Abstract
The ongoing COVID-19 pandemic highlights the need to tackle viral variants, expand the number of antigens, and assess diverse delivery systems for vaccines against emerging viruses. In the present study, a DNA vaccine candidate was generated by combining in tandem envelope protein domain III (EDIII) of dengue virus serotypes 1–4 and a dengue virus (DENV)-2 non-structural protein 1 (NS1) protein-coding region. Each domain was designed as a serotype-specific consensus coding sequence derived from different genotypes based on the whole genome sequencing of clinical isolates in India and complemented with data from Africa. This sequence was further optimized for protein expression. In silico structural analysis of the EDIII consensus sequence revealed that epitopes are structurally conserved and immunogenic. The vaccination of mice with this construct induced pan-serotype neutralizing antibodies and antigen-specific T cell responses. Assaying intracellular interferon (IFN)-γ staining, immunoglobulin IgG2(a/c)/IgG1 ratios, and immune gene profiling suggests a strong Th1-dominant immune response. Finally, the passive transfer of immune sera protected AG129 mice challenged with a virulent, non-mouse-adapted DENV-2 strain. Our findings collectively suggest an alternative strategy for dengue vaccine design by offering a novel vaccine candidate with a possible broad-spectrum protection and a successful clinical translation either as a stand alone or in a mix and match strategy.
Keywords: dengue, DNA vaccine, antibody-dependent enhancement, EDIII domain, NS1 protein, consensus sequence, dengue surveillance
Graphical abstract
Developing a successful vaccine against dengue has been challenging. Arun Sankaradoss and colleagues developed a genotype-specific DNA DENV vaccine candidate that elicits robust dengue immune responses. The low cost and thermostability of DNA vaccines should allow countries to implement large-scale vaccination programs, which could greatly reduce dengue cases.
Introduction
Vaccine development is an evolving process. The current coronavirus disease 2019 (COVID-19) pandemic has facilitated the development of numerous vaccine technologies to mitigate public health crises. Among other technologies, nucleic acid vaccines have emerged as a rapid and versatile platform for an emergency, which is why these are among the very first COVID-19 vaccines in human use.1 Nucleic acid vaccines require a short time from design to clinical trials; therefore, it may be possible to test together, in the same vaccine, different variants of antigens that cover circulating mutations.2 Nucleic acid vaccines are adaptable for mix and match vaccination approaches.3,4 In addition, they offer a more natural antigen presentation to the immune system, resulting in better T cell responses.5 An advantage of DNA over mRNA is that they are more versatile, temperature-stable, cost-effective, and cold-chain-free, which are essential features for delivery to resource-limited settings.6 After more than 3 decades of research on DNA vaccines, the world's first DNA vaccine against COVID-19 has been approved in India for emergency use, highlighting the evolution of low-cost vaccine platforms.7 Moreover, the low cost and thermostable aspect of the DNA vaccine should allow countries to drive any vaccination programs on a larger scale than is currently possible, especially in middle- and low-income countries.
Developing a successful vaccine against dengue has been challenging. In addition to the standard vaccine approaches (i.e., whole-genome, peptide, and virus-like particle vaccines) several nucleic-acid-based vaccines for dengue virus (DENV) are in the pipeline.8,9 DENV vaccine development is complicated by four antigenically diverse serotypes (DENV1–4) and their intraserotype (genotype) diversity at both local and global scales.10 Several studies have reported the association between the genotype shift and magnitude of the outbreak and disease severity.11,12 However, data on circulating genotype level information are limited in India. The recent data in India highlighted the emergence of DENV1 genotypes I and III, DENV3 genotype III in Southern India, and DENV2 cosmopolitan genotype dominated both in the south and in Delhi.13, 14, 15 Previous studies also indicated the cocirculation of DENV4 genotype I strains in Pune, along with DENV1, DENV2, and DENV3.16,17
The current DENV vaccine effort is also thought to be hampered by antibody-dependent enhancement (ADE), whereby cross-reactive antibodies against one serotype can enhance subsequent infection by a heterologous serotype.18 To circumvent such issues, regions or motifs of the antigen responsible for causing ADE must be eliminated from the vaccine design. The envelope protein domain III (EDIII) has been identified as the major target of highly neutralizing and protective serotype-specific antibodies, while, in contrast, precursor membrane (PrM)- and EDI–II-directed antibodies are reported to enhance infection via ADE.19 Recent studies have found that EDIII-based DENV vaccines could circumvent ADE of infection in mice, whereas the T cell response against EDIII DNA vaccine has been shown to play a role in disease protection through effective viral clearance.20,21 Several studies have also shown that EDIII-directed antibodies can inhibit the entry of the flavivirus into target cells. Mutations in EDIII may affect antibody binding as well as protein interaction with cellular receptors.22,23 Previous studies have reported that mutations in EDIII of DENV and other flaviviruses have lower virulence or have the ability to escape immune neutralization.24
DENV non-structural protein 1 (NS1) has also emerged as an attractive vaccine target. NS1, a highly conserved protein among flaviviruses, is involved in virus replication and immune evasion.25 However, it has also been reported to activate antibody Fc-mediated effector functions and provide protection against flaviviruses.26 NS1 has been demonstrated to trigger T cell responses in both experimental animals and humans.27 Previous studies have shown that NS1 induced T cell responses involved in protection against dengue infection. Furthermore, vaccination with DENV1, DENV3, or DENV4 NS1 protected against a heterologous DENV2 challenge,28 and the monoclonal antibodies directed against the DENV2 NS1 effectively protected mice against all serotypes of DENV infection.29 Taken together, combining EDIII and NS1 for vaccine design could be a possible way to circumvent immune interference among serotypes.
In addition, an effective DENV vaccine should be able to target circulating strain divergence; in this case, a consensus-based vaccine may be optimal. Consensus-based vaccines are an intriguing strategy that could minimize the sequence diversity across strains. Furthermore, recent studies on H1N1 and human immunodeficiency virus (HIV) in mice have suggested that consensus-immunogen-based vaccines may be critical to address the viral genetic divergence.30,31 Moreover, several studies have also shown that consensus prediction could outperform single-sequence determination methods in vaccine design.32,33
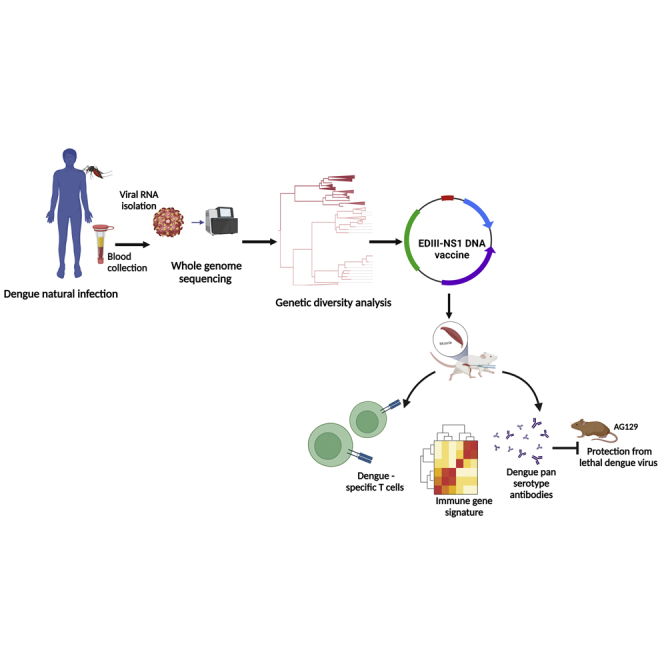
Here, we investigated DENV1–4 genotype diversity and their ED III and NS1 amino acid variants at both local and global scales. Further, we developed a DENV EDIII-based DNA vaccine (DDV) candidate by integrating the consensus sequence information from the circulating genotypes of each serotype instead of a single-strain genome sequence, incorporating the NS1 protein-coding region to induce broad T cell responses, and then optimizing the DNA vaccine construct for optimal protein expression. The immunogenicity of this vaccine candidate was evaluated by investigating its ability to induce tetravalent neutralizing antibodies and T cell responses as well as protective immunity in mice.
Results
DENV genome sequencing and genotype analysis
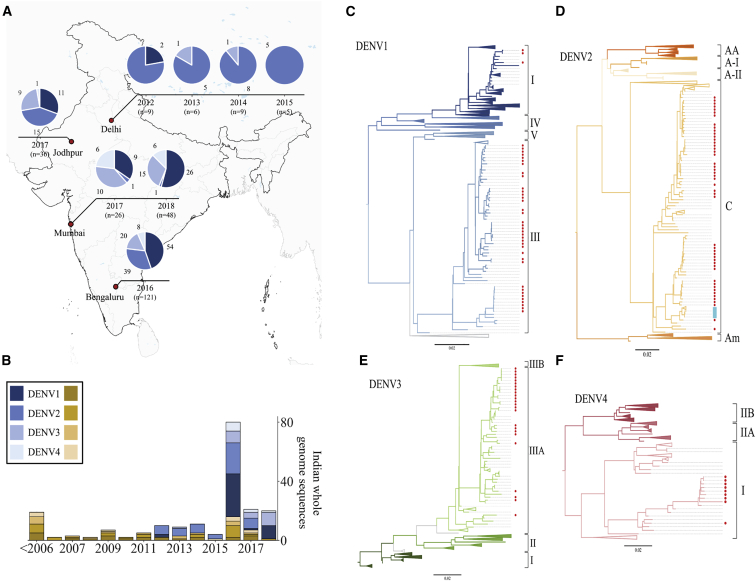
In this study, we investigated the genetic diversity of circulating DENV strains in India, designed a DNA vaccine based on circulating DENV genotypes, and evaluated the immunogenicity of this vaccine candidate in two murine models. In essence, we performed a retrospective analysis of 1,000 archived serum/plasma samples from NS1-positive patients with dengue clinical presentations collected from 2012 to 2018 from 4 hospitals in India. Whole-genome sequencing of DENVs was carried out from the serum of 319 patients. Table S1 summarizes the sequenced patient's sample clinical characteristics. Clinical and biochemical correlates of the sequenced samples of this study are consistent with earlier epidemiological observations.34,35 Of the 319 samples sequenced, we have obtained 120 whole-genome sequences as well as the serotype infection information of 217 patients. Among the 217 patients, 174 patients were infected with a single DENV serotype (mono-infection) and 43 patients had concurrent DENV coinfections. From the 174 patients, 68 (39.1%) of them typed as DENV1, 63 (36.2%) of them as DENV2, 29 (16.7%) of them as DENV3, and 14 (8%) of them as DENV4. The distribution of serotypes during the sample collection period is shown in Figure 1A. The number of whole-genome sequences available from India, including this study, is presented in Figure 1B. Our dengue surveillance data collectively suggest that India is endemic to all four serotypes; therefore, an effective vaccine must be tetravalent.
Figure 1.
DENV serotype distribution across sites
(A) Distribution of DENV serotypes 1–4 from the sites from which samples were received for this study. Serotype distribution for each site is shown as pie charts over time. Total numbers from the site are indicated, and each serotype is represented by a different color. (B) The number of whole-genome sequences available from India. Yellow represents sequences available before the study, and blue represents the sequences obtained in this study. Shades of yellow and blue represent different serotypes of dengue. (C–F) Phylogenetic trees for DENV1–4 using the maximum likelihood method are shown. All available complete DENV coding nucleotide sequences from human hosts were used for the tree construction. Sylvatic strains were used as outgroups to root the tree. Branches have been collapsed to aid in the visualization. Regions where Indian sequences are present are colored in red.
In addition, we included DENV genome surveillance data from Kenya. This includes three complete coding DENV sequences that we reported in a study conducted in 2016 and 2017 on 560 febrile patients in Kenya36 as well as all other available whole-genome sequences from Africa for further phylogenetic-based DENV genotype analysis.
The phylogenetic-based genotype analysis of our study revealed that all Indian DENV1–4 strains belong to a single genotype within the serotypes, except for DENV1 (Figures 1C–1F). DENV1 genotype III is the most dominant apart from a few sequences of genotype I. DENV2 sequences belong to the cosmopolitan genotype, whereas DENV3 and DENV4 sequences belong to genotypes III and I, respectively. Our data show that two different DENV2 lineages within the cosmopolitan genotype are propagating simultaneously in all four sites in this study. Both lineages emerged in the middle of the 1980s. Most of the neighboring sequences are from Asian countries. However, one cluster in the cosmopolitan-b lineage is found in Kenya (Figure S1) and one sequence is from a traveler from Ethiopia and Djibouti. Our findings are also consistent with a previous report from Kenya.36 All of the Indian sequences of DENV3 genotype III cluster with other Asian countries. Only genotype I of DENV4 is observed in India. All Indian DENV4-I sequences, along with one sequence from Pakistan, cluster separately within genotype I from other Southeast Asian sequences. These data highlight that even though there is an enormous intraserotype variation recorded all over the world, India has distinct genotypes for all four DENV serotypes with prominent intermixing between neighboring countries. Based on the implications of our findings on region-specific genotypes, we proposed a vaccine design approach against circulating strains, which may be a better representation, rather than strains not found here.
EDIII and NS1 global and local genetic diversity and target for vaccine design
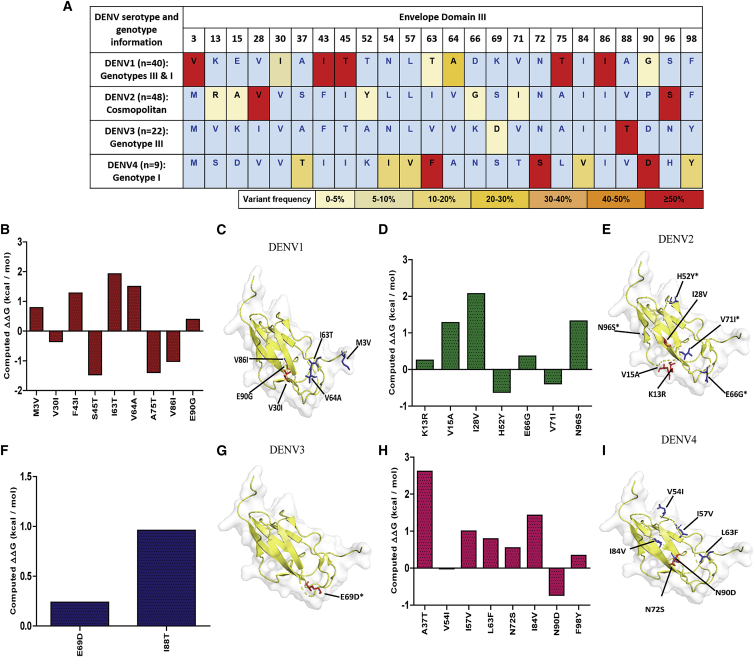
The E protein, primarily EDIII, stimulates host immune responses by evoking protective and neutralizing antibodies.20 Mutations in the EDIII region could potentially impact the neutralization of DENV and the host-receptor interaction. In addition to structural proteins, immune responses to DENV infections target non-structural protein NS1, which is currently used as a diagnostic candidate for dengue infection. We investigated EDIII and NS1 protein diversity in global strains, as well as how our study sequences diverge from reference strains. To assess the antigenic differences between the EDIII and NS1 of DENV1–4 strains from distinct parts of the world, we generated a maximum likelihood tree and selected representative genotype variants from each branch to comprehensively represent global diversity within the serotype. We performed a sequence alignment of these proteins and compared their EDIII and NS1 genetic diversity relative to the respective reference strains. Variable sites were designated when at least one virus showed an amino acid change at any of the amino acid positions in the alignment. While DENV genotypes are closely related, considerable genetic variation was observed across genotypes of the same serotype in the EDIII and NS1 region with a range of 0.97%–4.85% and 0.85%–4.9%, respectively (Tables S2A and S2B).
Further, EDIII and NS1 amino acid sequences of DENV1–4 clinical isolates from our study were compared for their similarity with the wild-type DENV strains. Even though India has its unique genotypes, considerable genetic variations were found in the EDIII and NS1 protein across genotypes of the same serotype. Our diversity analysis revealed 9, 7, 2, and 8 sites of EDIII variation (Figure 2A) and 24, 24, 8, and 17 variable sites within NS1 (Figure S2) in DENV1, DENV2, DENV3, and DENV4 among Indian genotypes, respectively. Among all four strains, DENV1 exhibited the highest EDIII diversity with a median of 4.85% (range, 2.91%–5.83%), followed by DENV4 and DENV2 with 2.91% (range, 2.91%–3.88%) and 1.94% (range, 0.97%–3.88%), respectively. In the case of NS1, the highest median diversity was 3.69%, which was observed in DENV1 (range, 1.99%–4.55%) and DENV4 (range, 3.12%–4.26%), followed by that of DENV2 with 2.27% (range, 1.99%–2.84%). The percentage diversity of DENV3 EDIII sequences was found to be limited, ranging from 0% to 0.97%. This was also observed for DENV3 NS1 with a median value of 1.42 (range, 1.14%–1.7%). A site was considered highly variable when greater than 50% of the study isolates showed a mutation at that position. The number of highly variable sites in EDIII and NS1 were 5, 3, 2, and 1 and 13, 13, 8, and 5 in DENV1, DENV4, DENV2, and DENV3 strains, respectively.
Figure 2.
DENV EDIII amino acid variation in our study clinical isolates for all four serotypes
(A) Frequency-based representation of all the variable sites for each serotype within our study sequences. Amino acid variant residues identified relative to NC_001477 (DENV1), NC_001474 (DENV2), NC_001475 (DENV3), and NC_002640 (DENV4). Colors were assigned based on the percentage of the frequency of mutations in the given clinical isolates. (B, D, F, and H) FoldX stability calculations for mutations in DENV1, DENV2, DENV3, and DENV4 strains, respectively. (C, E, G, and I) Amino acid variants on B cell and T cell epitopes are spotted on EDIII PDB structure (ribbon). The stick and ball representation on the PDB structure (PDB: 3IRC) indicates a mutation at the particular position. The presence of mutations in the B cell or T cell epitopes is shown by arrows. B cell mutations are shown in red, and T cell mutations are shown in blue. Asterisks (∗) signify mutations in both B and T cell epitopes.
An immunoglobulin G (IgG)-like fold present in the EDIII protein is typically associated with structures that have an adhesion function;8 hence, we investigated the effect of EDIII mutations on EDIII protein stability using FoldX. The predicted ΔΔG values for DENV1–4 mutations ranged from −1.4 to 2.6 kcal/mol (Figures 2B, 2D, 2F, and 2H). Except for DENV2 EDIII-I28V and DENV4 EDIII-A37T, all other EDIII non-synonymous mutations found in our clinical isolates had a minor or no effect on protein stability (Table S3). We also observed mutations in residues within known B cell and T cell epitope regions. These mutations were spotted on the EDIII PDB structure (Figures 2C, 2E, 2G, and 2I). Furthermore, some of the mutations (DENV1 EDIII-E90G, DENV3 EDIII-I88T) were observed in type-specific monoclonal antibody binding sites,37, 38, 39, 40 which could impact the antibody binding and neutralization. In addition to our study sequences, we also investigated EDIII mutations in all Indian DENV1–4 strain sequences deposited in the Virus Pathogen Database and Analysis Resource (ViPR) database. The frequency of amino acid variations is shown in Figure S3. In line with previous reports, our analysis implicates the EDIII amino acid residues as sites under immune pressure.41
DDV construction
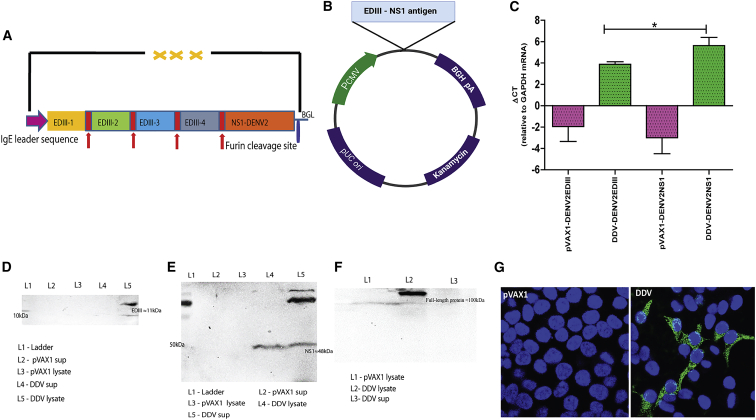
We developed the DDV candidate, which is more adapted to the strains of DENVs found in India and Africa. The consensus EDIII-NS1 vaccine sequence was designed by combining whole-genome sequences obtained from our study with published Indo-Africa-specific DENV sequences that were retrieved from the ViPR database. The generated consensus sequences were codon- and RNA optimized, synthesized commercially, and cloned into pVAX1 expression vectors, and the generated plasmid was designated as DDV (Figures 3A and 3B).
Figure 3.
DENV DNA vaccine construction
(A and B) Schematic representation of DENV DNA vaccine and cloning strategy. Synthetic DENV expression cassette was inserted into the pVAX1 expression vector between NheI and HindIII under the control of the cytomegalovirus (CMV) immediate-early promoter. (C) RT-PCR of RNA extracts from HEK293T cells transfected with DNA DENV vaccine or pVAX1. GAPDH is used as an internal expression normalization gene. Data are representative of three independent experiments. p values were determined by Student’s unpaired t test; ∗p ≤ 0.05. (D–F) Analysis of in vitro expression of EDIII (D), NS1 (E) protein, and full-length expressed protein (F) after transfection of 293T cells with DDV or plasmid control by western blot. (G) Immunofluorescence staining of 293T cells transfected with 5 μg/well DDV or plasmid control. Expression of antigen was measured using anti-DDV immune sera. Cell nuclei were counterstained with DAPI. The vector map was created with BioRender.com.
It is also noteworthy that approximately 26%–50% of Indian DENV1–4 strains exhibited 100% identity with consensus EDIII sequences represented in DDV. The remaining Indian sequences, for all serotypes, exhibited greater than 93% identity. Furthermore, DDV has 100% identity with African DENV2 and DENV3 strains, while African DENV1 and DENV4 strains exhibited >96% identity with their corresponding serotype DDV sequences. DDV also shares >95.15 identities with the EDIII of the top 1,000 international dengue sequences of the cognate serotype in the ViPR database (Table S4).
Epitope analysis for the EDIII construct
We predicted the structural stability of the EDIII constructs and checked for the 3D structural conservation at the predicted B cell discontinuous epitope regions (Table S5). The homology models for the EDIII constructs were subjected to energy minimization, RMSD (root-mean-square deviation with the template used for modelling) calculation with the PDB structures and Ramachandran map (https://saves.mbi.ucla.edu), and energy analysis (https://prosa.services.came.sbg.ac.at/prosa.php), which predicted that the constructs were stable structurally and energetically (Figure S4). In order to estimate the population coverage of the vaccine constructs, we also predicted the T cell epitopes, and the human leukocyte antigen (HLA) subtypes predicted to bind to each of the epitopes. This analysis revealed that 90%–98% of the world population could recognize the major histocompatibility complex (MHC) class I epitopes (using predicted strong binding epitopes) and that 90%–99% of the population can recognize the MHCII epitopes (using both strong and weak binding epitopes). We have chosen nine geographical regions with either frequent or sporadic dengue occurrence as per a CDC report (https://www.cdc.gov/dengue/areaswithrisk/around-the-world.html). These regions are South Asia, Southeast Asia, East Africa, West Africa, Central Africa, West Indies, Central America, South America, and Oceania. We see that the population coverage for epitopes is more than 75% of most of the regions except for the West Indies and Central America, which have a lower population coverage for some of the serotypes (Tables S6–S8).
In vitro antigen expression and localization
We first assessed encoded DENV EDIII and NS1 transgene expression at the RNA level in HEK293T cells transfected with DDV. Using the total RNA isolated from the transfected 293T cells, we confirmed EDIII and NS1 mRNA expression by qRT-PCR (Figure 3C). In vitro, EDIII and NS1 protein expression in HEK-293T cells was measured by western blot using anti-DDV immune sera on cell lysates. Western blots of the lysates of HEK-293T cells transfected with the DDV construct revealed bands near predicted molecular weights of ≈11 (Figure 3D) and ≈48 kDa (Figure 3E) for EDIII and NS1, respectively. We also detected secreted DENV NS1 in the culture supernatants and a full-length protein prior to furin cleavage of ≈100 kDa (Figure 3F) in lysates of transfected HEK-293T cells. EDIII and NS1 protein expressions were further validated using EDIII and NS1 monoclonal antibodies (Figure S5). In immunofluorescence studies, the EDIII and NS1 protein was detected in HEK-293T cells transfected with DDV and exhibited antigen staining of the expressed proteins mainly in the cytoplasm, which suggested the immune reactivity of the encoded protein (Figure 3G). In summary, in vitro studies revealed the expression of antigens at both the RNA and protein levels after transfection of cell lines with the candidate vaccine construct DDV.
Induction of humoral immune responses against DDV in BALB/c and C57BL/6J
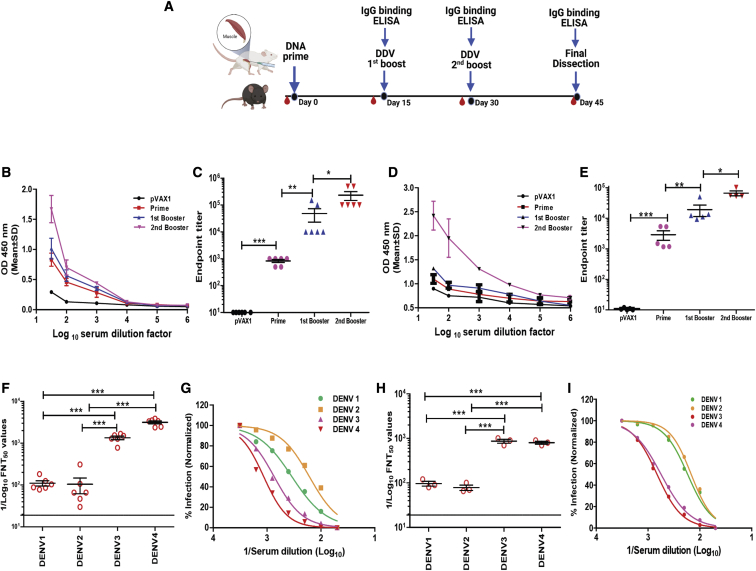
DENV-specific humoral responses following vaccination were characterized in two different murine strains, BALB/c and C57BL/6J. Mice (n = 6) were vaccinated three times 2 weeks apart with 50 μg of the DNA vaccines or control pVAX1 plasmid vector using tibialis anterior (TA) muscle delivery (Figure 4A). Vaccinated mice were bled at day 0 and 2 weeks after each vaccination to obtain sera, which were assayed for the presence of DENV antibodies by enzyme-linked immunosorbent assay (ELISA) against recombinant protein (Figure S6) as a capture protein. Binding antibody ELISA data revealed that the DDV induced DENV-specific antibody responses. The results showed that all mice developed anti-DENV antibodies after a single immunization. The anti-dengue IgG responses were significantly increased after 1–2 booster immunizations, appearing to peak 2 weeks after the second booster in both BALB/c and C57BL/6J strains (Figures 4B and 4D). Comparison between the serum IgG endpoint titers of DDV and plasmid control groups in both murine models showed robust titers elicited by DDV immunization. The endpoint tiers ranged from 1:1,000 to 1:500,000 in individual animals (Figures 4C and 4E). These findings demonstrated the ability of the DDV construct to potentially express in mammalian cells, and the antibodies induced by these constructs were able to react with the dengue vaccine antigens.
Figure 4.
Antibody response induced by DENV DNA vaccination in BALB/c and C57BL/6J mice
(A) Schedule of vaccination and antibody assays. BALB/c (n = 6/group) and C57BL/6J (n = 5/group) mice were immunized by TA injection of 50 μg DDV or plasmid vector at days 0, 15, and 30. Sera were collected at each time point. (B–E) Indirect ELISA reactivity against DDV antigens represented by an OD measured at 450 (OD450) nm (in pooled sera) and serum IgG binding endpoint titers (in individual animals), respectively, of BALB/c (B and C) and C57BL/6J (D and E) strains. Data shown represent mean OD450 nm values (mean ± SD) for each group of mice. (F–I) Sera neutralization titers against DENV1–4 laboratory prototype strains and recent clinical isolates. Sera were collected 14 days after the second booster dose and analyzed for neutralization of DENV by FNT assay. (G and I) Representative normalized percentage of infection curves are shown from laboratory prototype strains and recent clinical isolates. FNT50 values were calculated for individual animals and presented in (F) and (H) from prototype strains (n = 6) and clinical isolates (n = 3), respectively. Each point represents an individual animal, while horizontal lines indicate the mean ± SD. Data are representative of three independent experiments. p values were determined by Student’s unpaired t test; ∗p ≤ 0.05, ∗∗p ≤ 0.005, ∗∗∗p ≤ 0.0005. The study design schematic diagram was created with BioRender.com.
A flow-based virus neutralization assay was performed in U937-DC-SIGN cells to assess the levels of anti-DDV immune sera-induced neutralizing antibody titers against laboratory DENV strains. DDV-vaccinated mouse immune sera showed a clear neutralizing antibody titer against all four serotypes simultaneously, and the median FNT50 titers against DENV1–4 ranged from 182–3,500 (Figures 4F and 4G). As per WHO recommendation for DENV vaccines,42 we next investigated the neutralizing potency of anti-DDV immune sera against recent DENV1–4 clinical isolates. DENV1–4 serotypes were isolated from DENV NS1-positive patients in India, and a flow-cytometry-based neutralization test (FNT) was performed. Our data showed that anti-DDV immune sera also effectively neutralized clinical isolates, and FNT50 titers against DENV1–4 ranged from 150–900 (Figures 4H and 4I). These data provide evidence that DDV-induced neutralizing antibody responses cover major currently circulating strains as well.
Robust T cell responses induced by DDV
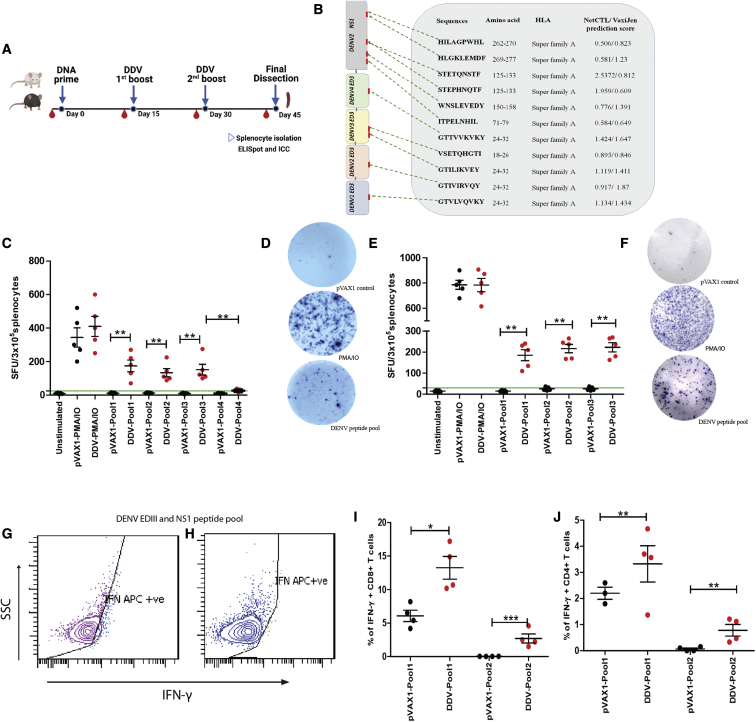
DENV-specific cellular responses were assessed in vaccinated animals by enzyme-linked immunosorbent spot (ELISpot). A total of 95 and 344 EDIII- and NS1-specific T cell peptides, respectively, were identified in the DDV construct. MHC class I binding predictions, peptide selection, and antigenicity of these peptides were analyzed by NetCTL 1.2 and Vaxijen 2.0. A total of 15 cytotoxic T lymphocyte (CTL) epitopes (9-mer peptides) were screened, of which 11 were found to possess antigenicity and were chosen for synthesis (Figure 5B). We assayed T cell responses against dengue antigens via IFN-γ ELISpot, as the IFN response has previously been shown to be associated with vaccine immunogenicity following yellow fever vaccination.43 Furthermore, IFN-γ has been described as a mediator of T cell responses and plays a distinctive role in antiviral activity against DENV.44 Mice were immunized as described before. Two weeks after the second booster, DDV- or pVAX1-immunized mice were euthanized, and splenocytes were isolated (Figure 5A). Single-cell suspensions were stimulated with the peptide pools (pool 1: EDIII peptide mixture; pool 2: NS1 peptide mixture; pool 3: EDIII and NS1 peptide mixture; pool 4: HIV-1 Nef peptide mixture), and the number of IFN-γ-producing cells was analyzed. Results show that both EDIII- and NS1-specific cellular responses (presented as IFN-γ spot-forming units [SFU]/cells) were detected by ELISpot in DDV-vaccinated animals (Figures 5C–5F). Furthermore, negligible or significantly low spots were detected in the pVAX1- or DDV-vaccinated BALB/c mice splenocytes, which were stimulated with the HIV-1 Nef T cell peptide pool (Figure 5C). This suggests that T cell responses elicited by DDV are antigen-specific rather than owing to non-specific T cell activation. Interleukin (IL)-4 is a strong Th2 cytokine known to suppress the production of antiviral cytokines and cell-mediated immune responses.45 Thus, in addition to IFN-γ, we evaluated IL-4-secreting cells upon DDV or pVAX1 vaccination via IL-4 ELISpot. Single-cell suspensions were stimulated with the peptide pools (pool 1: EDIII peptide mixture; pool 2: NS1 peptide mixture; pool 3: EDIII and NS1 peptide mixture), and the number of IL-4-producing cells was calculated. While a low number of spots was detected for IL-4 (Figure 6F), DDV-vaccinated animals showed significantly higher counts of IFN-γ spots. Phorbol myristate acetate and ionomycin (PMA/IO) was used as a non-specific positive control. As expected, stimulation with PMA/IO in all ELISpot assays performed with cells from DDV- or pVAX1-vaccinated animals induced high IFN-γ and IL-4 spots.
Figure 5.
Characterization of cellular immune response induced by DENV DNA vaccine in mice
(A) Schedule of vaccination and T cell assays. BALB/c (n = 6/group) and C57BL/6J (n = 5/group) were immunized with 50 μg DENV DNA vaccine or pVAX1 control and sacrificed at 2 weeks after the 2nd booster, and spleens were analyzed for T cell responses by ELISpot and flow cytometry. (B) Map of the DDV and predicted potential immunodominant peptides through NETCTL and VAXIJEN. (C–F) Antigen-specific T responses to pooled EDIII-NS1 and HIV-Nef peptides were measured by IFN-γ ELISpot after vaccination with DDV or pVAX1 in BALB/c (C and D) and C57BL/6J animals (E and F) (pool 1: EDIII peptide mixture; pool 2: NS1 peptide mixture; pool 3; EDIII and NS1 peptide mixture; pool 4: HIV-Nef peptide mixture). (G–J) Flow cytometric analysis of intracellular cytokine staining for IFN-γ in C57BL/6J mice (n = 5/group) splenocytes. (G and H) Representative image of intracellular IFN-γ staining in CD8+ and CD4+ T cells in DDV-vaccinated mice splenocytes, respectively. (I and J) Percentage of IFNγ+ CD8+ and IFNγ+ CD4+ T cells in DDV- and pVAX1-control-vaccinated animals determined through the intracellular cytokine staining (ICC; pool 1: ConA; pool 2: DENV EDIII and NS1 peptides). Data are representative of three independent experiments. Values are depicted are mean ± SD. The percentage of IFN-γ-producing T cells was compared between groups with Student’s unpaired t test; ∗p ≤ 0.05, ∗∗p ≤ 0.005, ∗∗∗p ≤ 0.0005. The study design schematic diagram was created with BioRender.com.
Figure 6.
DDV elicits Th1-biased immune responses
(A) Th1/Th2 assay schedule: BALB/c (n = 5/group) and C57BL/6J mice (n = 4/group) were immunized by TA injection of 50 μg DDV or pVAX1 at days 0, 15, and 30 2 weeks after the 2nd dose. Sera were collected and assayed for IgG subclass antibodies. (B and C) DENV-specific IgG subclasses and the ratio of IgG2a/IgG1 in BALB/c mice immunized with DDV or plasmid control. (D and E) DENV-specific IgG subclasses and the ratio of IgG2c/IgG1 in C57BL/6J mice immunized with DDV or plasmid control. (F) IL-4 responses to pooled EDIII-NS1 peptides were measured by ELISpot after vaccination with DDV or pVAX1 in BALB/c (n = 4/group) (pool 1: EDIII peptide mixture; pool 2; NS1 peptide mixture; pool 3: EDIII and NS1 peptide mixture). (G) Number of IFN-γ- and IL-4-secreting cell ratios. PMA/IO was used as a non-specific positive control. Data are representative of three independent experiments. Values are depicted are mean ± SD. p values were determined by Student’s unpaired t test; ∗∗∗p ≤ 0.0005. #, insignificant. The study design schematic diagram was created with BioRender.com.
We further analyzed intracellular IFN-γ in CD8+ and CD4+ T cells in both DDV and pVAX1 groups of animals. As a positive stimulus for T cell activation, PMA/IO or concanavalin A (ConA) were used. Exocytosis of cytokines was blocked by the addition of brefeldin A (10 μg/mL) during stimulation. Cells were permeabilized, labeled, and fixed for flow cytometry. IFN-γ CD8+ (Figure 5G) and IFN-γ CD4+ (Figure 5H) T cells were proportionately higher upon stimulation with non-specific stimulants as well as with the DENV EDIII and NS1 peptide pool, as found using intracellular staining with flow cytometry, in DDV-vaccinated compared with in pVAX1-vaccinated animals (Figures 5I and 5J). In summary, DENV DNA vaccine candidate DDV induces a robust T cell response in mice.
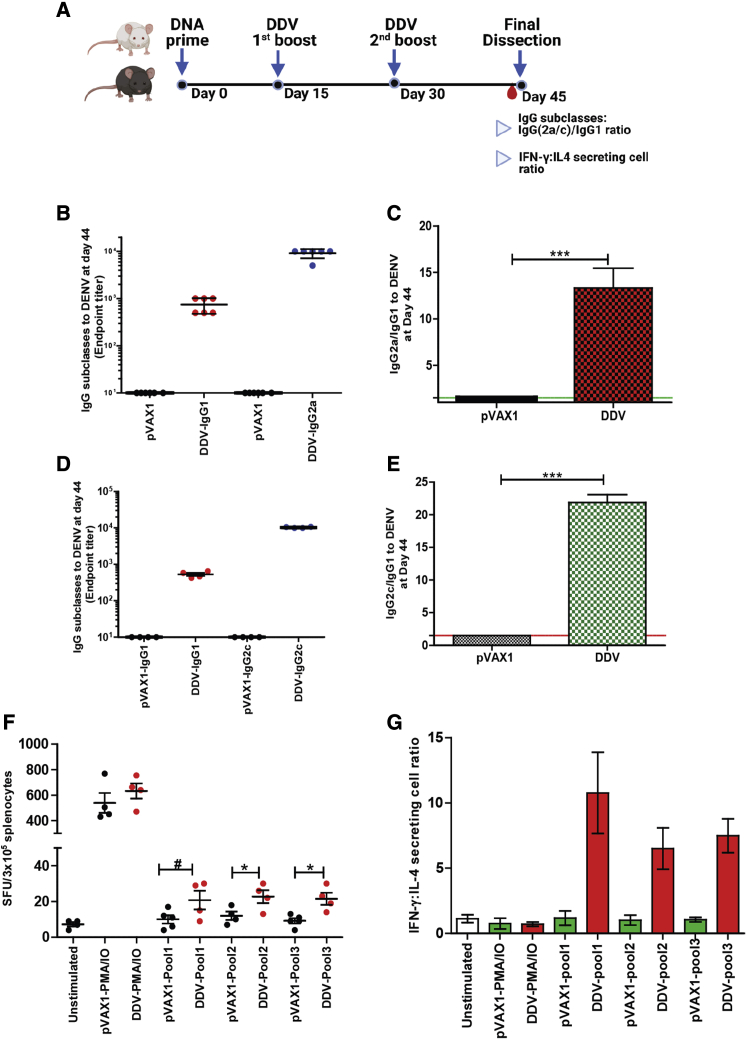
DDV vaccination skewed a Th1-dominant response
Th1 skewness has been shown to elicit a robust adaptive response in terms of cellular activation and antibody production. The induction of polyfunctional Th1 cells is an essential element of a protective vaccine response.46 The vaccine development of respiratory disease viruses such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle Eastern respiratory syndrome (MERS)-CoV have highlighted the importance of a Th1-skewed response in mitigating the risk of vaccine-induced disease enhancement.47 Thus, the Th1/Th2 balance elicited by vaccination with DDV was investigated (Figure 6A). The IgG subclass fate of plasma cells is highly governed by T helper (Th) cells. To determine whether DDV showed a skewing of Th1 over Th2 responses, we measured Th1-associated IgG subclasses IgG2a (BALB/c) (Figures 6B and 6C) and IgG2c (C57BL/6J) (Figures 6D and 6E) against the Th2-associated IgG1. C57BL/6J and BALB/c mice are prototypical Th1 and Th2 animal strains, with C57BL/6J producing high IgG2c and BALB/c producing mostly IgG2a. Hence, in C57BL/6J mice, the evaluation of IgG2c and IFN-γ is critical to correct the interpretation of Th1 immune responses. In both strains of mice, DDV vaccination induced Th1-skewed IgG subclass responses. We further calculated the number of IFN-γ- and IL-4-secreting ELISpot cell ratios to assess Th1/Th2 skewness. The number of IFN-γ-producing cells are ≥6 times higher than the number of IL-4-producing cells in DDV-vaccinated mice stimulated with dengue peptides (Figure 6G). This result is consistent with the observed IgG subclass antibody responses after DDV vaccination. Overall, our findings suggest that the DDV elicits Th1-dominant immune responses.
Immune gene expression following DDV vaccination
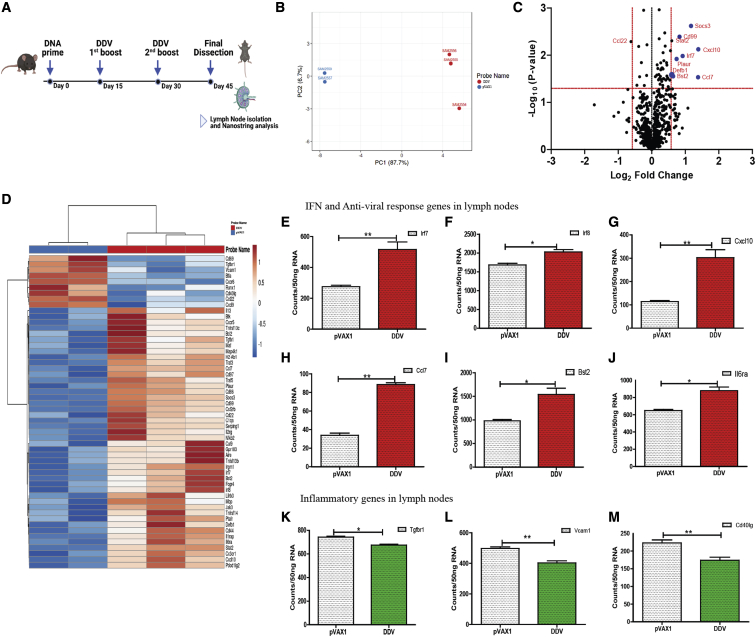
To determine how the DDV exerts its immunogenicity, C57BL/6J mice were vaccinated three times 2 weeks apart with 50 μg of the DDV, and we compared these with equivalent doses of the pVAX1 vector control. Since immune responses develop in germinal centers in draining lymph nodes, the inguinal lymph nodes of mice 2 weeks after the 2nd booster were isolated and analyzed with the Nanostring v.2 immunology panel (Figure 7A). Principal-component analysis (PCA) of immune gene expression showed a clustering of responses to DDV distinct from the pVAX1 plasmid controls, indicating clear differences in immune gene expression following DDV vaccination (Figure 7B).
Figure 7.
Transcriptomic analysis of immune genes after vaccination with DDV
(A) C57BL/6J (n = 5/group) mice were immunized with either pVAX1 or DDV, animals were sacrificed 14 days after 2nd booster dose, and lymph nodes were harvested. Gene expression of immune genes were measured in lymph nodes. (B) Principal-component analysis (PCA) of immune gene expression following vaccination with DDV or pVAX1 control. (C) Volcano plots of fold change of DDV versus pVAX1 control (x axis) and log10 p value of DDV versus pVAX1 control (y axis). A deviation of 1.5 log2 fold change in gene expression was set as the cut-off value. (D) Differentially expressed genes DDV and plasmid control in lymph nodes presented as heatmap of Z scores. (E–M) Lymph node transcriptomic data 14 days after 2nd booster immunization showing Nanostring counts per 50 ng RNA of selected IFN and inflammatory genes. Data are representative of three independent experiments. Values are depicted are mean ± SD. p values were determined by Student’s unpaired t test; ∗p ≤ 0.05, ∗∗p ≤ 0.005. The study design schematic diagram was created with BioRender.com.
Differentially expressed genes were examined in the lymph nodes of mice injected with DDV compared with those administered a similar dose of the pVAX1 plasmid control (Figure 7D). Volcano plot analysis identified a significant enrichment of various innate and adaptive immune responses, lymphocyte activation, cytokine, interferon signaling, and class I antigen presentation genes in DNA-vaccine-immunized animals (Figure 7C). Some of the most highly expressed genes included Cxcl10, Socs3, Ccl7, Plaur, and Defb1, which play roles in directing Th1 and Th2 effector responses. These genes have also been linked to antigen presentation, innate and adaptive immune cell recruitment, T cell stimulation, and dendritic cell maturation in the context of host immunity.48, 49, 50, 51, 52 Furthermore, there are several antiviral defense and IFN-Th1 responsive genes that were also activated upon DDV vaccination. These include Stat2 (IFN signaling), Irf7 (IFN-inducible genes expressed on Th1 cells), Bst2 (development of antiviral T cell distribution and function in addition to augmenting dendritic cell [DC] activation), and Cd99 (Th1-type cytokine response)53, 54, 55 (Figure 7E–7J). In addition, several inflammatory signaling genes such as Tgfbr1, Vcam1, and Cd40lg were downregulated in mice after the DDV vaccination (Figures 7K–7M). These genes have been shown to contribute to inflammatory and autoimmune diseases,56, 57, 58 and their downregulation following vaccination indicates the controlled immune response evoked by the DDV. These findings collectively suggest the development of an immune response in the inguinal lymph nodes of mice immunized with DDV.
Passive transfer of serum from DDV animals protects against lethal DENV2 challenge
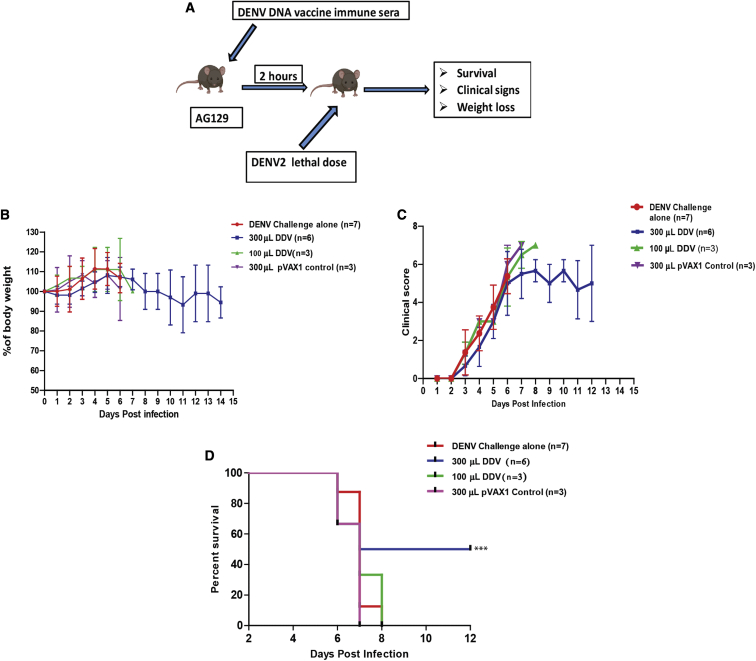
To assess the role of humoral immune responses in mediating protection from DENV challenge, we passively transferred serum from BALB/c mice immunized with either plasmid control or DDV into AG129 mice. Groups of AG129 mice received anti-pVAX1 sera at 300 μL per mouse or anti-DDV immune sera at two dosage levels, 100 and 300 μL per mouse. All mice were challenged 2 h after the passive transfer with a lethal dose of DENV-2 (105 plaque-forming units [PFU]/mouse). The control group did not receive immune sera but were subjected to the lethal challenge dose. All groups were tracked for body-weight changes, clinical signs, and survival for up to 14 days post-challenge (Figure 8A).
Figure 8.
The capacity of anti-DDV immune sera to confer protection against lethal DENV2 challenge in AG129 mice
(A) Schematic representation of the experimental design. Groups of AG129 mice were administered (intraperitoneally [i.p.]) BALB/c immune sera in two dosage levels, 100 (n = 4/group) and 300 (n = 6/group) μL per mouse. The pVAX1 group of animals received a 300 μL dosage (n = 3/group). Two hours after passive transfer, the mice were challenged with a lethal dose of DENV2 (105 FIU/mouse; n = 7/group). (B–D) All groups were monitored for body-weight changes (B), clinical symptoms (C), and survival (D). Data are representative of three independent experiments. Values are depicted are mean ± SD. The study design schematic diagram was created with BioRender.com.
The DENV-infection-only group showed an initial increase in body weight; however, there was a steep decrease from day 5 or 6. The maintenance of body weight or the percentage of increase were observed to be better in 300-μL-immunized mice compared with in the DENV-challenged group and the pVAX1 control group (Figure 8B). The mice started showing noticeable symptoms from the 3rd day post-infection starting with ruffled fur, which continued to become more aggressive and prominent with the disease progression. Despite the initial rise in symptoms, clinical scoring showed a reduced manifestation of the disease in the 300-μL-immunized group at later stages compared with in the infection control and pVAX1 control groups. However, the 100-μL-sera-immunized group did not show any significant difference in clinical symptoms from both the DENV-challenge group and the pVAX1 controls (Figure 8C). In the infection control group, mortality started on the 6th day post-infection, and a 100% mortality was observed by the 8th day post-infection (Figure 8D). The pVAX1 control group showed a similar mortality pattern as the infection control group. On the other hand, in the 300-μL-sera-injected group, three mice survived for at least 12 days post-infection out of the total six mice. The data from the survival curve indicate about a 50% better survival rate for the 300-μL-sera-vaccinated group compared with for the controls. Immunization with 100 μL immune sera failed to provide any advantage in survivability compared with that provided in the infection and pVAX1 control groups.
Discussion
DENV genetic and antigenic differences between genotypes of the same serotype were not considered to impact long-term protective immunity and vaccine efficacy. Several recent studies challenge this assumption. DENV homotypic reinfection, which has been reported in people in Nicaragua and Peru, is potentially driven by genotype differences between primary and secondary infecting viruses.59,60 Some studies have demonstrated that antigenic variation of genotypes can have a high impact on the breadth of antibody neutralization against different genotypes elicited by immune sera from people who have been exposed to natural infections or vaccination.61 Petty et al. and Shrestha et al. demonstrated that monoclonal antibody panels generated against DENV1 genotype II and DENV2 Southeast Asia genotypes have lower neutralizing efficacy against heterotypic genotypes of the same serotypes.62,63 A similar study on genotype cross-reactivity for DENV3 revealed that DENV3 genotype I neutralizing antibodies did not confer effective protection against DENV3 genotype IV.64 Moreover, the limited efficacy of Dengvaxia against DENV2 points toward discrepancies in vaccine-strain- and disease-causing DENV in the clinical trial sites.65 These findings suggest that while selecting vaccine strains, investigators and vaccine developers should consider circulating genotype variants.
Here, we used the whole-genome sequencing approach to investigate the sequence diversity of Indian dengue strains and their distribution patterns. Our sequencing efforts have increased the whole-genome sequences from India by 187%. The serotype distribution data of this study revealed a parallel evolution of India-specific genotypes for all four serotypes of DENV by genetic drift with a continuous exchange among various regions. Despite globalization and frequent traveling, genotypes are contained within the geographical boundaries, with prominent intermixing between neighboring countries. Since the genotypes are bound geographically, the vaccines should be tested at different locations before approval, or they can be restricted to areas that have similar genotypes.
EDIII is a potent immunogen known to elicit serotype-specific antibodies and is less likely to enhance dengue viral infection.66 Previous studies have indicated that the EDIII, rather than EDI or EDII, region has the highest sequence heterogeneity among DENV genotypes.41 Yang et al. and Wahala et al. demonstrated that DENV3-genotype-specific EDIII mutations in the type-specific monoclonal antibody (mAb) (8Ab and 1H9) binding sites lead to significant variation or loss of neutralization efficacy across DENV3 genotypes.67,68 Furthermore, mutation in DENV NS1 has been associated with greater dengue disease severity in mice.69 Sequencing data of our study revealed that there were multiple mutations in EDIII and NS1 antigens, both within and across genotypes of all four serotypes circulating in this region. Particularly, mutations were noticed in the major B cell and T cell epitope regions in both EDIII and NS1 proteins, which could impact the neutralization of viruses and, hence, need further investigation. Furthermore, the likelihood of cross-protection across genotypes following vaccination is debatable.
Despite promising results in preclinical trials, the low immunogenicity of DNA vaccines limits their development for human use.70 In this study, DDV was subjected to codon optimization, RNA optimization, and IgE leader sequence utilization, which was reported to be favorable to increase the immunogenicity in Chikungunya virus (CHIKV),71 Zika,72 MERS,73 and SARS-CoV274,75 DNA vaccines. In proof-of-principle studies, we showed that vaccination of mice with DDV delivered in vivo by intramuscular electroporation (EP) induced robust anti-DENV reactive IgG responses in both BALB/c and C57BL/6J strains. Antibody responses induced by the DDV broadly neutralized the infectivity of DENV1–4 clinical isolates as well as global strains of various genotypes. These data indicate that the polyclonal repertoire of antibodies induced by the DNA vaccine has an adequate breadth of antigenic specificity, encompassing multiple strains within each of the four serotypes. The preclinical results of Takeda’s TAK-003 vaccine showed high titers for DENV1–3, while neutralizing antibody (nAb) titers for DENV4 remained poor.76 This trend is recapitulated in Dengvaxia's non-human primate (NHP) studies where DENV3 has the least amount of titers (plaque reduction neutralization test [PRNT]50 20–640) compared with DENV1 (PRNT50 160–20,480).77 Our data are also consistent with this trend, as the FNT50 for DENV1 and DENV2 is comparatively lower than for DENV3 and DENV4. This could be due to viral or antigenic interference in tetravalent formulations or viral strains employed in the neutralization assay.
Antigen-specific T cell responses live longer than nAbs and provide durable protection, making them critical for DENV vaccine development.78 DDV was found to be capable of inducing antigen-specific T cell responses in mice, as evidenced by the generation of IFN-γ in vaccinated animal's splenocytes. It is well known that IFN signaling plays a key role in priming adaptive T cell responses and directly influences the fate of both CD4+ and CD8+ T cells during the initial phases of antigen presentation, thus shaping the effector and memory T cell pool.79 The production of IFN-γ cytokine has previously been shown to be associated with protection against DENV in experimental animals as well as in humans.80 Overall, our data indicate that antigen-specific T cell immunity is indeed robustly activated following immunization of DDV. Furthermore, like whole-virus-based vaccines, DDV elicits robust humoral and cellular immune responses, demonstrating the advantage of DNA vaccines.
Our results indicate that several innate and adaptive immune genes are induced by DDV and can be used to predict the strength of the immune response. In terms of adaptive immunity, DDV was able to elicit DENV-specific, Th1-predominant responses, as revealed by the IFN-γ and IL-4 ratio and IgG(2a/2c)/IgG1 subtype ratio of antibodies induced by DDV. Th1 skewness has been shown to elicit robust adaptive responses in terms of cellular activation and antibody production, while Th2 cells are largely responsible for generating the humoral response.46 Furthermore, Th1 memory cells, especially CD8+ populations, confer long-term protection through rapid expansion and viral clearance.81 One important advantage of using the DNA EDIII-NS1-based vaccine candidate is that DDV induced predominantly a Th1-biased T cell response, thereby reducing the risk of potential ADE. Additionally, immunological imbalance and exacerbated disease pathology due to Th2 skewness highlight the necessity of Th1-biased immunity in vaccination regimens.81
A nAb response is an established correlate of protection following vaccination against several flaviviruses, including Zika and yellow fever viruses,82,83 and likely contributes to protection following immunization with DDV. Our data indicate that the passive transfer of serum from DDV-immunized mice was sufficient to confer protection against lethal DENV2 challenge in mice. Our data are also comparable with the EDIII virus-like-particle (VLP)-based tetravalent DENV vaccine.20 The passive transfer of the immune serum protected AG129 mice challenged with a virulent, non-mouse-adapted DENV-2 strain, indicating that in spite of the lower neutralization level of the antibodies generated by the vaccine candidate against DENV-2, the antibodies are protective. To our knowledge, there is no epidemiological data on the magnitude of the nAb titers necessary for protection against DENV infection. Critically, it was also observed that the passive transfer of immune sera reduces clinical severity. These data support the quality of EDIII-specific antibodies, which could protect against DENV infection.
In summary, DENV genome surveillance confirms the serotypes and their genotype diversity in India. We have shown that immunization with DDV resulted in robust nAb responses against all four serotypes simultaneously and that the passive transfer of vaccinated immune sera confer protection against lethal DENV challenge. In addition, our DNA vaccine candidate induces multifunctional antigen-specific T cell responses. Taken together, DDV is a promising vaccine candidate that warrants further investigation. Further, our study emphasizes the utility of the consensus sequence approach, in conjunction with DNA delivery, for the development of vaccines against various emerging/re-emerging viruses.
Materials and methods
Samples and ethical consideration
Serum/plasma samples of DENV-NS1-positive patients were obtained from four hospitals in India (viz. St. John's Medical College and Hospital [SJRI], Bangalore; All India Institute of Medical Sciences [AIIMS], Jodhpur; Kasturba Hospital for Infectious Diseases [KHI], Mumbai; and AIIMS, Delhi) during 2012–2018. The Institutional Ethical Clearance Review Boards approved the study of all institutions participating in this study (SJRI, IRB: -236/2016, AIIMS, Jodhpur: AIIMS/IEC/2017/49 and AIIMS/IEC/2019-20/939, KIH, IRB: 07/2018, Ethics/THSTI/2011/2.1; AIIMS: IEC/NP:338/2011). Informed consent was obtained from enrolled patients. Clinical information of patients whose samples were included in this study was recorded in a preformed proforma, which included the patient's demographic details, signs and symptoms of illness, laboratory parameters, and treatment details.
DENV viral RNA sequencing
Serum/plasma were tested for DENV-specific IgM and IgG antibodies using Panbio Dengue IgM and IgG capture ELISA kits, respectively (catalog no. 01PE20/01PE21). Dengue infection was classified as primary or secondary, and further samples were categorized as per WHO 2009 guidelines. According to the manufacturer's instructions, viral RNA was extracted from 150 μL patient serum using QIAamp Viral RNA Mini Kit (Qiagen). Sequencing library preparation was performed with 1.2 ng normalized double-stranded (ds) cDNA using the Illumina Nextera XT sequencing kit and was sequenced on an Illumina Miseq platform, using 2 × 75 and 2 × 150 bp paired-end reads. Raw sequence reads were inspected for quality using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and reads were filtered and trimmed based on the quality score.
Reads were then mapped to the reference sequences obtained from the RefSeq database (NC_001477 [DENV1], NC_001474 [DENV2], NC_001475 [DENV3], and NC_002640 [DENV4]) using Geneious Assembler (3 iterations and medium-low sensitivity). Assembled reads were checked manually for errors. Serotypes were assigned manually, and coverage was assigned based on the following criteria: >10× coverage if the depth of the coverage at each base was >20 and the breadth of the coverage was >95%, 5–10× coverage if depth was <20 and breadth was >95, 1× if depth was <10 and breadth was >90. Consensus sequences were generated from the samples having >5× coverage based on the majority rule and were clipped to the length of reference sequences. iqtree v.1.6 × 1084 was used to construct maximum likelihood trees with 1,000 bootstrap replicates.85 All available DENV complete-coding nucleotide sequences (DENV1: n = 1,800, DENV2: n = 1,395, DENV3: n = 823, DENV4: n = 220) from human hosts were used for tree construction. Sylvatic strains GenBank: EF457905 (for DENV1), GenBank: EF105379 (for DENV2), GenBank: KT424097 (for DENV3), and GenBank: JF262779 and JF262780 (for DENV4) were used as outgroups to root the tree. Genotypes were assigned to our sequences based on their positions in the tree. Figtree v.1.4 (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the tree.86
DENV EDIII and NS1 genetic diversity analysis
Indian DENV sequences specific to the serotypes, including those sequenced during the study, were retrieved from the ViPR database. Sequence alignment was performed using MUSCLE and was visualized in AliView. EDIII and NS1 protein sequences were retained and used for downstream diversity analysis. EDIII and NS1 sequences were compared pairwise against respective DENV reference strains and analyzed for percentage diversity (diversity%) and percentage identity (identity%). Diversity% was given by (no. of variable sites/EDIII or NS1 length) × 100, while identity% was calculated as (100 – diversity%).
FoldX was used to investigate the effect of mutations on DENV-EDIII protein stability. The models were generated for wild-type DENV-EDIII serotypes (GenBank: NP_722460.2, NP_739583.2, YP_001531168.2, NP_740317.1) using PDB: 4GT0, 4UT6, 4GSX, and 5B1C as templates. After repairing the models with the FoldX RepairPDB module, models for each mutation for all four serotypes were generated with the BuildModel module. The Gibbs free energy of folding (in kcal mol−1) was calculated for each mutation and provided a threshold ΔΔG > −2 and ≤ −0.5 (mildly stabilizing), ΔΔG ≥ 0.5 and < 2 (mildly destabilizing) and ΔΔG ≤ −2 or ≥ 2 (stabilizing and destabilizing, respectively).
DDV construction
The polyvalent DDV construct encodes the EDIII of all four serotypes and the NS1 sequence of DENV2. The consensus gene sequences were constructed using the predicted consensus sequences from sequences obtained from our study (DENV1: n = 40, DENV2: n = 48, DENV3: n = 22, DENV4: n = 9) and Indo-Africa-specific sequences available in the ViPR. The Aliview and Geneious tools were used to align and select the consensus amino acid sequence. A consensus was generated from the most frequent residues at each site. Total sequences employed in the generation of the vaccine construct are as follows: DENV1: 182, DENV2: 406, DENV3: 131, and DENV4: 61. Due to the fact that African DENV2 and DENV3 genotypes are identical to Indian genotypes, these sequences were pooled directly with Indian DENV2 and DENV3 strains for consensus generation. Africa DENV1 and DENV4 genotypes differ from those in India; hence, consensus sequences were generated separately and compared with Indian DENV1 and DENV4 strains represented in the vaccine expression cassette. African DENV1 and DENV4 strains exhibited >96% identity with their respective serotype DDV sequences.
The DDV expression cassette was designed with the consensus sequences of the EDIII gene of all four serotypes and the consensus sequences of the NS1 gene of DENV2 linked together in a single construct with furin cleavage sites between the individual genes. Consensus sequences were optimized for EDIII and NS1 expression, including codon and RNA optimization. The Kozak sequences and human IgE leader sequence were added upstream to the DNA sequences, as were furin cleavage sites, to facilitate EDIII and NS1 processing. Finally, the synthetic DENV DNA expression cassette was inserted into the pVAX1 expression vector between NheI and HindIII under the control of the cytomegalovirus immediate-early promoter (Genscript Biotech, Piscataway, NJ, USA).
Epitope prediction and population coverage analysis
Homology modeling was carried out using Modeller tool (v)87, 100 models were predicted, and the model with the lowest discrete optimized protein energy (DOPE) score was used for the analysis. Energy minimization was carried out using Schrodinger module (v). B cell discontinuous epitopes were predicted for the PDB templates and the construct sequences from the DiscoTope webserver (Immune Epitope Database and Analysis Resource [IEDB]) using the default parameters.88 This tool uses the solvent-accessible surface area and contact distances for predicting structural B cell epitopes. T cell epitopes were predicted using NetMHCpan EL 4.1 (MHCI) and a combination of NetMHCIIpan 4.0, NN-align 2.3, and SMMalign (MHCII) from IEDB. The thresholds used were <0.5 for strong binders and <2 for weak binders for MHCI and <2 for strong binders and <10 for weak binders for MHCII.
In vitro RNA expression (qRT-PCR)
In vitro mRNA expression of DDV was demonstrated by the transfection of HEK293T cells with 5 μg plasmids followed by the analysis of the total RNA extracted from the cells using reverse transcription and PCR. The transfection of plasmids was performed using the X-tremeGENE HP DNA transfection reagent (Sigma). The transfection was performed in duplicate. Following 24 h of incubation, the total RNA was extracted using Trizol reagent according to the manufacturer's protocols (Invitrogen). 1 μg was used for reverse transcription using superscript III reverse transcriptase, and qRT-PCR was performed using SYBR green PCR master mix (Applied Biosystems) according to the manufacturer's protocols on the step one Real-Time PCR system (Applied Biosystems). Primers that are specific to the target are as follows: DENV2-EDIII forward 5′-ATCACCGCCAATCCTATC-3′; DENV2-EDIII reverse 5′-GCTCCGATCACGATGTAA-3′; DENV2-NS1 forward 5′-CCATCAAGGACAACAGAG-3′; DENV2-NS1 reverse 5′-CGATCTTCCATGTATCATT-3′; GAPDH forward 5′-CTGGGCTACACTGAGCACC-3′; GAPDH reverse 5′-AAGTGGTCGTTGAGGGCAATG-3′. The following cycling program was used: initial denaturation at 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 60 s, followed by a melt curve step. mRNA expression profiles were normalized to levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in each sample, and the fold change in expression was calculated relative to GAPDH mRNA.
In vitro expression and immunofluorescence assays
HEK293T cells were transfected with DDV using the X-tremeGENE HP DNA transfection reagent (Sigma-Aldrich) as per the manufacturer's instructions. The transfected cell lysate and supernatants were collected 36 h post-transfection, and the antigen expression was confirmed by western blot analysis. Cells were washed with phosphate-buffered saline (PBS) and lysed with NP40 supplemented with protease inhibitor cocktail (Roche), and 1 mM PMSF (Sigma) was used to make cell lysates. Protein lysates were separated on SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane (Bio-Rad), and blocked for 1 h in 5% skimmed milk. Subsequently, membranes were incubated in mouse anti-sera (1:200 dilution) against DDV. Secondary antibodies conjugated to horseradish peroxidase (HRP) were used at a dilution of 1:1,500. After washing with PBS/PBST, the blots were developed using an enhanced chemiluminescence system (Thermo Fisher).
For immunofluorescence, about 105 cells were plated on a coverslip. The next day, cells were fixed in ice-cold methanol and permeabilized with 0.1% Triton X-100 for 10 min, followed by blocking with 1% BSA for 30 min at room temperature (RT). Permeabilized cells were then incubated with anti-sera (1:100) for 1 h at RT and washed three times with 1× PBS, followed by incubation with goat anti-mouse IgG-AF488 at 37°C for 30 min. Cells were again washed three times with 1× PBS, mounted (ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole [DAPI], Invitrogen), air-dried, and visualized using an FV1000 confocal microscope.
Immunization of mice with DDV constructs with electroporation
To determine the immunogenicity of the DDV constructs, mice were immunized with 50 μg DNA in a total volume of 50 μL sterile water by a syringe into the anterior tibialis (TA) muscles then electroporated using BTX ECM 830 with 8 square 40-V electric pulses in alternating directions with a time constant of 0.05 s and an interpulse interval of 1 s. Each group received 2 booster doses at 2-week intervals, and mice were euthanized 2 weeks following the last immunization. All experimental procedures were approved by the Institutional Animal Ethics Committee (IAEC) of National Center for Biological Sciences (NCBS) and TheraIndx Lifesciences (NCBS-IAE-2019/12 (N) and IAEC/10/2019/119).
IgG and IgG subtype binding antibody titers by indirect ELISA
The DENV binding antibody titer was analyzed by indirect ELISA. 96-well plates (Thermo Scientific) were coated with recombinant protein in a coating buffer (0.1 M NaHCO3) and incubated overnight at 4°C. The following day, plates were blocked with 3% BSA in PBS for 2 h at RT. Triplicate samples of serially diluted plasma ranging from 1:100 to 1:500,000 were added to the plate and incubated for 2 h at RT or overnight at 4°C. After washing, secondary anti-IgG (Sigma), anti-IgG1(Invitrogen), and anti-IgG2a (Invitrogen) or IgG2c (Abcam) antibodies conjugated with HRP were added at 1:2,000 (IgG, IgG1, IgG2a) and 1:5,000 (IgG2c) dilution for 1 h at RT. The plates were then developed with 3,3',5,5'-tetramethylbenzidine (TMB) substrate (Sigma) for 15–20 min. The reaction was stopped with a stop buffer (Invitrogen), and the optical density (OD) measured at 450 nm. Cut-off values for each dilution were set using the OD of naive samples in the formula: naive OD at a dilution + (2.5 × standard deviation). Starting from the lowest dilution, the sample dilution prior to the one that was exceeded by the cut-off was considered to be the end titer value.89
FNT
The flow-cytometry-based neutralization assays were performed in triplicate in 96-well cell culture plates with flat-bottom wells. Each well contained 5 × 104 DCSIGN-expressing U937 cells. Immune sera were serially diluted, and the virus was pre-incubated with the sera for 1 h at 37°C. The cells were washed, and the virus and serum mixture was added to the cells for 1 h at 37°C. Next, the wells were filled with cell culture medium, and the plates were incubated for 24 to 48 h at 37°C in 5% CO2. The cells were prepared for flow cytometry analysis by washing them in PBS and transferring them to 96-well plates with round-bottom wells. The cells were fixed and permeabilized by using a Cytofix/Cytoperm kit (BD-PharMingen, San Diego, CA, USA) and stained with mAb 4G2, a mAb that recognizes the flavivirus E protein. The cells were analyzed with a FACScan flow cytometer. The serum dilution that neutralized 50% of the viruses was calculated by non-linear, dose-response regression analysis with Prism 4.0 software (GraphPad Software, San Diego, CA, USA).
IFN-γ and IL-4 ELISpot assays
ELISpot was performed with the Mouse IFN-γ and IL-4 ELISpot Kits (Mabtech IFN-γ-3321-4AST-2; IL-4-3311-4APW-2). In brief, freshly isolated 0.5 M splenocytes/animal were plated into polyvinylidene fluoride (PVDF)-coated 96-well plates containing IFN-γ or IL-4 capture antibodies. Cells were stimulated with immunogenic 9-mer T cell peptides predicted from DENV1–4 EDIII and NS1 antigens (5 μg/mL) and HIV-1 Nef peptide pool (5 μg/mL). Negative control wells contained no peptide. Fifty ng/mL PMA and 500 ng/mL IO or 2 μg/mL ConA were used as a positive control. After overnight stimulation, plates were washed and sequentially incubated with biotinylated IFN-γ detection antibody (R4-6A2), streptavidin-alkaline phosphatase (ALP), and finally 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT). Plates were imaged with ImmunoSpot Analyzer and quantified with ImmunoSpot software.
Intracellular cytokine staining
Intracellular cytokine staining was performed by stimulating freshly isolated splenocytes with PMA and IO or T cell peptides derived from DENV1–4 EDIII and NS1 antigens in the presence of Brefeldin for 5–6 h. After stimulation, surface staining of CD4 and CD8 was performed, followed by intracellular staining of IFN-γ (BioLegend). Data acquisition was performed on a BD LSRFortessa and analyzed with FlowJo.
Immune and inflammatory gene expression profile
Mice were sacrificed 2 weeks post-second immunization, and lymph nodes were collected for analysis of genes involved in immune responses. Lymph nodes were homogenized, and RNA was extracted with TRIzol LS. RNA (50 ng) from whole blood cells and lymph nodes were hybridized to the NanoString nCounter mouse inflammation and immunology v.2 panels (NanoString Technologies), respectively. RNA was hybridized with reconstituted with CodeSet and ProbeSet. Reactions were incubated for 24 h at 65°C and ramped down to 4°C. Hybridized samples were then immobilized onto a nCounter cartridge and imaged on a nCounter SPRINT (NanoString Technologies). Data were analyzed with nSolver Analysis software and PRISM. For normalization, samples were excluded when the percentage field of vision registration was <75, the binding density was outside the range 0.1–1.8, the positive control R2 value was <0.95, and the 0.5 fM positive control was %2 standard deviation (SD) above the mean of the negative controls. Background subtraction was performed by subtracting the estimated background from the geometric means of the raw counts of negative control probes. Probe counts less than the background were floored to a value of 1. The geometric mean of positive controls was used to compute positive control normalization parameters. Samples with normalization factors outside 0.3–3.0 were excluded. The geometric mean of housekeeping genes was used to compute the reference normalization factor. Samples with reference factors outside the 0.10–10.0 range were also excluded. To identify differentially expressing genes (DEGs) between groups, GraphPad Prism software was used to analyze variance with a cutoff of p < 0.05. Log2 fold changes generated were used for volcano plots constructed with Prism 5 software. DEGs were identified by a fold-change cutoff of 1.5. Unsupervised PCA was performed to visualize the variability between DDV and pVAX1 control animals.
Protective efficacy in AG129 mice
AG129 mice (deficient in IFN-α/β and -γ receptors) were obtained from B&K Universal (UK). The mice were bred and maintained under specific-pathogen-free conditions in individually ventilated cages. Three-to-four-week-old AG129 mice were injected intraperitoneally with 100 μL and 300 μL immune serum obtained from vaccinated BALB/c mice. After 2 h of passive immunization, the mice were subjected to DENV challenge with 1 × 104 plaque forming units (PFUs) of DENV-2 strain90 through a subcutaneous route. In the pVAX1 control group, the AG129 mice were passively immunized with serum from the pVAX1 control group of BALB/c mice. Mice with DENV-2 virus challenge alone were kept as the infection control. After infection, each mouse was observed for the development of clinical symptoms and body-weight changes until mortality or the 14th day post-infection. The clinical symptom scoring was done based on the following criteria: score 1: ruffled fur; score 2: 1 + hunched back; score 3: 2 + slow movements or lethargy; score 4: 3 + facial edema; score 5: 4 + facial edema with closed eyes; score 6: 5 + hemorrhage (bloody stool) or limb paralysis; and score 7: death. All experimental procedures were approved by the IAEC of the Rajiv Gandhi Centre for Biotechnology (RGCB) (IAEC/820/SREE/2020).
Statistical analysis
Immunological and virologic data analysi was performed using GraphPad Prism. Kruskal-Wallis or chi-square was used for the clinical data analysis. p values were determined by Student’s unpaired t test; ∗p ≤ 0.05, ∗∗p ≤ 0.005, ∗∗∗p ≤ 0.0005.
Acknowledgments
The authors thank Dr. Krishnamurthy, Ms. Raksha, Dr. Yogesh Chandra, and Dr. Swetha Reddy for their technical assistance. We acknowledge the staff of the BLiSC Animal Care and Resource Centre and TheraIndx Lifesciences Pvt., Ltd., for assistance with animal husbandry and immunizations. We also thank the RGCB central animal facility for carrying out AG129 mice challenge studies. The authors are grateful to the Bioassay lab at THSTI and TheraCUES Innovations Pvt., Ltd., for performing antibody neutralization and nanostring assays, respectively. We would like to thank the BLiSC Central Imaging and Flow Cytometry Facility for help with fluorescence-activated cell sorting (FACS) data curation and image acquisition and the BLiSC Mass spectrometry facility for protein sequence validation. We would also like to thank the C-CAMP sequencing facility and Genotypic India Pvt., Ltd., for the sequencing dengue viruses used in this study. We also want to express our gratitude to the C-CAMP incubation facility for their assistance. S.K acknowledges funding from philanthropist Mr. Narayana Murthy, Co-founder of Infosys (17X6777). S.K also received support from National Centre for Biological Sciences, Tata Institute of Fundamental Research (NCBS-TIFR) planned funds and Department of Biotechnology (DBT), India. E.S thanks the support from RGCB- Intramural grant and G.M acknowledges the TRC-BIRAC grant (BT/NBM0099/02/18). The authors would like to thank NCBS (TIFR) for infrastructural facilities. R.S. acknowledges funding and support provided by JC Bose Fellowship (SB/S2/JC-071/2015) from the Science and Engineering Research Board, India. R.S. and M.I. are thankful for the support from the Bioinformatics Centre Grant funded by the Department of Biotechnology, India (BT/PR40187/BTIS/137/9/2021).
Author contributions
A.S. conceptualized, designed, and characterized the DDV, performed the experiments, analyzed the results, and wrote the manuscript. S.J., R.R., and C.P. conceptualized and conducted the DENV next-generation sequencing (NGS) data analysis and interpretation. J.N. conducted EDIII diversity analysis, performed DDV in vitro mRNA and protein expression, and tested antibody T cell assays. S.-E.-M. conducted animal EP-TA immunizations and blood/organ collections. A.M. conducted AG129 mice challenge studies. J.F. conducted EDIII and NS1 diversity analyses and antibody and T cell assays. M.S. and M.I. conducted structural and HLA analyses under the supervision of R.S. J.S.S., M.D., R.G., A.A., M.V., and S.A. collected DENV samples and clinical data of the samples used in this study. A.P. conducted the DENV sequencing runs. A.N. and A.K. conducted DENV sequencing collected from Bangalore (Batch1) and Delhi sites. S.S.B. and T.D. analyzed and classified clinical data of sequenced samples. M.B. isolated viral RNA and collected clinical data from samples collected at the Bangalore site. B.S. performed plasmid isolation. M.M.M., P.M., and O.A. conducted an analysis of Africa DENV sequences. K.M. provided intellectual input on DNA vaccines and helped in study design. G.R.M. conducted DENV sample collection from the Delhi region, supervised FNT assays, and provided critical comments on the first draft of the manuscript and on the revisions. S.K. conceptualized and designed the DDV, designed the study, supervised and organized the manuscript structure, and acquired funding specific to this study. E.S. conducted and supervised the mouse challenge studies and provided critical comments on the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.01.013.
Contributor Information
Arun Sankaradoss, Email: asankaradoss@ncbs.res.in.
Easwaran Sreekumar, Email: esreekumar@rgcb.res.in.
Supplemental information
References
- 1.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silveira M.M., Moreira G.M.S.G., Mendonça M. DNA vaccines against COVID-19: perspectives and challenges. Life Sci. 2021;267:118919. doi: 10.1016/j.lfs.2020.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barros-Martins J., Hammerschmidt S., Cossmann A., Odak I., Stankov M.V., Ramos G.M., Jablonka A., Heidemann A., Ritter C., Friedrichsen M., Schultze-Florey C.R. Humoral and cellular immune response against SARS-CoV-2 variants following heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. medRxiv. 2021 doi: 10.1101/2021.06.01.21258172. [DOI] [Google Scholar]
- 4.Schmidt T., Klemis V., Schub D., Mihm J., Hielscher F., Marx S., Abu-Omar A., Ziegler L., Guckelmus C., Urschel R., et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat. Med. 2021;2021:1–6. doi: 10.1038/s41591-021-01464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Restifo N., Ying H., Hwang L., Leitner W. The promise of nucleic acid vaccines. Gene Ther. 2000;7:89. doi: 10.1038/sj.gt.3301117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flingai S., Czerwonko M., Goodman J., Kudchodkar S.B., Muthumani K., Weiner D.B. Synthetic DNA vaccines: improved vaccine potency by electroporation and co-delivered genetic adjuvants. Front. Immunol. 2013;4:354. doi: 10.3389/fimmu.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momin T., Kansagra K., Patel H., Sharma S., Sharma B., Patel J., Mittal R., Sanmukhani J., Maithal K., Dey A., Chandra H. Safety and immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine. 2021;38:101020. doi: 10.1016/j.eclinm.2021.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanathan M.P., Kuo Y.C., Selling B.H., Li Q., Sardesai N.Y., Kim J.J., Weiner D.B. Development of a novel DNA SynCon™ tetravalent dengue vaccine that elicits immune responses against four serotypes. Vaccine. 2009;27:6444–6453. doi: 10.1016/j.vaccine.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 9.Pinto P.B.A., Assis M.L., Vallochi A.L., Pacheco A.R., Lima L.M., Quaresma K.R.L., Pereira B.A.S., Costa S.M., Alves A.M.B. T cell responses induced by DNA vaccines based on the DENV2 e and NS1 proteins in mice: importance in protection and immunodominant epitope identification. Front. Immunol. 2019;10:1522. doi: 10.3389/fimmu.2019.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drumond B.P., Mondini A., Schmidt D.J., Bronzoni R.V.D.M., Bosch I., Nogueira M.L. Circulation of different lineages of Dengue virus 2, genotype American/Asian in Brazil: dynamics and molecular and phylogenetic characterization. PLoS ONE. 2013;8:e59422. doi: 10.1371/journal.pone.0059422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manakkadan A., Joseph I., Prasanna R.R., Kailas L., Sreekumar E. Lineage shift in Indian strains of Dengue virus serotype-3 (Genotype III), evidenced by detection of lineage IV strains in clinical cases from Kerala. Virol. J. 2013;10:1–11. doi: 10.1186/1743-422X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahamed S.F., Rosario V., Britto C., Dias M., Nayak K., Chandele A., Kaja M.K., Shet A. Emergence of new genotypes and lineages of dengue viruses during the 2012–15 epidemics in southern India. Int. J. Infect. Dis. 2019;84:S34–S43. doi: 10.1016/j.ijid.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Dias M., Pattabiraman C., Siddappa S., Gowda M., Shet A., Smith D., Muehlemann B., Tamma K., Solomon T., Jones T., Krishna S. Complete assembly of a dengue virus type 3 genome from a recent genotype III clade by metagenomic sequencing of serum. Wellcome Open Res. 2018;3:44. doi: 10.12688/wellcomeopenres.14438.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kar M., Nisheetha A., Kumar A., Jagtap S., Shinde J., Singla M., Saranya M., Pandit A., Chandele A., Kabra S.K., Krishna S. Isolation and molecular characterization of dengue virus clinical isolates from pediatric patients in New Delhi. Int. J. Infect. Dis. 2019;84:S25–S33. doi: 10.1016/j.ijid.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alagarasu K., Patil J.A., Kakade M.B., More A.M., Yogesh B., Newase P., Jadhav S.M., Harmanmeet K., Nivedita G., Neetu V., et al. Serotype and genotype diversity of dengue viruses circulating in India: a multi-centre retrospective study involving the Virus Research Diagnostic Laboratory Network in 2018. Int. J. Infect. Dis. 2021;111:242–252. doi: 10.1016/j.ijid.2021.08.045. [DOI] [PubMed] [Google Scholar]
- 17.Shrivastava S., Tiraki D., Diwan A., Lalwani S.K., Modak M., Mishra A.C., Arankalle V.A. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS ONE. 2018;13:e0192672. doi: 10.1371/journal.pone.0192672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization Dengue vaccine: WHO position paper, September 2018-Recommendations. Vaccine. 2019;37:4848–4849. doi: 10.1016/j.vaccine.2018.09.063. [DOI] [PubMed] [Google Scholar]
- 19.Fahimi H., Mohammadipour M., Haddad Kashani H., Parvini F., Sadeghizadeh M. Dengue viruses and promising envelope protein domain III-based vaccines. Appl. Microbiol. Biotechnol. 2018;1027:2977–2996. doi: 10.1007/s00253-018-8822-y. [DOI] [PubMed] [Google Scholar]
- 20.Ramasamy V., Arora U., Shukla R., Poddar A., Shanmugam R.K., White L.J., Mattocks M.M., Raut R., Perween A., Tyagi P., et al. A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice. Plos Negl. Trop. Dis. 2018;12:e0006191. doi: 10.1371/journal.pntd.0006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H.W., Hu H.M., Wu S.H., Chiang C.Y., Hsiao Y.J., Wu C.K., Hsieh C.H., Chung H.H., Chong P., Leng C.H., Pan C.H. The immunodominance change and protection of CD4+ T-cell responses elicited by an envelope protein domain III-based tetravalent dengue vaccine in mice. PLoS ONE. 2015;10:e0145717. doi: 10.1371/journal.pone.0145717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallichotte E.N., Young E.F., Baric T.J., Yount B.L., Metz S.W., Begley M.C., De Silva A.M., Baric R.S. Role of zika virus envelope protein domain III as a target of human neutralizing antibodies. MBio. 2019;10:e01485. doi: 10.1128/mBio.01485-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C.W., Wu S.C. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 2003;77:2600–2606. doi: 10.1128/JVI.77.4.2600-2606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gromowski G.D., Roehrig J.T., Diamond M.S., Lee J.C., Pitcher T.J., Barrett A.D. Mutations of an antibody binding energy hot spot on domain III of the dengue 2 envelope glycoprotein exploited for neutralization escape. Virology. 2010;407:237–246. doi: 10.1016/j.virol.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 25.Chen H.R., Lai Y.C., Yeh T.M. Dengue virus non-structural protein 1: a pathogenic factor, therapeutic target, and vaccine candidate. J. Biomed. Sci. 2018;25:1–11. doi: 10.1186/s12929-018-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan S.W., Lu Y.T., Huang C.H., Lin C.F., Anderson R., Liu H.S., Yeh T.M., Yen Y.T., Wu-Hsieh B.A., Lin Y.S. Protection against dengue virus infection in mice by administration of antibodies against modified nonstructural protein 1. PLoS ONE. 2014;9:e92495. doi: 10.1371/journal.pone.0092495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubor-Bauk B., Wijesundara D.K., Masavuli M., Abbink P., Peterson R.L., Prow N.A., Larocca R.A., Mekonnen Z.A., Shrestha A., Eyre N.S., et al. NS1 DNA vaccination protects against Zika infection through T cell–mediated immunity in immunocompetent mice. Sci. Adv. 2019;5:eaax2388. doi: 10.1126/sciadv.aax2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beatty P.R., Puerta-Guardo H., Killingbeck S.S., Glasner D.R., Hopkins K., Harris E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 2015;7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 29.Lai Y.C., Chuang Y.C., Liu C.C., Ho T.S., Lin Y.S., Anderson R., Yeh T.M. Antibodies against modified NS1 wing domain peptide protect against dengue virus infection. Sci. Rep. 2017;7:1. doi: 10.1038/s41598-017-07308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weaver E.A., Rubrum A.M., Webby R.J., Barry M.A. Protection against divergent influenza H1N1 virus by a centralized influenza hemagglutinin. PLoS ONE. 2011;6:e18314. doi: 10.1371/journal.pone.0018314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyerhoff R.R., Scearce R.M., Ogburn D.F., Lockwood B., Pickeral J., Kuraoka M., Anasti K., Eudailey J., Eaton A., Cooper M., et al. HIV-1 consensus envelope-induced broadly binding antibodies. AIDS Res. Hum. Retroviruses. 2017;33:859–868. doi: 10.1089/aid.2016.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R., Zheng X., Sun J., Feng K., Gao N., Fan D., Chen H., Jin X., An J. Vaccination with a single consensus envelope protein ectodomain sequence administered in a heterologous regimen induces tetravalent immune responses and protection against dengue viruses in mice. Front. Microbiol. 2019;0:1113. doi: 10.3389/fmicb.2019.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.H., Grey H., Sette A. A consensus epitope prediction approach identifies the breadth of murine T CD8+-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 34.Singla M., Kar M., Sethi T., Kabra S.K., Lodha R., Chandele A., Medigeshi G.R. Immune response to dengue virus infection in pediatric patients in New Delhi, India—association of viremia, inflammatory mediators and monocytes with disease severity. Plos Negl. Trop. Dis. 2016;10:e0004497. doi: 10.1371/journal.pntd.0004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shastri J., Williamson M., Vaidya N., Agrawal S., Shrivastav O. Nine year trends of dengue virus infection in Mumbai, Western India. J. Lab. Physicians. 2017;9:296–302. doi: 10.4103/JLP.JLP_169_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masika M.M., Korhonen E.M., Smura T., Uusitalo R., Vapalahti K., Mwaengo D., Jääskeläinen A.J., Anzala O., Vapalahti O., Huhtamo E. Detection of dengue virus type 2 of Indian origin in acute febrile patients in rural Kenya. Plos Negl. Trop. Dis. 2020;14:e0008099. doi: 10.1371/journal.pntd.0008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renner M., Flanagan A., Dejnirattisai W., Puttikhunt C., Kasinrerk W., Supasa P., Wongwiwat W., Chawansuntati K., Duangchinda T., Cowper A., et al. Characterization of a potent and highly unusual minimally-enhancing antibody directed against dengue virus. Nat. Immunol. 2018;19:1248. doi: 10.1038/s41590-018-0227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahala W.M.P.B., Donaldson E.F., Alwis R., Accavitti-Loper M.A., Baric R.S., Silva A.M. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLOS Pathog. 2010;6:e1000821. doi: 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y., Austin S.K., Fremont D.H., Yount B.L., Huynh J.P., de Silva A.M., Baric R.S., Messer W.B. The mechanism of differential neutralization of dengue serotype 3 strains by monoclonal antibody 8A1. Virology. 2013;439:57–64. doi: 10.1016/j.virol.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukupolvi-Petty S., Brien J.D., Austin S.K., Shrestha B., Swayne S., Kahle K., Doranz B.J., Johnson S., Pierson T.C., Fremont D.H., et al. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J. Virol. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao D.Y., King C.C., Wang W.K., Chen W.J., Wu H.L., Chang G.J.J. Strategically examining the full-genome of dengue virus type 3 in clinical isolates reveals its mutation spectra. Virol. J. 2005;2:1–10. doi: 10.1186/1743-422X-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roehrig J.T., Hombach J., Barrett A.D. Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 2008;21:123–132. doi: 10.1089/vim.2008.0007. [DOI] [PubMed] [Google Scholar]
- 43.Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., Kennedy K. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diamond M.S., Roberts T.G., Edgil D., Lu B., Ernst J., Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 2000;74:4957. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma D.P., Ramsay A.J., Maguire D.J., Rolph M.S., Ramshaw I.A. Interleukin-4 mediates down regulation of antiviral cytokine expression and cytotoxic T-lymphocyte responses and exacerbates vaccinia virus infection in vivo. J. Virol. 1996;70:7103. doi: 10.1128/jvi.70.10.7103-7107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenthal K.S., Zimmerman D.H. Vaccines: all things considered. Clin. Vaccin. Immunol. 2006;13:821–829. doi: 10.1128/CVI.00152-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., Yau C., Low J.G., Kalimuddin S., Matsuda D., Allen E.C. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021;29:1970–1983. doi: 10.1016/j.ymthe.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krathwohl M.D., Anderson J.L. Chemokine CXCL10 (IP-10) is sufficient to trigger an immune response to injected antigens in a mouse model. Vaccine. 2006;24:2987–2993. doi: 10.1016/j.vaccine.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 49.Inoue H., Kubo M. SOCS proteins in T helper cell differentiation: implications for allergic disorders? Expert Rev. Mol. Med. 2004;6:1. doi: 10.1017/S1462399404008348. [DOI] [PubMed] [Google Scholar]
- 50.Ford J., Hughson A., Lim K., Bardina S.V., Lu W., Charo I.F., Lim J.K., Fowell D.J. CCL7 is a negative regulator of cutaneous inflammation following Leishmania major infection. Front. Immunol. 2019;10:3063. doi: 10.3389/fimmu.2018.03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahmood N., Mihalcioiu C., Rabbani S.A. Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front. Oncol. 2018;8:24. doi: 10.3389/fonc.2018.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andresen E., Günther G., Bullwinkel J., Lange C., Heine H. Increased expression of beta-defensin 1 (DEFB1) in chronic obstructive pulmonary disease. PLoS ONE. 2011;6:e21898. doi: 10.1371/journal.pone.0021898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loyet K.M., Ouyang W., Eaton D.L., Stults J.T. Proteomic profiling of surface proteins on Th1 and Th2 Cells. J. Proteome Res. 2005;4:400–409. doi: 10.1021/pr049810q. [DOI] [PubMed] [Google Scholar]
- 54.Urata S., Kenyon E., Nayak D., Cubitt B., Kurosaki Y., Yasuda J., de la Torre J.C., McGavern D.B. BST-2 controls T cell proliferation and exhaustion by shaping the early distribution of a persistent viral infection. Plos Pathog. 2018;14:e1007172. doi: 10.1371/journal.ppat.1007172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salem M., Mony J.T., Løbner M., Khorooshi R., Owens T. Interferon regulatory factor-7 modulates experimental autoimmune encephalomyelitis in mice. J. Neuroinflammation. 2011;8:1. doi: 10.1186/1742-2094-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vander Ark A., Cao J., Li X. TGF-β receptors: in and beyond TGF-β signaling. Cell Signal. 2018;52:112–120. doi: 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Kong D.H., Kim Y.K., Kim M.R., Jang J.H., Lee S. Emerging roles of vascular cell adhesion molecule-1 (VCAM-1) in immunological disorders and cancer. Int. J. Mol. Sci. 2018;19:1057. doi: 10.3390/ijms19041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peters A.L., Stunz L.L., Bishop G.A. October. CD40 and autoimmunity: the dark side of a great activator. Semin. Immunol. 2009;21:293–300. doi: 10.1016/j.smim.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waggoner J.J., Balmaseda A., Gresh L., Sahoo M.K., Montoya M., Wang C., Abeynayake J., Kuan G., Pinsky B.A., Harris E. Homotypic dengue virus reinfections in nicaraguan children. J. Infect. Dis. 2016;214:986–993. doi: 10.1093/infdis/jiw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forshey B.M., Reiner R.C., Olkowski S., Morrison A.C., Espinoza A., Long K.C., Vilcarromero S., Casanova W., Wearing H.J., Halsey E.S., et al. Incomplete protection against dengue virus type 2 re-infection in Peru. Plos Negl. Trop. Dis. 2016;10:e0004398. doi: 10.1371/journal.pntd.0004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinez D.R., Yount B., Nivarthi U., Munt J.E., Delacruz M.J., Whitehead S.S., Durbin A.P., de Silva A.M., Baric R.S. Antigenic variation of the dengue virus 2 genotypes impacts the neutralization activity of human antibodies in vaccinees. Cell Rep. 2020;33:108226. doi: 10.1016/j.celrep.2020.108226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sukupolvi-Petty S., Austin S.K., Engle M., Brien J.D., Dowd K.A., Williams K.L., Johnson S., Rico-Hesse R., Harris E., Pierson T.C., et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 2010;84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shrestha B., Brien J.D., Sukupolvi-Petty S., Austin S.K., Edeling M.A., Kim T., O'Brien K.M., Nelson C.A., Johnson S., Fremont D.H., Diamond M.S. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brien J.D., Austin S.K., Sukupolvi-Petty S., O’Brien K.M., Johnson S., Fremont D.H., Diamond M.S. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J. Virol. 2010;84:10630. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juraska M., Magaret C.A., Shao J., Carpp L.N., Fiore-Gartland A.J., Benkeser D., Girerd-Chambaz Y., Langevin E., Frago C., Guy B., et al. Viral genetic diversity and protective efficacy of a tetravalent dengue vaccine in two phase 3 trials. Proc. Natl. Acad. Sci. U S A. 2018;115:E8378–E8387. doi: 10.1073/pnas.1714250115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frei J.C., Wirchnianski A.S., Govero J., Vergnolle O., Dowd K.A., Pierson T.C., et al. Engineered dengue virus domain III proteins elicit cross-neutralizing antibody responses in mice. J. Virol. 2018;92 doi: 10.1128/JVI.01023-18. e01023–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Y., Austin S.K., Fremont D.H., Yount B.L., Huynh J.P., Silva A.M., Baric R.S., Messer W.B. The mechanism of differential neutralization of dengue serotype 3 strains by monoclonal antibody 8A1. Virology. 2013;439:57. doi: 10.1016/j.virol.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wahala W.M.P.B., Donaldson E.F., De Alwis R., Accavitti-Loper M.A., Baric R.S., De Silva A.M. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. Plos Pathog. 2010;6:1000821. doi: 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan K.W.K., Watanabe S., Jin J.Y., Pompon J., Teng D., Alonso S., Vijaykrishna D., Halstead S.B., Marzinek J.K., Bond P.J., et al. A T164S mutation in the dengue virus NS1 protein is associated with greater disease severity in mice. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aat7726. [DOI] [PubMed] [Google Scholar]
- 70.Hobernik D., Bros M. DNA vaccines—how far from clinical use? Int. J. Mol. Sci. 2018;19:3605. doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muthumani K., Lankaraman K.M., Laddy D.J., Sundaram S.G., Chung C.W., Sako E., Wu L., Khan A., Sardesai N., Kim J.J., et al. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine. 2008;26:5128–5134. doi: 10.1016/j.vaccine.2008.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tebas P., Roberts C.C., Muthumani K., Reuschel E.L., Kudchodkar S.B., Zaidi F.I., White S., Khan A.S., Racine T., Choi H., et al. Safety and immunogenicity of an anti–zika virus DNA vaccine — preliminary report. N. Engl. J. Med. 2017 doi: 10.1056/NEJMoa1708120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., Reuschel E.L., Robb M.L., Racine T., Oh M., et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dey A., Rajanathan T.M.C., Chandra H., Pericherla H.P.R., Kumar S., Choonia H.S., Bajpai M., Singh A.K., Sinha A., Saini G., et al. Immunogenic potential of DNA vaccine candidate, ZyCoV-D against SARS-CoV-2 in animal models. Vaccine. 2021;39:4108. doi: 10.1016/j.vaccine.2021.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osorio J.E., Brewoo J.N., Silengo S.J., Arguello J., Moldovan I.R., Tary-Lehmann M., Powell T.D., Livengood J.A., Kinney R.M., Huang C.Y.-H., et al. Efficacy of a tetravalent chimeric dengue vaccine (DENVax) in cynomolgus macaques. Am. J. Trop. Med. Hyg. 2011;84:978. doi: 10.4269/ajtmh.2011.10-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guirakhoo F., Pugachev K., Zhang Z., Myers G., Levenbook I., Draper K., Lang J., Ocran S., Mitchell F., Parsons M., et al. Safety and efficacy of chimeric yellow fever-dengue virus tetravalent vaccine formulations in nonhuman primates. J. Virol. 2004;78:4761. doi: 10.1128/JVI.78.9.4761-4775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hellerstein M. What are the roles of antibodies versus a durable, high quality T-cell response in protective immunity against SARS-CoV-2? Vaccin. X. 2020;6:100076. doi: 10.1016/j.jvacx.2020.100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huber J.P., David Farrar J. Regulation of effector and memory T-cell functions by type I interferon. Immunology. 2011;132:466–474. doi: 10.1111/j.1365-2567.2011.03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costa V.V., Fagundes C.T., Souza D.G., Teixeira M.M. Inflammatory and innate immune responses in dengue infection: protection versus disease induction. Am. J. Pathol. 2013;182:1950–1961. doi: 10.1016/j.ajpath.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 81.Foulds K.E., Wu C.Y., Seder R.A. Th1 memory: implications for vaccine development. Immunol. Rev. 2006;211:58–66. doi: 10.1111/j.0105-2896.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- 82.Wieten R.W., Jonker E.F.F., Leeuwen E.M.M., Remmerswaal E.B.M., Berge I.J.M., Visser A.W., Genderen P.J.J., Goorhuis A., Visser L.G., Grobusch M.P., et al. A single 17D yellow fever vaccination provides lifelong immunity; characterization of yellow-fever-specific neutralizing antibody and T-cell responses after vaccination. PLoS ONE. 2016;11:e0149871. doi: 10.1371/journal.pone.0149871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maciejewski S., Ruckwardt T.J., Morabito K.M., Foreman B.M., Burgomaster K.E., Gordon D.N., Pelc R.S., DeMaso C.R., Ko S.-Y., Fisher B.E., et al. Distinct neutralizing antibody correlates of protection among related Zika virus vaccines identify a role for antibody quality. Sci. Transl. Med. 2020;12:9066. doi: 10.1126/scitranslmed.aaw9066. [DOI] [PubMed] [Google Scholar]
- 84.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rambaut A. Institute of Evolutionary Biology, University of Edinburgh; 2010. FigTree v1.3.1.https://www.scirp.org/(S(lz5mqp453edsnp55rrgjct55))/reference/ReferencesPapers.aspx?ReferenceID=1661474 [Google Scholar]
- 87.Malavige G.N., McGowan S., Atukorale V., Salimi M., Peelawatta M., Fernando N., Jayaratne S.D., Ogg G. Identification of serotype-specific T cell responses to highly conserved regions of the dengue viruses. Clin. Exp. Immunol. 2012;168:215. doi: 10.1111/j.1365-2249.2012.04566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012;8:e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bagarazzi M.L., Yan J., Morrow M.P., Shen X., Parker R.L., Lee J.C., Giffear M., Pankhong P., Khan A.S., Broderick K.E., Knott C. Immunotherapy against HPV16/18 generates potent TH1 and cytotoxic cellular immune responses. Sci. Translational Med. 2012;4:155ra138. doi: 10.1126/scitranslmed.3004414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh S., Anupriya M.G., Sreekumar E. Comparative whole genome analysis of dengue virus serotype-2 strains differing in trans-endothelial cell leakage induction in vitro. Infect. Genet. Evol. 2017;52:34–43. doi: 10.1016/j.meegid.2017.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.