Abstract
Introduction
Understanding the different transmission routes of SARS-CoV-2 is crucial in planning effective interventions in healthcare institutions. This study aimed to evaluate the presence of SARS-Cov-2 genome on inanimate surfaces in COVID-19 intensive care unit and emergency care cohorts.
Methods
This is a prospective cross-sectional study. Samples of the environmental surface of objects and furniture were collected between July 15 and October 15, 2020, at COVID-19 intensive and emergency care units. The presence of SARS-CoV-2 genome was determined by quantitative RT-qPCR. The positivity rate for SARS-Cov-2 genome is presented as the arithmetic mean of the sum of the values obtained in each collection. Values of 1.0, 0.5, and 0.0 were assigned for positive, indeterminate, and negative events, respectively.
Results
In the intensive care unit, 86% of samples collected at the stethoscope and bed rail surfaces were positive. In the emergency care unit, 43% of bathroom tap, bed rails, and bedside table samples were positive. SARS-CoV-2 genome was not detected at the computer mouse and keyboard. At the emergency care unit, 14.3% of the samples from the collection room armchair were positive.
Conclusions
SARS-CoV-2 genome can be found at the environmental surface of objects and furniture at COVID-19 care units. They can represent a potential source of indirect transmission pathway for COVID-19, especially within health service institutions.
Keywords: Surface contamination, SARS-CoV-2 patients, Environment samples, Molecular epidemiology, Public health
Introduction
In early 2020, the world went on alert due to an outbreak of pneumonia caused by a new coronavirus whose first case was reported in December 2019, in the city of Wuhan, capital of Hubei province, China [1]. The rapid increase in the number of cases was recognized as a major outbreak, leading the World Health Organization (WHO) to declare the situation as a public health emergency of international interest in late January 2020 [1].
The first coronavirus family member (infectious bronchitisvirus) was isolated in 1937, with subsequent viral isolations in rodents, domestic animals, and humans. In 1965, it was described as coronavirus due to its profile under microscopy, similar to a crown [2]. Between 2002 and 2003, WHO reported 774 deaths due to severe acute respiratory syndrome-related coronavirus (SARS-CoV) and, in 2012, 858 deaths caused by the middle east respiratory syndrome-related coronavirus (MERS-CoV) in Saudi Arabia, both complications caused by members of the coronavirus family [2]. Eight years later, in 2019–2020, a novel coronavirus, named SARS-CoV-2, was identified as the cause of deaths due to acute respiratory failure and other complications that have milder symptoms than SARS-CoV and MERS-CoV-infected patients. However the transmissibility of SARS-CoV-2 is greater, and thus, the disease cause by it, the COVID-19, had considerable impact on health systems [3–5].
In this scenario, WHO declared COVID-19 as a pandemic on March 11, 2020, and instituted effective preventative and coping measures to be adopted by the population, health institutions, in addition to national, international, and local public health authotities [1].
The suggested measures included hand hygiene, social distancing, wearing masks, breathing etiquette, self-isolation, contact precautions in health institutions and surfaces cleaning and disinfection.
Although it is not the main route of virus propagation and contamination, the persistence of SARS-CoV-2 on inanimate surfaces can lead to indirect transmission. It can be relevant in health institutions, where cross-contamination between healthcare professionals, patients, and multi-touch surfaces, such as a stethoscope, bed rail, switches, bathroom tap, computer mouse, and keyboard, can easily occur [3–8].
Experimental studies have evaluated the persistence of SARS-CoV-2 on inanimate surfaces in intensive care units or cohort units for COVID-19 using the polymerase chain reaction (PCR-detectable SARS-CoV-2) technique [5–7].They concluded that, despite the positivity common among studies of the isolated patient’s stethoscope, bathroom taps, bed rails, door handles, monitors, and light switches, the challenge was not the effectiveness of cleaning, and appropriate disinfection techniques but the direct touch contamination by either the patient or healthcare workers after contact with infected respiratory fluids [4–7]. However, the challenge and the gap were the lack of internal policies and guidelines to prevent cross-contamination and risk of contamination among health professionals on surfaces that are frequently touched [4–7]. Understanding the different transmission routes of SARS-CoV-2 is crucial in planning effective interventions in healthcare institutions. In view of this reality, contradictions, and limited data that indicate spreads of COVID-19 through indirect contact with contaminated environments, the following questions are raised: What is the possibility of environmental contamination of SARS-CoV-2 on inanimate surfaces in intensive care unit and emergency care cohort? Data regarding the hospital levels of contamination can inform hospital cleaning and disinfection protocols to reduce the risk of healthcare-associated disease. This study evaluated the presence of SARS-Cov-2 genome on inanimate surfaces in COVID-19 intensive care unit and emergency care cohorts.
Methods
Study type
Prospective cross-sectional study carried out in the ICU of a philanthropic institution and in an Emergency Care Unit (ECU) cohort for COVID-19 in the countryside of Minas Gerais.
Surface sample sites
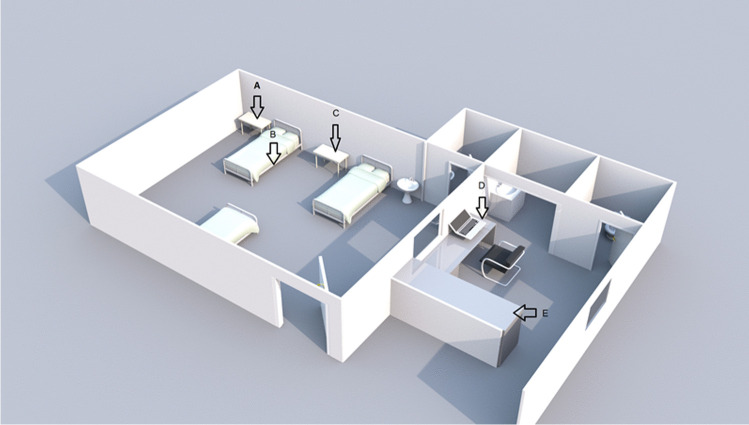
Samples were collected from equipment furniture and utensils that are frequently touched and thus considered contributing routes of propagation and indirect transmission of SARS-CoV-2 [8]. Five surfaces were selected in the ICU: bed rails, bedside table, isolated bed stethoscope, prescription room bench, and the computer mouse in the prescription room (Fig. 1). In the ECU, 9 surfaces were selected: reception chair, table and armchair in the screening room, the screening room door handle, computer keyboard, bathroom tap, stethoscope, bedside table, bed rails (Fig. 2). Only one sample was collected from the emergency service (ES): the waiting chair.
Fig. 1.
Distribution of collection sites in the ICU. A—stethoscope; B—patient bed protection rail; C—patient bedside table; D—prescription room computer mouse; E—prescription room bench
Fig. 2.
Distribution of collection sites in the ECU: A—stethoscope; B—bedside table; C—bed rails; D—bathroom tap; E—computer keyboard; F—reception chair; G—screening room door handle; H—collection table; I—collection armchair
Sample collection
Samples of objects and furniture’s environmental surfaces were collected between July 15 and October 15, 2020, at intervals of fifteen days, totaling 7 collections. The samples were collected with sterile nylon swabs pre-moistened in the virus Universal Transport Medium (UTM™). The standard size of the sampling surface area was 30 × 10 cm, where a horizontal sweep was performed, followed by vertical and diagonal sweeps. In the case of smaller surfaces, the largest possible area of the item surface was collected. All environmental swabs were obtained at 7:30 am in the morning, before the first daily disinfection of the units. For surface disinfection, quaternary ammonium compounds and polyhexamethylene biguanide (0.50%w/v) were prepared after cleaning with detergent every 12 h or if visible dirt as recommended by the institution. These agents were diluted with sterilized distilled water according to the manufacturer’s instructions.
After collection, tubes containing the swabs immersed in UTM were transported to the laboratory, packaged in a thermal box, with controlled temperature (4 to 8 °C). In the laboratory, the samples were stored at −80 °C for testing.
Viral RNA extraction and detection
The qualitative detection of viral RNA included the nucleic acid extraction (Maxwell® RSC 16 Promega), reverse transcription and amplification by real-time polymerase chain reaction (RT-qPCR; Applied Biosystems StepOne™System or StepOnePlus™ System; ThermoFisher Scientific, Waltham, USA).
To viral RNA extraction, samples were thawed and homogenized for 2 min in a vortex mixer. RNA extraction was performed according to the manufacturer instructions, using the Maxwell® System: Maxwell® RSC simply RNA Cells Kit and Maxwell® RSC Instrument (Promega, Wisconsin, USA). Briefly, 200 μL of the sample was added to 250 μL of Lysis Buffer (Lysis Buffer plus Proteinase K). RNA was eluted on 80 μL of water.
Detection of the SARS-CoV-2 RNA was performed using a real-time RT-PCR kit, according to the CDC 2019-nCoV Real-Time RT-qPCR Diagnostic Panel [9]. This panel targets the N1 and N2 genes of SARS-CoV-2. The panel also targets a human RP (Human RNase P Combined Primer/Probe Mix) constitutive gene, to monitor sample adequacy and integrity. Primers and probes set sequences are indicated in Table 1.
Table 1.
CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic Panel: primers and probes sequence
| Description | Oligonucleotide sequence | Label1 |
|---|---|---|
| N1 forward primer | GAC CCC AAA ATC AGC GAA AT | None |
| N1 reverse primer | TCT GGT TAC TGC CAG TTG AAT CTG | None |
| N1 probe | FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1 | FAM, BHQ-1 |
| N2 forward primer | TTA CAA ACA TTG GCC GCA AA | None |
| N2 reverse primer | GCG CGA CAT TCC GAA GAA | None |
| N2 probe | FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1 | FAM, BHQ-1 |
| RNAse P forward primer | AGA TTT GGA CCT GCG AGC G | None |
| RNAse P reverse primer | GAG CGG CTG TCT CCA CAA GT | None |
| RNAse P probe | FAM – TTC TGA CCT GAA GGC TCT GCG CG – BHQ-1 | FAM, BHQ-1 |
1TaqMan® probes are labeled at the 5′-end with the reporter molecule 6-carboxyfluorescein (FAM) and with the quencher, Black Hole Quencher 1 (BHQ-1) (Biosearch Technologies, Inc., Novato, CA) at the 3′-end
Briefly, for the test, a reaction mix of 20 μL final volume was prepared by adding: RNAse-free water, 3.1 μL; pimer/probe (N1, N2 or RP), 1.5 μl; GoTaq® qPCR Master Mix 2X, 10 μL; Enzyme (GO®Taq) 0.4 μL. The mix was prepared in 48- or 96-well plates. The RNA sample volume used was 5 μL. For negative controls, sample volume was substitute by RNase-free water. A pool of viral RNA from positive samples was used as positive control. Thermal cycle conditions included reverse transcription at 45 °C for 15 min, inactivation of the reverse transcriptase and activation of the DNA polymerase at 95 °C for 2 min, followed 45 cycles of denaturation (15 s at 95 °C) and annealing and extension (1 min at 60 °C).
Data analysis
Positive detection for the virus was recorded as long as the amplification of both N1 and N2 genes was observed, using the amplification cycle threshold (Ct) as a parameter. The sample was considered positive when the Ct was less than, or equal to 40 cycles, and negative when the Ct was between 40 and 45 cycles. Samples were considered indeterminate when the amplification of only one of the two SARS-CoV-2 targets (N1 or N2) was observed. The positivity rate for SARS-CoV-2 genome was presented as the arithmetic mean of the sum of the values obtained in each collection, assigning a value of 1.0 for positive events, 0.5 for indeterminate, and 0.0 for negatives. The data were analyzed descriptively, and the figures were drawn using GraphPad Prism® 8.0.
Results
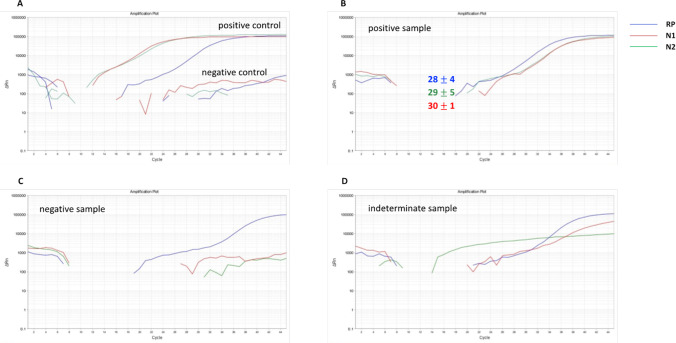
Table 2 shows the distribution of collections and the materials and equipment from the ICU and ECU that are frequently touched and were analyzed in this study. Illustrative amplification plots of positive, negative, and indeterminate samples and the mean Ct values of the positive samples are presented on Fig. 3.
Table 2.
SARS-CoV-2 genome persistence at COVID-19 in intensive care units and emergency care objects and furniture
| Objects/furniture | Sampling period | |||
|---|---|---|---|---|
| July (1 collection) | August (2 collections) | September (2 collections) | October (2 collections) | |
| ECU | ||||
| Reception chair | – | −/− | −/− | −/− |
| Door handle | + | −/− | −/− | ± |
| Collection room table | – | −/− | −/− | −/− |
| Collection room armchair | – | ?/− | −/? | ± |
| Computer keyboard | – | −/− | −/− | −/− |
| Bathroom tap | – | +/+ | −/? | −/+ |
| Bedside table | – | +/+ | ± | −/− |
| Bed rails | – | +/+ | +/− | −/− |
| Stethoscope | – | ± | −/+ | −/− |
| ICU | ||||
| Prescription bench | – | −/− | −/? | −/− |
| Prescription room mouse | – | −/− | −/− | −/− |
| Stethoscope | – | ± | +/+ | −/− |
| Bed rails | – | ± | −/+ | +/− |
| Bedside table | – | ?/− | −/− | −/− |
| ES | ||||
| Waiting chair | – | −/− | −/− | −/− |
Persistence of SARS-CoV-2 genome in each evaluation are qualitative expressed as negative (−), positive (+), or indetermined (?). EUC emergency unit cohort for Covid-19, ICU intensive care unit, ES emergency service
Fig. 3.
Illustrative amplification plots. Positive and negative controls (A) and SARS-CoV-2 genome positive (B), negative (C) and indeterminate (D) samples. Mean Ct values ± SD of positive samples are indicated on panel B for RP (blue), N1 (red), and N2 (green) genes. SD = standard deviation
Of the fifteen sites examined, the viral genome was detected in nine of them (60%). In addition, over the collection period, most sites had altered between SARS-CoV-2 genome detection and non-detection. The SARS-CoV-2 genome was not detected in any collections in ECU reception chair and collection room table, ES waiting chair, and ICU prescription room mouse.
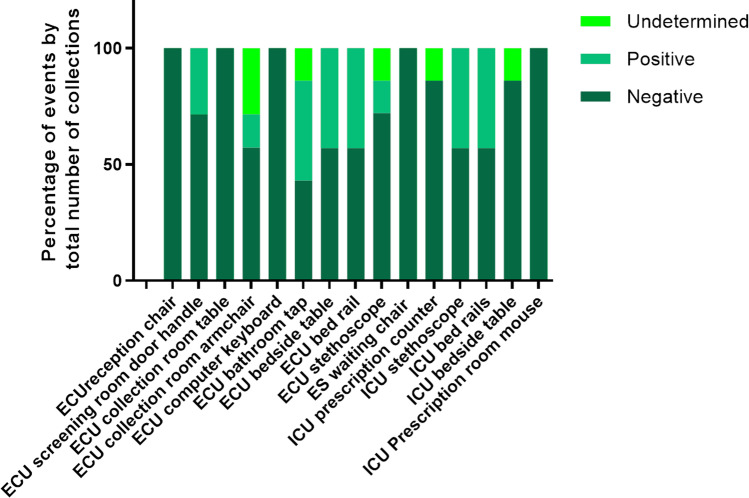
Figure 4 shows the percentage of positive, negative, and undetermined events, for SARS-CoV-2 genome detection on samples collected in the ICU and ECU. ECU bathroom tap, bedside table, bedrail and ICU stethoscope and bed rail presented the higher percentage of SARS-CoV-2 genome positivity (40%).
Fig. 4.
Percentage of positive, negative, and undetermined events, for SARS-CoV-2 virus, measured by RT-PCR, in samples of surfaces collected in the Intensive Care Units (ICU), Emergency Service (ES), and Emergency Care Unit (ECU)
For the quantitative analysis SARS-CoV-2 genome positivity in samples collected from ICU, the negative, indeterminate, and positive events were considered, per sample, in each dosage, as verified in Fig. 5. The higher positivity rate was detected in ECU bedside table.
Fig. 5.
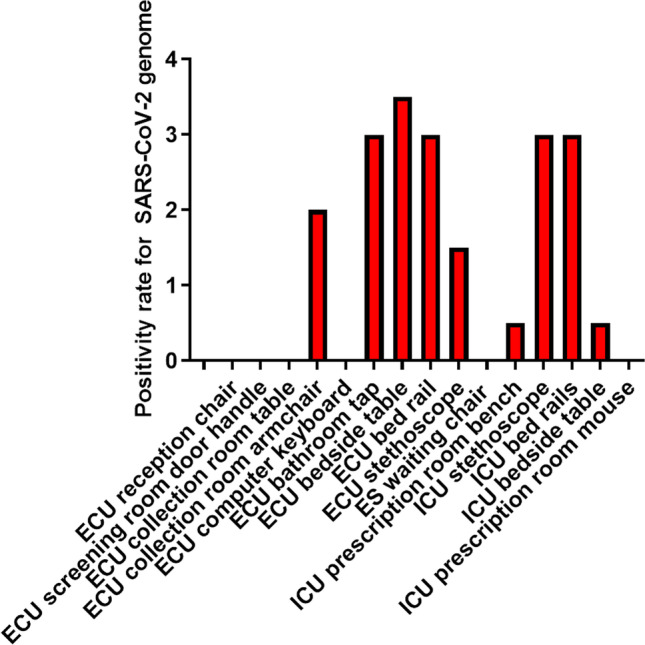
Positivity rate for SARS-CoV-2 genome. Negative, indeterminate, and positive events were considered, by sample, in each dosage. Values presented as the arithmetic mean of the sum of the values obtained in each collection, with a value of 1.0 for positive events, 0.5 for undetermined events, and 0.0 for negative ones. ICU: intensive care unit, ES: emergency service; ECU: emergency care unit
Discussion
According to the results of this study, both in the ICU and in the ECU, the stethoscope and bed rails had more than two SARS-CoV-2 genome positive samples collected at different times (Table 1). The stethoscope is used among many different health professionals involved in the patient care. It is submitted to systematic disinfection, by rubbing the equipment with the disinfectant recommended by the institution, in one direction, from the earpiece to the drum, before and after contact with patients. The bed rails, on the other hand, are a common contact surface for different health professionals. It is also a place frequently touched by the patient that is not immediately disinfected after patient contact, which increases the risk of indirectly spreading SARS-CoV-2. Phan et al. [10] presented an observational study in which the surfaces of the bed rails and the stethoscope of isolated patients also stood out as frequently touched areas, by both patients and health professionals.
Another common collection site in which three samples were positive was the ECU bathroom tap, a site considered of risk as this is not an isolated place used only by infected individuals, but also by patients suspected of COVID-19. In this case, strict isolation measures should be implemented to strengthen prevention training and infection precautions, not only during the COVID-19 outbreak but also in the daily practice of health services. Investigations of environmental contamination identified that the bed and bathroom rails are heavily contaminated by SARS-CoV-2 [10, 11], and that bathrooms were the most contaminated space by the new coronavirus. These findings are probably due to the presence of the virus in the air, on surfaces and in fecal material from infected patients. Another experimental study showed that SARS-CoV-2 can survive 3 to 4 days in the feces and urine of a COVID-19 patient [12]. These data suggest that the bathroom, as a risk area in health institutions, should be monitored regarding the hygiene of users’ hands and correct surface disinfection. In this context, several studies have proven that different types of disinfectants are active in reducing viral infectivity [10–14]. However, surface contamination is more associated with cross-contamination than with the efficacy of cleaning and disinfection in removing pathogen contamination [10–14]. In addition, in this study, samples were collected before the cleaning process. This led us to infer that the time for surfaces disinfection recommended by the institution may have been short, in face to a high frequency of cross-contamination between health professionals and frequently touched environment surfaces. It could mean that prolonged potential transmission of coronavirus might be existed via environment surface contact.
Health professional can contribute to virus dissemination by their hands, personal protection equipment (PPE) and contaminated equipment. The recognition of this potential risk is important to reduce contamination by indirect contact at health institutions and services. The bedside table and the door handle were also positive for SARS-CoV-2 genome in this study. Such data corroborate with other studies which found the SARS-CoV-2 genome in environmental surfaces in health institutions [6, 10–12]. Unlike other studies [3, 6, 12, 13] in this work, SARS-CoV-2 genome was not found on the computer mouse and keyboard. As frequently touched areas among health professionals, this equipment is generally at risk to facilitate indirect contamination [7]. The CDC and WHO recently announced that, although frequently touched surfaces are not the main route of COVID-19 transmission, indirect transmission can occur if there is no appropriate preventive care in health institutions [8, 14].
In view of the limitations and uncertainties regarding the contamination of SARS-Cov-2 on environmental surfaces, some considerations are important to better define the risk of indirect contamination. It will be important, for example, to identify viable infectious doses of SARS-CoV-2 in different types of materials [15, 16]. In this study, N1 and N2 Ct values of positive samples ranged from 18 to 35 and, in most of the samples, it was approximately double the value of the positive control. On the other hand, the mean RP Ct value of the positive samples was similar to the one of the positive controls. This suggests that although the environmental surfaces we evaluated in this study were contaminated with human genomic material, as indicated by the RP amplification, the relatively higher N1 and N2 Ct values suggest that the presence of the virus genome in surfaces frequently touched was not so abundant. However, the presence of the virus genome in the surfaces indicates it can be a potential source of contamination. As reported by Zhou et al. [3], viable virus can be recovered from nonporous surfaces inoculum with a Ct value <30. Of the surfaces sampled in this study, 5 (bed rail, bedside table, and isolated bed stethoscope at ICU and bathroom tap and bed rails at ECU) reached, at least once, a Ct value <30 for both N1 and N2 SARS-CoV-2 genes, and thus, these can be considered a source of viral cross-contamination.
Also, the standardization of the methods used to access surface contamination, to allow data comparison can greatly contribute to this field. Factors as material type, and the number of times it is touched and exposed to SARS-CoV-2, can potentially affect for how long the virus can remain viable. This knowledge can help to guide changes in the daily practice of health professionals, as data on virus viability in environmental surfaces and its quantification can affect the risk of new coronavirus transmission. As shown in this study, evidence suggests that environmental contamination by SARS-CoV-2 is probably high and is supported by recent research that focused on the transmission of COVID-19 by environmental contamination [4, 5, 10, 14, 16].
This contamination can presumably occur by indirect inhalation of droplets through the mouth, and penetration of microparticles from secretions in the eyes and nose [11, 13]. An experimental study collected samples from high-frequency contact sites by professionals and patients with COVID-19, at a hospital that admitted symptomatic and asymptomatic patients [11]. Samples collected from toilets, taps, remote controls, chairs, patient room tables, pillows, and bedding were positive for SARS-CoV-2. Also, a greater number of surface/environmental positive samples were detected in sites frequented by asymptomatic COVID-19 patients. This suggests that infection prevention care and active surveillance by health professionals over asymptomatic patients in the daily practice of the institution is low. It is also considered that, in some types of PPE, such as masks and gloves, SARS-CoV-2 can remain viable for up to seven days and promote the spread of the virus in the health institution, in addition to remaining active on surfaces made of plastics, stainless steel, glass, and ceramics for up to seven days [15].
In addition, Coil et al. [16] published the recovery of the SARS-CoV-2 genome sequences directly from environmental surface swabs by means of a phylogenetic analysis and microscopy of the new isolated genomes of the new coronavirus. The data confirmed that the virus can serve as a means of indirect propagation and contribute to the contamination of the hospital environment.
This study has some limitations. Most notably was the limited number of samples. It was not possible to implement a consistent organized monitoring protocol to test hypotheses for the routes of transmission in the specific outbreak context. Moreover, no surface samples were tested in the area for passage and lockers of medical staff, and in the dressing room. However, these findings emphasized that in a hospital environment, the contamination of inanimate surfaces can exist and, would increase the actual risk of infection, mainly due to the cross-contamination carried out by healthcare works.
Conclusion
The results of this study showed the potential persistence of SARS-CoV-2 on surfaces at health care services. It can represent a potential source for and act as an indirect transmission of COVID-19. Thus, our finding suggests that the hospitals should seek to examine multiple interventions to increase education and to ensure adequate environmental cleaning, strengthen infection prevention training, and improve infection prevention precautions among the healthcare works. Further studies can investigate the role of environmental contamination in the disease transmission via analytical epidemiological studies and the extent that indirect transmission can have.
Declarations
Ethical approval
This research does not involve human beings and it was not necessary to go through the research ethics committee.
Competing interests
The authors declare no competing interests.
Footnotes
Etel Rocha-Vieira and Danilo Bretas de Oliveira are joint senior authors.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Word Health Organization (2020) Considerations for quarantine of individuals in the context of containment for coronavirus disease (COVID-19).Geneva: WHO. Available at: https://apps.who.int/iris/handle/10665/331299
- 2.Le C, Yan Y, Wang L. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 2020;34:75–80. doi: 10.1016/j.tmrv.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J, Otter JA, Price JR, Cimpeanu C, Garcia DM, Kinross BP, Mason S, Bolt F, Holmes AH, Barklay W. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin Infect Dis. 2021;73(7):e1870–e1877. doi: 10.1093/cid/ciaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong JCC, Hapuarachchi HC, Arivalan S, Tien WP, Koo C, Mailepessov D, Kong M, Nazeem M, Lim M, Ng LC. Environmental contamination of SARS-CoV-2 in a non-healthcare setting revealed by sensitive nested RT-PCR. In J Environ Res Public Health. 2020;18(1):117. doi: 10.3390/ijerph18010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye G, Lin H, Chen S, Whang S, Zeng Z, Zhang WS, Remann T, Li Y, Pan Z, Wang ZYY, Wang F, Qian Z, Wang X. Environmental contamination of the SARS-CoV-2 in healthcare premises: an urgent call for protection for healthcare workers. J Inf Secur. 2020;81(2):e1–e5. doi: 10.1016/j.jinf.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Z, Qian H, Xu B, Huang Y, Te M, Yen HL, Xiao S, Cui L, Wu X, Shao W, Song Y, Li S, Zhou L, Xu Y, Zhu B, Li Y. Toilets dominate environmental detection of SARS-CoV-2 virus in a hospital. Sci Total Environ. 2021;753:141710. doi: 10.1016/j.scitotenv.2020.141710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, Lim XF, Lim SA, Sutjipto SS, Lee PH, Son TT, Young BE, Milton DK, Gray GC, Schuster S, Barkham T, Marimuthu K. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Interim infection prevention and control recommendations for healthcare personnel during the coronavirus disease 2019 (COVID-19) pandemic. Atlanta: CDC;2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control-recommendations.html
- 9.Centers for Disease Control and Prevention. Novel Coronavirus (2019-nCoV) Real-time RT-PCR diagnostic panel. Atlanta:CDC;2020. Available at: https://www.fda.gov/media/134922/download
- 10.Phan LT, Marita D, Mortiz DC, Bleasedale SC, Jones RM. Environmental contact and self-contact patterns of healthcare workers: implications for infection prevention and control. Clin Infect Dis. 2019;13:S178–S184. doi: 10.1093/cid/ciz558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo L, Liu D, Zhang H, Zhihao L, Zhen R, Zhang X, Huaping X, Song W, Liu J, Hang Q, Liu J, Yang X, Chen Z, Mao C. Air and surface contamination in non-healthcare settings among 641 environmental specimens of 39 COVID-19 cases. PLoS Negl Trop Dis. 2020;14:e0008570. doi: 10.1371/journal.pntd.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Li T, Deng S, Liu S, Zhang D, Li H, Wang X, Han JJ, Bei Z, Li L, Li J. Stability of SARS-CoV-2 on environmental surfaces and in human excreta. J Hosp Infect. 2021;107:105–107. doi: 10.1016/j.jhin.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y, Wang H, Chen Y, He J, Chen L, Liu Y, Hu X, Li A, Liu S, Zhang P, Zou H, Hua S. Clinical data on hospital environmental hygiene monitoring and medical staff protection during the coronavirus disease 2019 outbreak. BMJ Yale Preprint 2020; Posted March 02, 2020. 10.1101/2020.02.25.20028043
- 14.World Health Organization (2020) Transmission of SARS-CoV-2: implications for infection prevention precautions. Geneva:WHO, Available at: https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions
- 15.Van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, Wit E, Munster VJ. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coil DA, Albertson T, Banerjee S, Brennan G, Campbell AJ, Cohen SH, Dandekar S, Díaz-Muñoz SL. SARS-CoV-2 detection and genomic sequencing from hospital surface samples collected at UC Davis. PLoS One. 2021;16(6):e0253578. doi: 10.1371/journal.pone.0253578. [DOI] [PMC free article] [PubMed] [Google Scholar]