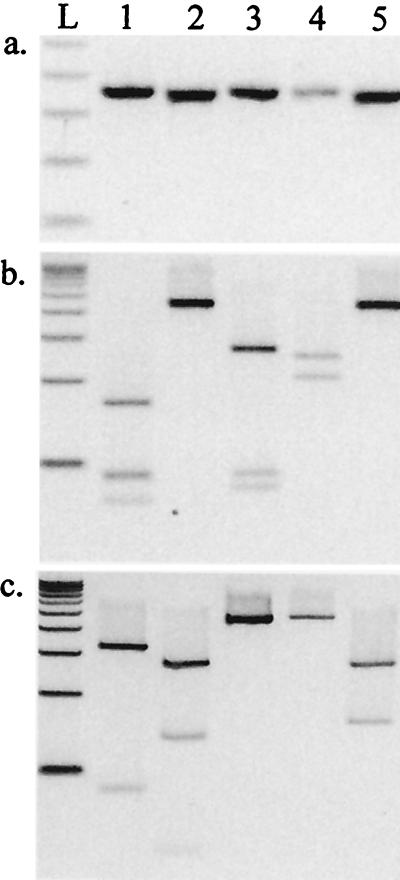
FIG. 2.
PCR products (a) and RFLP profiles for HpaII (b) and TaqI (c) obtained with genes nimA to nimD and a novel nim gene from strain ARU 6881 (lanes 1 to 5, respectively). L, 100-bp ladder.
The four different nim gene PCR products from control strains produced unique digestion profiles with HpaII and TaqI (Fig. 2). PCR products from nim genes in 20 clinical strains were identified by comparison of digestion patterns with those from the four nim genes from control strains (Table 2).
Five clinical strains (B. thetaiotaomicron ARU 3690 and ARU 10769, B. fragilis ARU 6881 and ARU 11563, and B. ovatus ARU 11564) that gave MTZ MIC values of ≥32 mg/liter had HpaII PCR-RFLP profiles consistent with nimB, but the TaqI PCR-RFLP profiles were inconsistent with reported nim genes (Table 2 and Fig. 2). Direct sequencing of nim gene PCR products indicated that these strains contained a novel gene, which is most similar to nimB but which exhibited only 75% DNA sequence similarity. The predicted amino acid sequence exhibited 82% identity and 92% similarity with nimB.
DISCUSSION
Bacteroides spp. are isolated frequently from human clinical material, but routine phenotypic identification can be laborious and is often unsuccessful with commercial kits. The variable biochemistry of strains in a species (Table 1) (12) and the dependence of cellular physiological responses upon the precise media and environmental conditions offers considerable opportunity for inaccurate identification and poor reproducibility. In the present study it was evident, after repetition, that initial phenotypic identification was accurate only 72% of the time. The 16S rDNA PCR-RFLP approach offered an alternative to conventional methods, permitting accurate grouping of strains and identification of Bacteroides strains to the species level. In addition, the present method produced results similar to those of direct 16S rDNA sequencing (where applied) for the identification of Bacteroides spp. and is comparatively less expensive. However, as the cost of direct sequencing falls and access to autosequencers increases, sequencing may eventually prove to be the method of choice for identification.
In addition to the inaccuracies and inconsistencies of phenotypic identification, any potential new species may also be overlooked because of the inherent variation tolerated by identification schemes (Table 1). Five atypical strains were, in initial phenotypic studies, assigned to known species. However, these strains could not be identified by 16S rDNA PCR-RFLP, and 16S rDNA sequencing has indicated that they may be members of a new species.
The emergence of MTZ resistance in Bacteroides spp. in France (1, 2) and a case of treatment failure in Kuwait have been reported recently (19). However, there is a lack of susceptibility data regarding MTZ resistance in Bacteroides spp. elsewhere in the world. In clinical microbiology laboratories in the United Kingdom, it is common practice to assume that only colony morphotypes for which growth is inhibited near a 5-μg MTZ disk on solid media are obligate anaerobes (4). This practice overlooks the possibility of MTZ resistance in anaerobic organisms and may result in an underestimation of anaerobic infection and MTZ resistance because colonies growing within the “zone of susceptibility” are presumed to be facultative organisms. This procedure may not present a problem with non-Bacteroides gram-negative anaerobic bacilli because MTZ resistance has been reported to be very rare in this group (14). However, the clinical staff should be fully aware of the potential for Bacteroides spp. to be resistant to MTZ and other antimicrobial agents (1, 2, 9, 17). The present study has shown that 20 of 22 Bacteroides strains that were clinically resistant to MTZ (NCCLS criteria indicate that an MIC of >16 mg/liter is clinically significant) possessed nim genes. A further five of six strains that exhibited decreased sensitivity to MTZ with an MIC of between 3 and 16 mg/liter produced nim gene PCR products. No PCR products were obtained with MTZ-sensitive strains (MIC, <1 mg/liter).
Resistance to 5-nitroimidazoles has not been reported to be a problem in the United Kingdom, but 23 of the 28 MTZ-resistant strains analyzed in the present study were referred to the ARU for identification by hospitals in the United Kingdom. The fact that the current study has analyzed referred strains precludes the possibility of epidemiological analysis because the data may be heavily biased. However, increasing referrals of MTZ-resistant Bacteroides spp. as a percentage of referrals to the ARU over the past 5 years presents a worrisome trend (1995, 1.9%; 1996, 4%; 1997, 3.8%; 1998, 7.5%; first 6 months of 1999, 14%).
RFLP analysis of nim gene PCR products permitted identification of all four reported nim genes (17). Clinical strains possessing plasmid encoded genes (nimA, nimC, and nimD) tended to exhibit lower MICs against MTZ than those with nimB (Table 2), confirming the observation of Trinh and Reysset (22). However, no association between particular nim genes and individual species was evident, suggesting that transfer of these genes between species may be common. A novel nim gene (deposited in EMBL as nimE) was present in five MTZ-resistant (MIC, >32 mg/liter) strains identified as B. fragilis (n = 2), B. thetaiotaomicron (n = 2), and B. ovatus (n = 1). Minor variations in published nim gene sequences have been reported recently (15). However, the novel gene found in the present study exhibited only 75% DNA sequence similarity with the closest nim gene (nimB). The origin and characteristics of this novel nitroimidazole resistance determinant are the subject of further study in our laboratory.
Nitroimidazole resistance (nim) genes were not detected in three MTZ-resistant B. fragilis strains (MIC range, 6 to >32 mg/liter). It is possible that an alternative mechanism such as that of NCTC strain 11295 is involved in the resistance of these strains (17) or that additional nim genes may exist that were not recognized by the PCR primers used (22).
The present study has described a combined PCR-RFLP technique using two restriction enzymes to characterize two different PCR products. It is hoped that this accurate, reliable, and relatively inexpensive method will permit improved identification of Bacteroides spp. and facilitate the recognition and epidemiology of nitroimidazole resistance determinants.
ACKNOWLEDGMENTS
We thank G. Reysset (Institut Pasteur, Paris, France) and H. N. Shah (CPHL, Colindale, London, United Kingdom) for kindly supplying a number of bacterial strains.
REFERENCES
- 1.Breuil J, Dublanchet A, Truffaut N, Sebald M. Transferable resistance in the Bacteroides fragilis group. Plasmid. 1989;21:151–154. doi: 10.1016/0147-619x(89)90060-7. [DOI] [PubMed] [Google Scholar]
- 2.Breuil J, Burnat C, Patey O, Dublanchet A. Survey of Bacteroides fragilis susceptibility patterns in France. J Antimicrob Chemother. 1989;24:69–75. doi: 10.1093/jac/24.1.69. [DOI] [PubMed] [Google Scholar]
- 3.Brook I. Bacteroides infections in children. J Med Microbiol. 1995;43:92–98. doi: 10.1099/00222615-43-2-92. [DOI] [PubMed] [Google Scholar]
- 4.Bruce D, Bradley P. Metronidazole discs on anaerobic neomycin blood agar plates. An aid to diagnosis of anaerobic infections. Med Lab Sci. 1977;34:273–275. [PubMed] [Google Scholar]
- 5.Carlier J P, Sellier N, Rager M N, Reysset G. Metabolism of a 5-nitroimidazole in susceptible and resistant isogenic strains of Bacteroides fragilis. Antimicrob Agents Chemother. 1997;41:1495–1499. doi: 10.1128/aac.41.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerden B I. The isolation and identification of Bacteroides spp. from the normal human faecal flora. J Med Microbiol. 1980;13:69–78. doi: 10.1099/00222615-13-1-69. [DOI] [PubMed] [Google Scholar]
- 7.Finegold S M. Anaerobic bacteria in human disease. New York, N.Y: Academic Press, Inc.; 1977. [Google Scholar]
- 8.Finegold S M. General aspects of anaerobic infections. In: Finegold S M, George W L, editors. Anaerobic infections in humans. San Diego, Calif: Academic Press, Inc.; 1989. pp. 137–153. [Google Scholar]
- 9.García-Rodrígeuz J A, García-Sánchez J E, Muñoz-Bellido J L. Antimicrobial resistance of anaerobic bacteria: current status. Anaerobe. 1995;1:69–80. doi: 10.1006/anae.1995.1001. [DOI] [PubMed] [Google Scholar]
- 10.Haggoud A, Reysset G, Sebald M. Cloning of a Bacteroides fragilis chromosomal determinant coding for 5-nitroimidazole resistance. FEMS Microbiol Lett. 1992;74:1–5. doi: 10.1016/0378-1097(92)90728-7. [DOI] [PubMed] [Google Scholar]
- 11.Hall V, O'Neill G L, Magee J T, Duerden B I. Development of amplified 16S ribosomal DNA restriction analysis for identification of Actinomyces species and comparison with pyrolysis-mass spectrometry and conventional biochemical tests. J Clin Microbiol. 1999;37:2255–2261. doi: 10.1128/jcm.37.7.2255-2261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holdeman L V, Cato E P, Moore W E C, editors. Anaerobe laboratory manual. 4th ed. (and updates) Blacksburg, Va: Virginia Polytechnic Institute; 1977. [Google Scholar]
- 13.Hutson R A, Thompson D E, Collins M D. Genetic interrelationships of assacharolytic Clostridium botulinum types B, E and F and related clostridia as revealed by small-subunit rRNA sequencing. FEMS Microbiol Lett. 1993;108:103–110. doi: 10.1111/j.1574-6968.1993.tb06081.x. [DOI] [PubMed] [Google Scholar]
- 14.King A, Downes J, Nord C-E, Phillips I. Antimicrobial susceptibility of non-Bacteroides fragilis group anaerobic gram-negative bacilli in Europe. Clin Microbiol Infect. 1999;5:404–416. doi: 10.1111/j.1469-0691.1999.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 15.Lubbe M M, Stanley K, Chalkley L J. Prevalence of nim genes in anaerobic/facultative anaerobic bacteria isolated in South Africa. FEMS Microbiol Lett. 1999;172:79–83. doi: 10.1111/j.1574-6968.1999.tb13453.x. [DOI] [PubMed] [Google Scholar]
- 16.Phillips K D. A simple and sensitive technique for determining the fermentation reactions of non-sporing anaerobes. J Appl Bacteriol. 1976;41:325–328. doi: 10.1111/j.1365-2672.1976.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 17.Reysset G. Genetics of 5-nitroimidazole resistance in Bacteroides species. Anaerobe. 1996;2:59–69. doi: 10.1006/anae.1996.0008. [DOI] [PubMed] [Google Scholar]
- 18.Reysset G, Haggoud A, Su W J, Sebald M. Genetic and molecular analysis of pIP417 and pIP419: Bacteroides plasmids encoding 5-nitroimidazole resistance. Plasmid. 1992;27:181–90. doi: 10.1016/0147-619x(92)90020-b. [DOI] [PubMed] [Google Scholar]
- 19.Rotimi V O, Khoursheed M, Brazier J S, Jamal W Y. Bacteroides species highly resistant to metronidazole: an emerging problem. Clin Microbiol Infect. 1999;5:166–169. doi: 10.1111/j.1469-0691.1999.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 20.Shah H N, Collins M D. Proposal to restrict the genus Bacteroides (Castellani and Chalmers) to Bacteroides fragilis and closely related species. Int J Syst Bacteriol. 1989;39:85–87. [Google Scholar]
- 21.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 22.Trinh S, Reysset G. Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J Clin Microbiol. 1996;34:2078–2084. doi: 10.1128/jcm.34.9.2078-2084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaneechoutte M, Cartwright C P, Williams E C, Jäger B, Tichy H V, De Baere T, De Rouke A, Verschraegen G. Evaluation of 16S rRNA gene restriction analysis for the identification of cultured organisms of clinically important Clostridium species. Anaerobe. 1996;2:249–256. [Google Scholar]