Abstract
Epstein-Barr virus (EBV) is the most common cause of infectious mononucleosis and is associated with the development of several human malignancies. A closely related herpesvirus in the same lymphocryptovirus (LCV) genera as EBV naturally infects rhesus monkeys and provides an important animal model for studying EBV pathogenesis. We cloned the small viral capsid antigen (sVCA) homologue from the rhesus LCV and developed a peptide enzyme-linked immunosorbent assay (ELISA) to determine whether epitopes in the rhesus LCV sVCA are a reliable indicator of rhesus LCV infection. In order to define a “gold standard” for rhesus LCV infection, we also cloned the EBV-encoded small RNA 1 (EBER1) and EBER2 homologues from rhesus LCV and developed a reverse transcription (RT)-PCR assay to detect persistent LCV infection in rhesus monkey peripheral blood lymphocytes. Animals from a conventional and a hand-reared colony were studied to compare the prevalence of rhesus LCV infection in the two groups. There was a 100% correlation between the peptide ELISA and EBER RT-PCR results for rhesus LCV infection. In addition, specificity for LCV infection and exclusion of potential cross-reactivity to the rhesus rhadinovirus sVCA homologue could be demonstrated using sera from experimentally infected animals. These studies establish two novel assays for reliable diagnosis of acute and persistent rhesus LCV infections. The rhesus LCV sVCA peptide ELISA provides a sensitive and reliable assay for routine screening, and these studies of the hand-reared colony confirm the feasibility of raising rhesus LCV-naive animals.
Epstein-Barr virus (EBV) is a human gammaherpesvirus in the lymphocryptovirus (LCV) genera which is associated with the development of several different malignancies, including Burkitt's lymphoma, B-cell lymphomas in immunosuppressed hosts, nasopharyngeal carcinomas, Hodgkin's disease, and gastric carcinomas (16). Closely related herpesviruses in the same LCV genera naturally infect a number of Old World primate species, and experimental infection of rhesus monkeys with the rhesus LCV is the only animal model which accurately reproduces the pathogenesis of acute and persistent EBV infections (24). Simian LCV infection is also associated with B-cell lymphomas in many Old World primate species and can cause lethal malignant disease in macaques experimentally infected and immunosuppressed with simian immunodeficiency virus (SIV) (8, 26). Thus, accurate diagnostic assays for LCV infection in Old World primates, and rhesus monkeys in particular, would be an important tool for primate care and for identifying LCV-naive animals for experimental studies.
The simian LCV and EBV genomes are colinear and appear to contain a similar repertoire of lytic and latent infection genes. The rhesus LCV latent infection genes have shown a surprising amount of sequence divergence from EBV, with only 20 to 50% amino acid identity (3, 4, 9, 10, 25, 28). The few lytic infection genes from simian LCV cloned to date have demonstrated better homology, with 50 to 90% amino acid identity (15a, 23, 46). Currently rhesus LCV diagnosis is made by detecting simian antibodies which are cross-reactive with EBV-lytic infection proteins. The cross-reactivity with EBV serologic assays can be useful for identifying animals with high antibody titers but can be difficult for excluding infection and identifying truly seronegative, naive animals. Serologic screening for herpes B virus infection has been successfully used to establish specific-pathogen-free colonies (6, 13). A simple, sensitive, and specific serologic screening assay for LCV infection would be a valuable tool for breeding LCV-naive macaque colonies. LCV-naive animals would have less risk of LCV-induced malignancies associated with experimentally induced immunosuppression, e.g., transplant and SIV experiments, and would provide a reliable source of animals for experimental infection and pathogenesis studies.
EBV infection is associated with a lifelong antibody response to lytic infection viral capsid antigens (VCA) and EBV latent infection nuclear antigens (EBNA) (27). An immunoglobulin G (IgG) antibody response to either is a reliable indicator of previous infection in humans. There are six different EBNA expressed in EBV-immortalized cells which are recognized by antibodies in EBV-immune human sera. The highest antibody titers are usually generated against EBNA-1 (12, 21), but these are still quite low compared to titers of antibody against VCA (34, 41–43). In addition, antibodies to EBNA-1 may not appear for several weeks or months after acute infection and may be low or difficult to detect in patients with immunodeficiency (12).
Three different EBV open reading frames are known to code for viral capsid proteins, BcLF1 (p135), BdRF1 (p40), and BFRF3 (p18 or p21) (31, 41, 43). Epitopes within EBV BFRF3, or small VCA (sVCA), are known to be immunodominant for the humoral response in EBV-infected humans and are routinely used in diagnostic serologic assays for EBV infection (34, 40–42). High sVCA antibody titers can be detected relatively early in acute primary EBV infection, i.e., infectious mononucleosis, and typically persist at high levels even in immunosuppressed hosts (20, 41). Therefore, we cloned the sVCA homologue from the rhesus LCV and developed a peptide enzyme-linked immunosorbent assay (ELISA) to determine whether antibody responses to the rhesus LCV sVCA are a sensitive and reliable indicator of rhesus LCV infection.
MATERIALS AND METHODS
Cell lines.
LCL 8664 is a rhesus monkey LCV (cercopithicine herpesvirus 15)- infected B-cell line derived from retro-orbital B-cell lymphoma in a rhesus monkey (26). LCL 8664 was grown in RPMI medium supplemented with 10% fetal bovine serum. COS-7 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum.
Cloning of the rhesus LCV BFRF3 and EBER homologues.
Genomic DNA from LCL 8664 cells was digested with a SalI-constructed cosmid library and screened as described previously (28). Rhesus LCV sVCA was cloned by PCR amplification from the rhesus LCV cosmid clone QA15, using EBV-specific primers (5′ GAGGTAGAATTGCCACCTGG 3′ and 5′ TTCGTGAGCCAGCTTCGCCG 3′) at reduced stringency. Multiple PCR products were cloned to derive the nucleotide sequence, and the open reading frame was cloned into pSG5 (Stratagene) for eukaryotic expression. The rhesus LCV EBV-encoded small RNA 1 (EBER1) and EBER2 homologues were identified by screening subclones of the rhesus LCV CC1 cosmid by cross-hybridization with the EBV EcoRI J DNA fragment encoding the EBERs.
Expression of rhesus LCV sVCA and Western blot analysis.
Recombinant sVCA expression constructs (30 μg) were transfected into COS-7 cells by electroporation. Twenty-four hours after transfection, cells were washed with phosphate-buffered saline (PBS) and lysed in 1× Laemmli buffer. Total cellular proteins were resolved by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis, transferred to nitrocellulose filters (Schleicher & Schuell), and immunoblotted with a 1:100 dilution of rhesus sera. A cassette Mini-protean II system (Bio-Rad) was used to screen multiple rhesus sera at one time. Filters were subsequently incubated with a goat anti-human IgG reagent coupled to horseradish peroxidase and detected with enhanced luminol and oxidizing reagents (NEN Life Science).
Peptide synthesis.
Two peptides representing the carboxy-terminal domains of rhesus LCV sVCA were synthesized by standard fluorenylmethoxycarbonyl chemistry. Peptide 1, AASAPAATPAVSSSISSLRAATSGAAASSA, corresponds to amino acids (aa) 117 to 146 of the rhesus LCV sVCA. Peptide 2, AVDTGSGGGAQPQDTSTRGARKKQ, corresponds to aa 147 to 170 of rhesus LCV sVCA. Peptides were purified to >80% purity by reverse-phase high-performance liquid chromatography.
Peptide ELISA.
Peptides 1 and 2 were resuspended in bicarbonate buffer (50 mM; pH 9.6) and used (0.5 μg in 200 μl) for overnight coating of Immulon 1 microtiter wells (Dynatech Laboratories, Inc., Chantilly, Va.) at 4°C. Unbound peptides were washed with PBS containing 0.1% Tween 20 and blocked with PBS (0.1% Tween 20, 3% bovine serum albumin [BSA]) for 2 h at room temperature. Rhesus monkey serum was diluted 1:100 in PBS (0.1% Tween 20, 3% BSA) before incubation and added to 96-well plates. After 1 h at room temperature the wells were washed and incubated with horseradish peroxidase-conjugated antiserum to human IgG diluted in PBS (0.1% Tween 20, 3% BSA). Peroxidase activity of bound conjugated antibodies was developed using O-phenylenediamine dihydrochloride tablets (Sigma Biosciences, St. Louis, Mo.). The absorbance at 450 nm was read after a 30-min incubation using a Bio-Rad microplate reader. Cutoff values were set at 3 standard deviations above the mean absorbance from triplicate wells with secondary antibody and no sera.
Real-time RT-PCR.
A two-step reverse transcription (RT)-PCR was optimized for the quantitation of rhesus LCV EBER1 using TaqMan technology (Roche Molecular Systems, Inc.) and a GeneAmp sequence detector (model 5700) during the PCR. The EBER1 forward and reverse primer sequences were 5′ GGAGGAGATGAGTGTGACTTAAATCA 3′ and 5′ TGAACCGAAGAGAGCAGAAACC 3′, and the probe sequence was labeled with a fluorescent reporter (FAM) at the 5′ end and a fluorescent quencher (TAMRA) at the 3′ end (5′ CCCGTCTTCACCACCCGGGA 3′).
Total RNA was isolated from 5 million Ficoll-Hypaque-purified peripheral blood mononuclear cells (PBMC) using TRIzol reagent (GIBCO BRL). The first step of the RT reaction was carried out using random hexamers and 400 ng of total RNA in a volume of 50 μl according to the manufacturer's protocol (PE Applied Biosystems). The PCR mixture was prepared as follows: 2× TaqMan universal PCR master mix, 10 μl of cDNA, 200 nM EBER1 probe, and a 75 nM concentration of each primer were mixed in a final volume of 50 μl and PCR amplified for 40 cycles (15 s at 94°C, 30 s at 60°C, 30 s at 72°C). Serial dilutions of a recombinant rhesus LCV EBER plasmid of known concentration were used as a PCR standard. The limit of detection for the real-time RT-PCR assay was determined to be 50 copies. Rhesus EBER1 RT-PCR products were also analyzed by electrophoresis through a 3% agarose gel. The DNA was transferred to a nylon membrane and probed with a 32P-labeled EBER1 oligonucleotide at 65°C. The blots were washed with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (with 0.5% sodium dodecyl sulfate) at 65°C three times for 15 min.
Nucleotide sequence accession numbers. The nucleotide sequences for the rhesus LCV sVCA, EBER1, and EBER2 homologues have been submitted to GenBank (accession no. AF227123, AF227124, and AF227125).
RESULTS
Cloning of rhesus LCV sVCA.
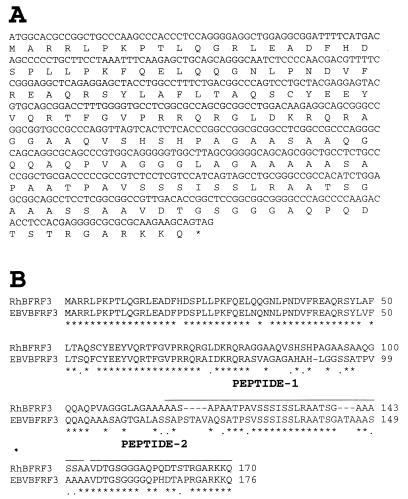
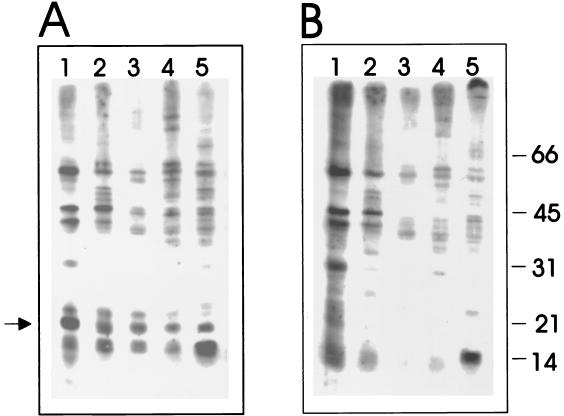
The rhesus LCV homologue for the EBV sVCA was cloned by PCR amplification using EBV-specific primers at reduced stringency from a rhesus LCV cosmid clone, QA15. Three PCR clones were sequenced to derive the nucleotide and amino acid sequence shown in Fig. 1A. Overall the rhesus LCV sVCA shows 69% amino acid identity with the EBV sVCA (Fig. 1B). To determine whether the antibody response in rhesus LCV-immune animals is directed at the sVCA gene product, the rhesus LCV sVCA was expressed after transfection of COS-7 cells, and lysates of vector control- or rhesus LCV sVCA-transfected cells were used for Western blotting with sera from five randomly selected rhesus monkeys in the conventional colony at the New England Regional Primate Research Center (NERPRC). As shown in Fig. 2, a unique 18- to 21-kDa band is detected by all five sera in rhesus LCV sVCA-transfected cells (Fig. 2A) but not in vector control-transfected cells (Fig. 2B). Thus, the rhesus LCV sVCA appears to be immunogenic in rhesus monkeys. However, the high background levels relative to the specific signal in these Western blots were emblematic of the difficulties that may arise in distinguishing between naive animals and those with lower antibody responses.
FIG. 1.
Sequence analysis of the rhesus LCV sVCA. (A) Nucleotide and predicted amino acid sequence of the rhesus LCV sVCA. (B) Amino acid comparison between rhesus LCV sVCA and EBV VCA p18 (p21). Identical amino acids are represented by an asterisk, similar amino acids are represented by a period, and gaps are indicated with a hyphen.
FIG. 2.
Expression of rhesus LCV sVCA in COS-7 cells. (A) Immunoblot strips with cell lysates of pSG5 rhesus LCV sVCA-transfected cells; (B) cell lysates of pSG5-transfected cells. Immunoblots containing cell lysates positive (A) and negative (B) for rhesus LCV sVCA were probed with five different LCV-immune rhesus sera (lanes 1 to 5) using a cassette Mini-protean II system (Bio-Rad).
Rhesus LCV sVCA ELISA.
In order to develop a more rapid, less labor-intensive, and more discriminating assay, we tested the potential utility of a rhesus LCV peptide ELISA. EBV sVCA is known to be an important target for the human antibody response to EBV infection, and the immunodominant epitopes are known to reside in the carboxy terminus (aa 119 to 176) (42). Therefore, we synthesized two peptides from similar regions of the rhesus LCV sVCA carboxy terminus (peptide 1, aa 117 to 146, and peptide 2, aa 147 to 170) (Fig. 1B). We used these peptides individually and in combination to test serologic responses in two populations of rhesus monkeys. The first population consisted of 20 randomly selected animals from the conventional colony at the NERPRC. Rhesus LCV infection was known to be highly prevalent in this colony (4). The second population was selected from a colony of hand-reared animals (6). These animals were separated early from their mother, reared by hand, and then raised in isolated colonies. This colony was serologically screened to exclude herpes B and D retrovirus infection. These animals were not specifically screened for LCV infection, but we hypothesized that these handling procedures were likely to result in a low prevalence of rhesus LCV infection. The average age of these animals was 1.5 years.
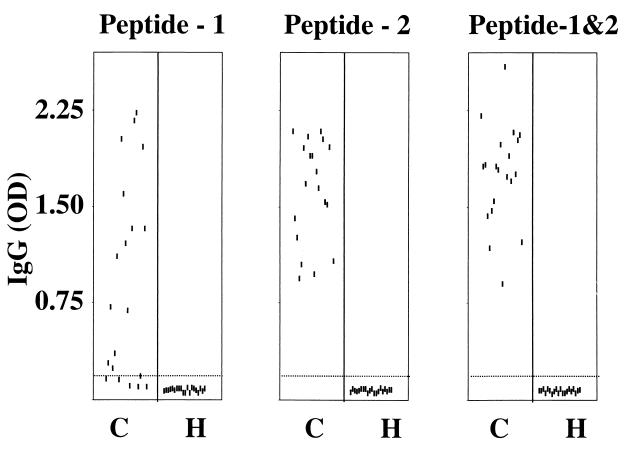
As shown in Fig. 3, none of the hand-reared animals had a detectable rhesus LCV sVCA serologic response by ELISA using peptides 1 and 2 alone or in combination. These animals were also tested for sVCA antibodies by Western blotting using sVCA protein expressed in COS-7 cells, and the results were negative (data not shown). In the conventional colony, sera from 14 of 20 animals reacted to peptide 1 with an optical density greater than 3 standard deviations above background levels. Sera from all 20 animals from the conventional colony had stronger reactivity to peptide 2 and could be easily distinguished from those from the hand-reared colony. The combination of peptides 1 and 2 did not provide any significant advantage over the results with peptide 2 alone. Previous studies of the rhesus LCV strains present in the oropharyngeal washes of the animals from the conventional colony showed an equal prevalence of type 1 and type 2 rhesus LCV (4). Thus, the carboxy terminus of the rhesus LCV sVCA contains immunodominant antibody epitopes independent of viral type.
FIG. 3.
Rhesus LCV sVCA peptide ELISA results for serum samples from animals in the conventional (C) and hand-raised (H) colonies. The dotted line near the bottom of each panel represents the cutoff value in the ELISA. ELISA optical density values shown are the average of triplicate values in a representative assay.
Currently, there is no “gold standard” for identifying rhesus LCV infection, making it difficult to validate the sensitivity and specificity of these peptide ELISA results. LCV infection persists for life. However, oropharyngeal shedding of virus is episodic, and PCR detection from oropharyngeal washes is positive for only a fraction of adult, EBV-seropositive humans (45). EBV infection also persists in 1 per 105 to 106 peripheral blood B cells, a level which is difficult to detect by DNA PCR of PBMC (22). However, the EBERs are expressed at a high copy number, 106 to 107 copies per infected cell, and could potentially increase the sensitivity of detecting persistent viral infection in the peripheral blood (37). Therefore, we cloned the rhesus LCV EBER homologues and tested whether an EBER RT-PCR could effectively detect persistent LCV infection in rhesus PBMC as an independent test for rhesus LCV infection.
RT-PCR for rhesus LCV EBER expression in rhesus monkey PBMC.
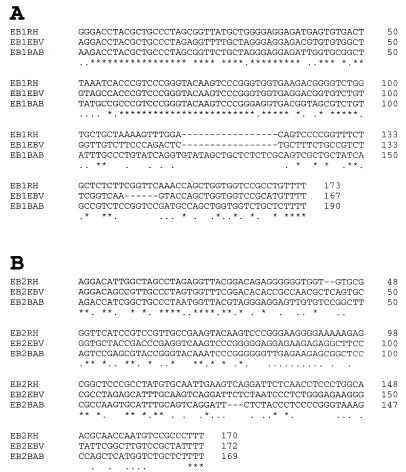
The rhesus LCV EBER1 has 73 and 68% nucleotide identity with the EBV and the baboon LCV EBER1 genes, respectively. The EBER2 genes are less well conserved, with only 42 and 38% identity with the EBV and baboon LCV EBER2 (Fig. 4). EBER1 was targeted since EBER1 is more abundant than EBER2 in EBV- and baboon LCV-infected cells (5, 14, 17) and better conserved. PCR primers were designed from EBER1 sequences conserved among all three species to minimize the potential effect of strain-dependent sequence variation.
FIG. 4.
Comparison of the EBV, rhesus LCV, and baboon DNA (14) sequences encoding EBER1 (A) and EBER2 (B). Nucleotides conserved among all viruses are represented with an asterisk, well conserved nucleotides are represented with a period, and gaps are indicated with a hyphen.
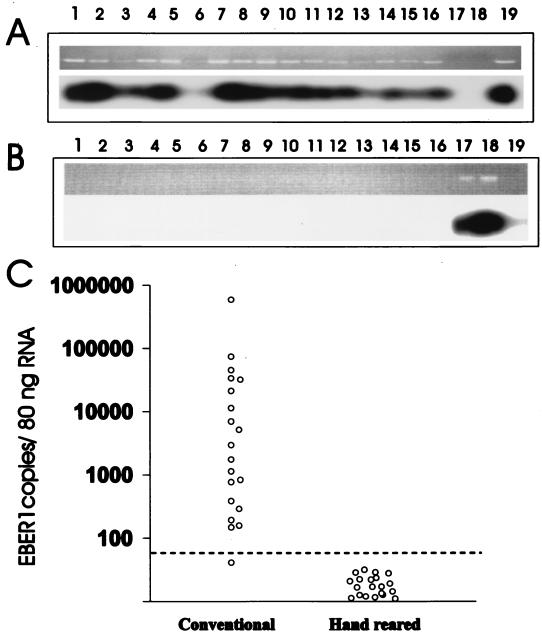
RT-PCR amplification for EBER1 RNA followed by Southern blot hybridization with an internal oligonucleotide probe was positive for 20 of 20 animals from the conventional colony (results from 16 animals shown in Fig. 5A, lanes 1 to 16). Signal intensity varied among these samples, and the signal was weakly detected in one animal on repeated assays (Fig. 5A, lane 6). All hand-reared animals were negative for EBER1 expression by RT-PCR amplification from peripheral blood lymphocytes (results from 16 animals shown in Fig. 5B, lanes 1 to 16). These results were identical to the rhesus LCV sVCA peptide ELISA results and suggested a nearly 100% prevalence of rhesus LCV infection in the conventional colony and absence of rhesus LCV infection in the hand-reared colony.
FIG. 5.
RT-PCR for EBER1 expression in rhesus monkey PBMC. (A) Southern blot analysis of RT-PCR-amplified products from conventional colony animals. Lanes 1 to 16 represent 16 animals randomly selected from the conventional colony. Positive (lane 19) and negative (lanes 17 and 18) controls are shown. (B) Southern blot analysis of RT-PCR-amplified products from hand-reared colony animals. Lanes 1 to 16 represent 16 randomly selected hand-reared animals. Positive (lanes 17 and 18) and negative (lane 19) controls are shown. (C) Semiquantitation of rhesus LCV EBER1 by real-time RT-PCR. The EBER1 copy number per 80 ng of total cellular RNA is shown. The limit of detection (50 copies/80 ng of total RNA) is shown by the dotted line.
In order to get a better appreciation of the relative magnitude of EBER RNA expression, samples were quantified by real-time PCR. As shown in Fig. 5C, 19 out of 20 animals were positive for EBER expression, with levels above the cutoff value. The relative EBER copy number determined by real-time PCR differed by almost 4 logs. One animal had undetectable EBER RNA by real-time PCR, and this was the same animal whose sample was weakly positive by Southern blot hybridization of the RT-PCR products and strongly positive by peptide ELISA. Therefore, this animal was likely rhesus LCV infected, and the weak or absent EBER signal by Southern blot hybridization and real-time PCR may be due to low viral load or sequence variation.
In order to determine the potential effect of strain-dependent sequence variation, RT-PCR products from four animals were cloned and sequenced. The sequences from two animals known to be infected with the type 1 rhesus LCV (4) were identical to the EBER1 sequence derived from the type 1 rhesus LCV cosmid clone. The sequences from two animals known to be infected with the type 2 rhesus LCV (4) were identical to each other and differed from the type 1 rhesus LCV sequence in 7 of 80 nucleotides. Since both rhesus LCV strains are equally prevalent in this colony (4), these sequence data and the ability to detect EBERs in nearly all animals suggest that strain-dependent sequence variation is unlikely to be a major problem.
Sensitivity and specificity of the rhesus LCV sVCA ELISA during acute rhesus LCV and RRV infection.
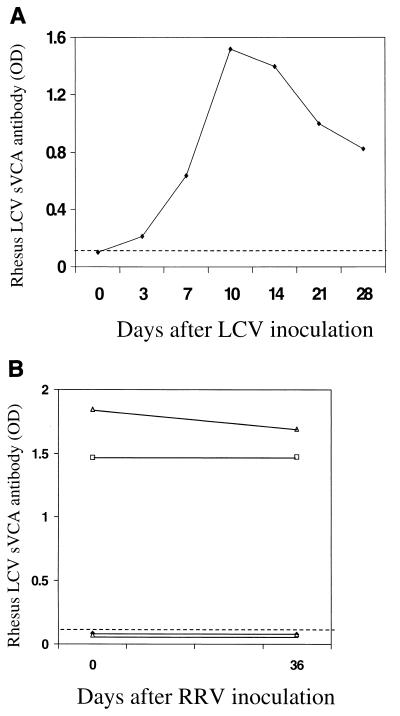
In order to further test the sensitivity and specificity of the rhesus LCV sVCA peptide ELISA, we examined the serologic response in an experimentally infected rhesus macaque. As shown in Fig. 6A, a rhesus macaque experimentally inoculated with rhesus LCV in the oropharynx (24) developed a brisk antibody response to the rhesus LCV sVCA, with a peak response developing by day 10. Rhesus macaques are also commonly infected with rhesus rhadinovirus (RRV), a gammaherpesvirus in the rhadinovirus subgroup (19, 44). Therefore, to rule out the remote possibility that serologic responses detected with the rhesus LCV sVCA might be due to cross-reactive antibodies specific for the RRV capsid protein, we tested sera from macaques experimentally inoculated with RRV. We obtained acute- and convalescent-phase sera from four animals with documented RRV seroconversion after experimental RRV inoculation (19) and tested these samples in the rhesus LCV sVCA peptide ELISA. Two animals were seropositive for rhesus LCV sVCA antibodies prior to infection, and the antibody titers did not change after RRV inoculation (Fig. 6B). Two animals were rhesus LCV seronegative prior to RRV inoculation and remained negative as shown by the rhesus LCV sVCA peptide ELISA. The previously documented RRV seroconversion in these specimens (19) confirmed that antibodies to the RRV VCA do not cross-react with the rhesus LCV sVCA.
FIG. 6.
Rhesus LCV sVCA peptide ELISA results from an animal experimentally inoculated with rhesus LCV (A) and from four animals experimentally inoculated with RRV (B). The dotted line represents the ELISA cutoff value.
DISCUSSION
We have developed highly sensitive and reproducible assays for acute and persistent rhesus LCV infections. As in humans, LCV infection is highly prevalent in nonhuman primates and is associated with B-cell malignancies in immunosuppressed hosts (8, 26). In addition, experimental infection of rhesus monkeys with the rhesus LCV provides the only animal model which accurately reproduces many aspects of acute and persistent EBV infections in humans (24). As these studies demonstrate, it is possible to breed a rhesus LCV-naive colony which would reduce the risk of LCV-induced malignancies in immunosuppressed animals and provide naive animals for pathogenesis and vaccine studies after experimental rhesus LCV infection.
The rhesus LCV sVCA is moderately well conserved with the EBV sVCA (69% amino acid identity), falling approximately halfway between the most and least well conserved lytic genes studied to date (90 to 50% amino acid identity) (15a, 23). The greatest sequence divergence is in the middle of the protein and overlaps positionally with our peptide 1 (aa 117 to 146), whereas the region represented by peptide 2 was more well conserved. In rhesus macaques the antibody responses to peptide 2 were clearly more prevalent in rhesus LCV-infected animals, whereas human antibody responses to the regions represented by peptides 1 and 2 are equally prevalent (42). The observed sequence divergence and potential differences in immunodominant epitopes may contribute to a loss of sensitivity when using the EBV sVCA as a cross-reactive antigen for detecting rhesus LCV-immune sera. The rhesus LCV VCA peptide 2 epitope was particularly useful since it not only identified all positive animals but also was associated with an extremely robust signal, clearly separating positive and negative results.
The sensitivity and specificity of the VCA peptide ELISA were also tested using sera from experimentally infected animals. First, a brisk antibody response was detected in an animal experimentally infected with rhesus LCV, similar to that previously detected by immunoblotting (24). Second, the specificity of the assay was checked using sera from animals experimentally infected with RRV. Rhadinoviruses are the herpesviruses most closely related to the LCV subgroup, and RRV infection is also highly prevalent in rhesus monkeys (19, 30, 44). The RRV VCA homologue shares only 24% amino acid identity (30), and the absence of any cross-reactivity could be confirmed by studying two animals who remained rhesus LCV seronegative after experimental RRV infection and documented RRV seroconversion (19). This specificity is similar to the lack of serologic cross-reactivity reported between the sVCAs of Kaposi's sarcoma-associated herpesvirus and EBV (18).
Since we had no gold standard for rhesus LCV infection, we also cloned the rhesus LCV EBER1 and EBER2 homologues and developed an EBER1 RT-PCR assay as an independent parameter for rhesus LCV infection. These two assays measuring different aspects of rhesus LCV lytic and latent infections gave identical results which further validate the accuracy of these assays. The ability to directly measure rhesus LCV RNA in the peripheral blood will also be an important tool for studying LCV pathogenesis in experimental infections. The RT-PCR assay is a sensitive indicator for persistent infection since EBER RNA expression could be detected in at least 95% of the animals. This rapid assay will be an important tool for experimental pathogenesis studies in which viral mutants with specific genetic lesions will be tested for their ability to establish persistent infection. The relatively broad range of EBER RNA expression levels was somewhat surprising and probably reflects a combination of different precursor frequencies of LCV-infected cells in the peripheral blood and different amounts of EBER RNA per quantity of infected cells. Therefore, it may be difficult to identify quantitative effects on viral persistence using simple quantitation of EBER RNA levels in the peripheral blood, given this broad range of EBER expression levels. Detecting more subtle quantitative effects on viral persistence will likely require limiting dilution analysis scored by an EBER RT-PCR assay to determine the precursor frequency of virus-infected cells (22).
Conservation of the EBER genes also suggests that they play an important role for the pathogenesis of LCV infection in vivo. The EBERs are expressed in LCV-immortalized B-cell lines in vitro, but genetic studies demonstrate that they can be deleted from the EBV genome with no detectable effect on virus replication or B-cell immortalization in tissue culture (36). Thus, one predicts that the EBERs must provide a function which is important for successful LCV infection in vivo but not necessarily in vitro. The EBERs have sequence similarity and can functionally replace the adenovirus VA RNAs (29), which block interferon-induced responses by inhibiting activation of an interferon-induced protein kinase and phosphorylation of the protein synthesis initiation factor eIF2 alpha (1, 2). The EBERs have also been reported to bind the interferon-inducible protein kinase PKR (33), the ribosomal protein L22 (7, 38, 39), cellular La protein (11, 15, 17), and (2′-5′) oligoadenylate synthetase (32). However, there is no obvious differential phenotype observed when B cells infected with wild-type EBV or EBER deletion mutant EBV are challenged with interferon in vitro (35, 36). The cloning and identification of the EBER genes from rhesus LCV are the first step towards generating an EBER deletion mutant of rhesus LCV and using the rhesus animal model to test the hypothesis that the EBERs are essential for successful LCV infection in vivo.
ACKNOWLEDGMENTS
This work was funded by grants from the American Heart Association and U.S. Public Health Service (CA68051 and CA65319). Animal resources were supported by the NERPRC (USPHS P51RR00168).
We thank Ronald Desrosiers and Sue Czajak for kindly providing sera from rhesus macaques experimentally infected with RRV. We thank Ashok Khatri and the peptide core facility at the Partners Center of AIDS Research for assistance with the peptide synthesis.
REFERENCES
- 1.Bhat R A, Thimmappaya B. Two small RNAs encoded by Epstein-Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc Natl Acad Sci USA. 1983;80:4789–4793. doi: 10.1073/pnas.80.15.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat R A, Thimmappaya B. Construction and analysis of additional adenovirus substitution mutants confirm the complementation of VAI RNA function by two small RNAs encoded by Epstein-Barr virus. J Virol. 1985;56:750–756. doi: 10.1128/jvi.56.3.750-756.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake N W, Moghaddam A, Rao P, Kaur A, Glickman R, Cho Y G, Marchini A, Haigh T, Johnson R P, Rickinson A B, Wang F. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J Virol. 1999;73:7381–7389. doi: 10.1128/jvi.73.9.7381-7389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho Y G, Gordadze A V, Ling P D, Wang F. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J Virol. 1999;73:9206–9212. doi: 10.1128/jvi.73.11.9206-9212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke P A, Sharp N A, Clemens M J. Expression of genes for the Epstein-Barr virus small RNAs EBER-1 and EBER-2 in Daudi Burkitt's lymphoma cells: effects of interferon treatment. J Gen Virol. 1992;73:3169–3175. doi: 10.1099/0022-1317-73-12-3169. [DOI] [PubMed] [Google Scholar]
- 6.Desrosiers R C. The value of specific pathogen-free rhesus monkey breeding colonies for AIDS research. AIDS Res Hum Retrovir. 1997;13:5–6. doi: 10.1089/aid.1997.13.5. [DOI] [PubMed] [Google Scholar]
- 7.Dobbelstein M, Shenk T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol. 1995;69:8027–8034. doi: 10.1128/jvi.69.12.8027-8034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feichtinger H, Li S L, Kaaya E, Putkonen P, Grunewald K, Weyrer K, Bottiger D, Ernberg I, Linde A, Biberfeld G, et al. A monkey model for Epstein Barr virus-associated lymphomagenesis in human acquired immunodeficiency syndrome. J Exp Med. 1992;176:281–286. doi: 10.1084/jem.176.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franken M, Annis B, Ali A N, Wang F. 5′ coding and regulatory region sequence divergence with conserved function of the Epstein-Barr virus LMP2A homolog in herpesvirus papio. J Virol. 1995;69:8011–8019. doi: 10.1128/jvi.69.12.8011-8019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J Virol. 1996;70:7819–7826. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glickman J N, Howe J G, Steitz J A. Structural analyses of EBER1 and EBER2 ribonucleoprotein particles present in Epstein-Barr virus-infected cells. J Virol. 1988;62:902–911. doi: 10.1128/jvi.62.3.902-911.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henle W, Henle G, Andersson J, Ernberg I, Klein G, Horwitz C A, Marklund G, Rymo L, Wellinder C, Straus S E. Antibody responses to Epstein-Barr virus-determined nuclear antigen (EBNA)-1 and EBNA-2 in acute and chronic Epstein-Barr virus infection. Proc Natl Acad Sci USA. 1987;84:570–574. doi: 10.1073/pnas.84.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilliard J K, Ward J A. B-virus specific-pathogen-free breeding colonies of macaques (Macaca mulatta): retrospective study of seven years of testing. Lab Anim Sci. 1999;49:144–148. [PubMed] [Google Scholar]
- 14.Howe J G, Shu M D. Isolation and characterization of the genes for two small RNAs of herpesvirus papio and their comparison with Epstein-Barr virus-encoded EBER RNAs. J Virol. 1988;62:2790–2798. doi: 10.1128/jvi.62.8.2790-2798.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howe J G, Steitz J A. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci USA. 1986;83:9006–9010. doi: 10.1073/pnas.83.23.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Jiang H, Cho Y, Wang F. Structural, functional, and genetic comparisons of Epstein-Barr virus nuclear antigen 3A, 3B, and 3C homologues encoded by the rhesus lymphocryptovirus. J Virol. 2000;74:5921–5932. doi: 10.1128/jvi.74.13.5921-5932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff E, Liebowitz D. Epstein-Barr virus and its replication. In: Knipe D, Fields B, Chanock R, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 1889–1920. [Google Scholar]
- 17.Lerner M R, Andrews N C, Miller G, Steitz J A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi's sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansfield K G, Westmoreland S V, DeBakker C D, Czajak S, Lackner A A, Desrosiers R C. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol. 1999;73:10320–10328. doi: 10.1128/jvi.73.12.10320-10328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margalith M, Sarov B, Sarov I, Rinaldo C, Detels R, Phair J, Kaslow R, Ginsberg H, Saah A. Serum IgG and IgA antibodies specific to Epstein-Barr virus capsid antigen in a longitudinal study of human immunodeficiency virus infection and disease progression in homosexual men. AIDS Res Hum Retrovir. 1990;6:607–616. doi: 10.1089/aid.1990.6.607. [DOI] [PubMed] [Google Scholar]
- 21.Milman G, Scott A L, Cho M S, Hartman S C, Ades D K, Hayward G S, Ki P F, August J T, Hayward S D. Carboxyl-terminal domain of the Epstein-Barr virus nuclear antigen is highly immunogenic in man. Proc Natl Acad Sci USA. 1985;82:6300–6304. doi: 10.1073/pnas.82.18.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 23.Moghaddam A, Koch J, Annis B, Wang F. Infection of human B lymphocytes with lymphocryptoviruses related to Epstein-Barr virus. J Virol. 1998;72:3205–3212. doi: 10.1128/jvi.72.4.3205-3212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 25.Peng R, Gordadze A V, Fuentes Panana E M, Wang F, Zong J, Hayward G S, Tan J, Ling P D. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J Virol. 2000;74:379–389. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangan S R, Martin L N, Bozelka B E, Wang N, Gormus B J. Epstein-Barr virus-related herpesvirus from a rhesus monkey (Macaca mulatta) with malignant lymphoma. Int J Cancer. 1986;38:425–432. doi: 10.1002/ijc.2910380319. [DOI] [PubMed] [Google Scholar]
- 27.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 28.Rivailler P, Quink C, Wang F. Strong selective pressure for evolution of an Epstein-Barr virus LMP2B homologue in the rhesus lymphocryptovirus. J Virol. 1999;73:8867–8872. doi: 10.1128/jvi.73.10.8867-8872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosa M D, Gottlieb E, Lerner M R, Steitz J A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serio T R, Angeloni A, Kolman J L, Gradoville L, Sun R, Katz D A, Van Grunsven W, Middeldorp J, Miller G. Two 21-kilodalton components of the Epstein-Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21) J Virol. 1996;70:8047–8054. doi: 10.1128/jvi.70.11.8047-8054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp T V, Raine D A, Gewert D R, Joshi B, Jagus R, Clemens M J. Activation of the interferon-inducible (2′-5′) oligoadenylate synthetase by the Epstein-Barr virus RNA, EBER-1. Virology. 1999;257:303–313. doi: 10.1006/viro.1999.9689. [DOI] [PubMed] [Google Scholar]
- 33.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C G, Hilse K, Clemens M J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shedd D, Angeloni A, Niederman J, Miller G. Detection of human serum antibodies to the BFRF3 Epstein-Barr virus capsid component by means of a DNA-binding assay. J Infect Dis. 1995;172:1367–1370. doi: 10.1093/infdis/172.5.1367. [DOI] [PubMed] [Google Scholar]
- 35.Swaminathan S, Huneycutt B S, Reiss C S, Kieff E. Epstein-Barr virus-encoded small RNAs (EBERs) do not modulate interferon effects in infected lymphocytes. J Virol. 1992;66:5133–5136. doi: 10.1128/jvi.66.8.5133-5136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toczyski D P, Matera A G, Ward D C, Steitz J A. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci USA. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toczyski D P, Steitz J A. EAP, a highly conserved cellular protein associated with Epstein-Barr virus small RNAs (EBERs) EMBO J. 1991;10:459–466. doi: 10.1002/j.1460-2075.1991.tb07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tranchand-Bunel D, Gras-Masse H, Bourez B, Dedecker L, Auriault C. Evaluation of an Epstein-Barr virus (EBV) immunoglobulin M enzyme-linked immunosorbent assay using a synthetic convergent peptide library, or mixotope, for diagnosis of primary EBV infection. J Clin Microbiol. 1999;37:2366–2368. doi: 10.1128/jcm.37.7.2366-2368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Grunsven W M, Nabbe A, Middeldorp J M. Identification and molecular characterization of two diagnostically relevant marker proteins of the Epstein-Barr virus capsid antigen complex. J Med Virol. 1993;40:161–169. doi: 10.1002/jmv.1890400215. [DOI] [PubMed] [Google Scholar]
- 42.van Grunsven W M, Spaan W J, Middeldorp J M. Localization and diagnostic application of immunodominant domains of the BFRF3-encoded Epstein-Barr virus capsid protein. J Infect Dis. 1994;170:13–19. doi: 10.1093/infdis/170.1.13. [DOI] [PubMed] [Google Scholar]
- 43.van Grunsven W M, van Heerde E C, de Haard H J, Spaan W J, Middeldorp J M. Gene mapping and expression of two immunodominant Epstein-Barr virus capsid proteins. J Virol. 1993;67:3908–3916. doi: 10.1128/jvi.67.7.3908-3916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong S W, Bergquam E P, Swanson R M, Lee F W, Shiigi S M, Avery N A, Fanton J W, Axthelm M K. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J Exp Med. 1999;190:827–840. doi: 10.1084/jem.190.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao Q Y, Rickinson A B, Epstein M A. Oropharyngeal shedding of infectious Epstein-Barr virus in healthy virus-immune donors. A prospective study. Chin Med J. 1985;98:191–196. . (In English.) [PubMed] [Google Scholar]
- 46.Yates J L, Camiolo S M, Ali S, Ying A. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]