Abstract
The novel coronavirus disease (COVID-19), which emerged in Wuhan, China, is continuously spreading worldwide, creating a huge burden on public health and economy. Ayurveda, the oldest healing schema of Traditional Indian Medicinal (TIM) system, is considered as a promising CAM therapy to combat various diseases/ disorders. To explore the regulatory mechanisms of 3038 Ayurvedic herbs (AHs) against SARS-CoV-2, in this study, multi-targeting and synergistic actions of constituent 34,472 phytochemicals (APCs) are investigated using a comprehensive approach comprising of network pharmacology and molecular docking. Immunomodulatory prospects of antiviral drug-alike potentially effective phytochemicals (PEPs) are presented as a special case study, highlighting the importance of 6 AHs in eliciting the antiviral immunity. By evaluating binding affinity of 292 PEPs against 24 SARS-CoV-2 proteins, we develop and analyze a high-confidence “bi-regulatory network” of 115 PEPs having ability to regulate protein targets in both virus and its host human system. Furthermore, mechanistic actions of PEPs against cardiovascular complications, diabetes mellitus and hypertension are also investigated to address the regulatory potential of AHs in dealing with COVID-19-associated metabolic comorbidities. The study further reports 12 PEPs as promising source of COVID-19 comorbidity regulators.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10352-x.
Keywords: Ayurveda, COVID-19, SARS-CoV-2, Network pharmacology, Immunomodulators, Comorbidity, Antiviral drugs
Introduction
In December 2019, a novel coronavirus caused an outbreak of pneumonia in Wuhan, Hubei Province of China [1], Since then, it has rapidly transmitted across the world [2], leading to the situation of Public Health Emergency of International Concern (PHEIC). The pathogen for leading coronavirus related pneumonia disease (COVID-19) has been classified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses. Compared to SARS-CoV responsible for the outbreak of SARS in 2003, the current risk of COVID-19 pandemic is mainly due to the high-transmission rate of SARS-CoV-2.
The concept of drug repurposing has become an attractive proposition for the identification of potentially active drugs against various diseases. With the time-consuming process, substantial costs and high failure rates of the development of new drugs, the reuse of existing drugs for other diseases offers an attractive schema for its lower developmental costs and shorter developmental timeline. The notion of drug repurposing is based on the multi-targeting ability of drugs which can be used to deal with various other diseases as disease pathogenesis is multi-factorial in nature [3]. The concept has been used for past several years to repurpose existing drugs against various other diseases than the disease they have been originally developed for [4, 5]. For COVID-19 also, the concept has been exploited to suggest potential existing drugs as there is an urgent requirement of drugs (single or combination based) to combat the disease. Recently, the antiviral drug repurposing approach has been implicated to a great extent to deal with SARS-CoV-2 [6].
In response to the current demand for a suitable vaccine, the research community has jumped into the race to find a cure. To find an answer to that, China has turned its way toward traditional therapies by promoting TCM (Traditional Chinese Medicine) as a common prescription against COVID-19 [7, 8]. The underlying mechanism lies in the multi-targeting nature of natural herbs that in addition to providing strong immunity support targets various ribosomal proteins, thereby inhibiting the viral replication event [7]. Ministry of AYUSH, Government of India, has also issued an advisory to use the Ayurveda, Siddha, Unani and Homeopathy as preventive measures [9]. Ayurveda, the traditional Indian knowledgebase of TIM system (Traditional Indian Medicine) which translates to "knowledge of life," is considered as the oldest healing schema originated more than four thousand years ago. Historical background of these medicines is also supporting the use of this system of medicine as preventive measures against variety of diseases and disorders including viral infection [10]. However, there are no controlled supporting data available for the use of any of these traditional medicines, and their efficacy for COVID-19 is unknown. Hence, the research scope of Ayurvedic medicines with valid scientific evidence is much worthy to combat the pandemic of COVID-19.
In recent years, a novel paradigm that integrates the concepts of network science and pharmacology, namely network pharmacology, has made its headway in the research of drug discovery and development [11]. The approach of network pharmacology has proved to be a promising strategy toward next-generation approach of drug discovery for traditional medicines [12, 13]. In this study, the information of Ayurvedic herbs was collected for their phytochemical composition and studied for their efficacies against COVID-19 using the approach of network pharmacology. A comprehensive dataset of phytochemicals was prepared for each herb utilizing the information available at public domain databases. The therapeutic relevance of the phytochemicals was estimated using several protein target prediction algorithms. The prioritization of phytochemicals effective in managing COVID-19 was performed using the multi-step strategy involving similarity analysis with antiviral drugs, binding affinity analysis against SARS-CoV-2 proteins, immune regulatory potential, comorbidity analyses, etc. We believe that the comprehensive methodology adopted in this study can serve as a powerful tool in deciphering the possible mechanism of action of Ayurvedic herbs of TIM origin for their management toward the global pandemic caused by novel coronavirus. Furthermore, the study may also serve as a universal guide toward illuminating the mechanisms of prescription of TIM against various other diseases and disorders. The complete workflow of the study is presented in Fig. 1.
Fig. 1.
Detailed workflow of the present study
Material and methods
Dataset of Ayurvedic herbs
The information of the Ayurvedic herbs was collected from Indian medicinal plants database (IMPD) (http://www.medicinalplants.in/), which enlists the information of 7258 unique herbs used in Indian medicinal system of Ayurveda as on March 2018. The scientific names of the herbs available at IMPD can be checked in Supplementary Table 1.
Phytochemical dataset of Ayurvedic herbs
A comprehensive list of the phytochemicals present in each Ayurvedic herb was developed using six database sources IMPPAT (Indian Medicinal Plants, Phytochemistry And Therapeutics) [14], TCM-MeSH [15], PCIDB (PhytoChemical Interactions DB) (https://www.genome.jp/db/pcidb), NPASS (Natural Product Activity and Species Source database) [16], CMAUP (Collective Molecular Activities of Useful Plants) [17] and Duke’s phytochemical database (https://phytochem.nal.usda.gov/phytochem/search). For this, the genus and species name of each herb was selected and inspected for their presence in the aforementioned databases. Out of 7258 herbs in IMPD, we considered only those herbs in this study for which we could identify at least one phytochemical in the aforementioned databases. Two chemical databases, namely PubChem [18] and ChEMBL [19], were used to map the phytochemicals for their chemical information. The Ayurvedic phytochemicals (APCs) for which no chemical mapping could be obtained were not considered in this study. Following these steps, a dataset of 3038 herbs and their varieties was prepared and used in further studies.
The hierarchical chemical classification of APCs was performed using “Classyfire” which utilizes the chemical ontology-based information of 4,825 organic and inorganic compounds to predict the chemical class of query molecule [20]. For clustering of APCs, cluster services available at ChemMine tools were chosen [21]. The ChemMine-algorithm was used to calculate atom pair descriptors (i.e., features) of each subjected query compound. Using the set of unique and common features, a similarity matrix was constructed, and the matrix was presented in the Newick tree format. The obtained tree format was displayed using iTOL (Interactive Tree Of Life), an interactive tree viewer and annotator. iTOL offers an online service which supports multiple phylogenetic tree formats for their display, annotation and manipulation [22]. The chemical information obtained from Classyfire server was also added to the tree format to display complete information associated with each APC molecule. The chemical classification was restricted to the APCs screened-in at the stage of “Anti-viral drug similarity calculations” (described in detail in the Material and Methods Section “Anti-viral drug dataset and similarity index calculation”).
Protein target identification of phytochemicals
The information of human proteins targeted by APCs was compiled from STICH5.0, SwissTargetPrediction and BindingDB. STITCH utilizes the information of manually curated and experimental data for cataloguing chemical target pairs [23]. For accessing high-confidence interaction pairs, the STITCH data were compiled at the confidence score of ≥ 0.4. SwissTargetPrediction is accessible through a web-based tool available at http://www.swisstargetprediction.ch/ and offers predictions based on similarity principle through reverse screening approach [24]. For each APC, only top-15 predictions from SwissTargetPrediction were incorporated for the analysis. BindingDB is a web-accessible public platform containing the binding information of about 7493 proteins and 820,433 chemical entities [25]. BindingDB RESTful API services specific to “getTargetByCompound” were used to derive the information of protein targets. The service allows the user to screen the compound of interest for other compounds in BindingDB of similar structure within the user-defined similarity cutoff. For each similar compounds returned, the information of their binding targets was also included. For the study, the targets were screened corresponding to molecules having chemical similarity ≥ 0.85.
Anti-viral drug dataset and similarity index calculation
DrugBank database (https://www.drugbank.ca/) was used to collect the information of currently used antiviral drugs (AVDs). Only AVDs corresponding to the class of small molecules were used in this study. For assessing the similarity between AVDs and APCs, a similarity measure based on Tanimoto coefficient (Tc) was calculated for each pair of 34,472 APCs and 125 AVDs. For the calculation, the chemical structure of input molecule was encoded in binary digits using molecular fingerprints. A path-based molecular fingerprint, namely FP2 which indexes the input molecule up to the length of seven atoms, was used for Tc calculation using OpenBabel [26]. Tc between two chemical compounds A and B is given by
where and represent the number of molecular fingerprints associated with chemical compounds A and B, respectively. The number of molecular fingerprints common to both the chemical compounds is represented by [27]. The value of the ranges in between 0 and 1, with 0 representing no similarity and 1 representing maximum similarity between the compounds. The Tc values between APCs and AVDs (obtained from DrugBank) are listed in Supplementary Table 2. The information of AVDs is also given in Supplementary Table 2.
To screen APCs capable of providing similar regulatory effects to existing antiviral drugs, the two-condition-based selection criterion was adopted [28]. Of these two conditions, first involves the selection of APCs whose Tc similarity is greater than 0.85, and the second one includes the APCs whose Tc value ≠ 1 and SMILES exactly similar against any of the 125 AVDs. Using this criterion, 292 APCs referred to as “potentially effective phytochemicals” (PEPs) could be identified.
Disease association of the protein targets
DisGeNET, a repository containing the information of gene–disease associations linked to Homo sapiens, was used to investigate the association of protein targets into various disease classes [29].
In silico molecular docking and interaction analysis
Molecular docking and binding energy (B.E.) calculations were used to assess the favorable conformation of ligand onto the protein active site. The 3D structures of 24 SARS-CoV-2 proteins were obtained from the I-TASSER platform available at https://zhanglab.ccmb.med.umich.edu/COVID-19/, and their molecular interactions with PEPs were studied using Autodock v4.2 [30] and AutoDock Vina packages [31]. While AutoDock utility of MGLTools [30] was used for obtaining .pdbqt files of receptor and ligand molecules, the obtained .pdbqt files were subjected for B.E calculations using Autodock Vina. The AutoDock Vina utilizes the semi-empirical force field to predict the best conformation of a ligand inside the protein cavity. The B.E. values were calculated for each PEP molecules against the active site of each SARS-CoV-2 protein considered in the study. To screen and prioritize the list of PEPs against each protein of SARS-CoV-2, a screening cutoff was decided for each SARS-CoV-2 protein on the basis of B.E. values distribution obtained from interactions with 292 PEPs. In order to select ligands with their best conformation inside the cavity of a SARS-CoV-2 protein, the high scoring SARS-CoV-2 protein–PEP pairs with B.E. values < (μ−σ) were considered for further studies, where μ is the mean of the 292 B.E. values and σ is their standard deviation. In this manner, out of 292 PEPs, 129 were screened-in against 24 SARS-CoV-2 proteins and were referred to as PEPcov2 i.e., potentially effective phytochemicals against SARS-CoV-2 proteins. The list of 129 PEPcov2 and their B.E. values with SARS-CoV-2 proteins is given in Supplementary Table 3.
Network construction and analysis
The complex relationship among the herb, phytochemicals, protein targets, pathways and disease classes was explored via contruction of networks and their analysis using Cytoscape v3.7, an open-source software that offers a versatile and interactive visualization interface for exploring molecular interaction networks. For the analysis of the generated networks, NetworkAnalyzer utility of Cytoscape was used [32].
The detailed description of each database source used in the current study can be checked in “Detailed Material & Methods section” given in Supplementary Data.
Results and discussion
Phytochemical dataset of Ayurvedic herbs
Of 7258 botanical names of Ayurvedic herbs mentioned in the Indian medicinal plants database, exhaustive mining from five databases could result in the compilation of 34,472 APCs (Ayurvedic phytochemicals). The Tanimoto-based similarity screening (as mentioned in Material and Methods Section “Anti-viral drug dataset and similarity index calculation”) of these APCs against antiviral drugs resulted in the selection of 292 APCs referred to as PEPs, and the further study focuses on the detailed examination of these PEPs. The detailed description of these PEPs with their phytochemical ID and chemical identifier is listed in Supplementary Table 3. Our previous work enlists the complete herb-wise data and detailed information of 34,472 APCs, which can be checked for any relevant information [33].
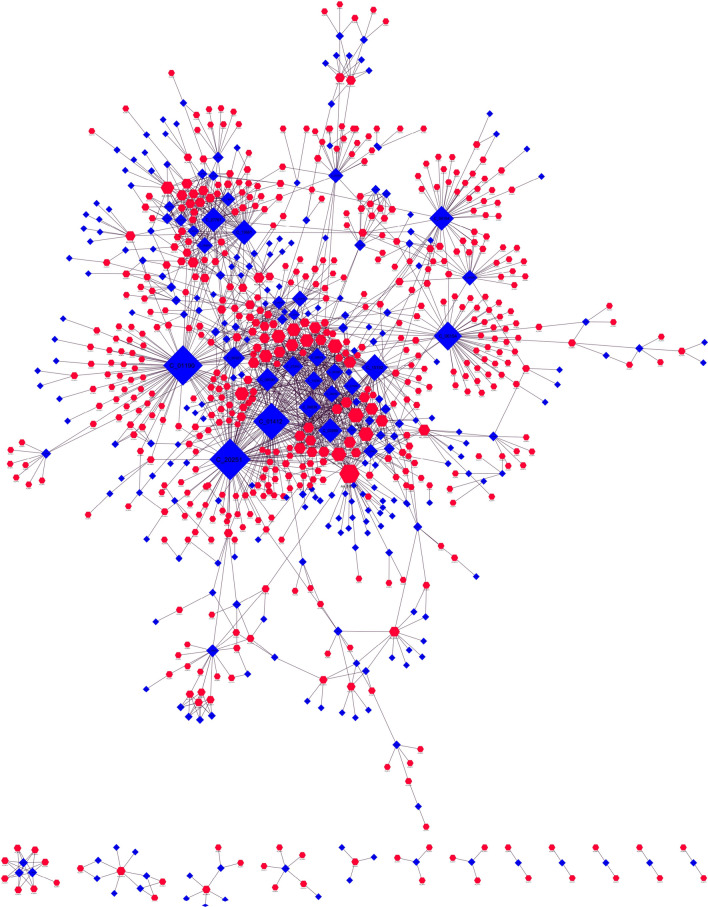
When checked for the presence of these phytochemicals in the Ayurvedic herbs, 292 PEPs were found to be distributed among 558 herb varieties. The detailed mapping of PEPs onto their respective herb can be checked in Supplementary Table 4. The information was used as input to construct the Ayurvedic herb–phytochemical network (AH-PEPs network) with network size of 850 nodes (558 herbs + 292PEPs) and 1685 edges (Fig. 2). Examining the distribution of PEPs among 558 herbs helped us identify that AH_0303-v1 contributes maximally to the PEPs category with 35 of its phytochemicals. The Ayurvedic herb AH_0303-v1 corresponds to Artemisia annua and earlier reported studies on the herb shows that the plant possess antiviral activity against SARS-CoV [34]. The alcoholic extract of the plant was one of the most potent herbal medicines used against SARS-CoV in 2005. Based on its antiviral properties, researchers across the globe are also trying to explore the effectiveness of this herb against novel coronavirus disease, COVID-19 [35].
Fig. 2.
AH-PEP network: The AH-PEP network representing associations of 292 PEPs (blue-colored triangles) with 558 herb varieties (red-colored octagons). Herb AH_0303-v1 (Artemisia annua) contributes maximally to the PEPs category with 35 of its phytochemicals in the AH-PEP Network, as seen with largest node size in the network where the size of nodes varies according to its degree centrality
In addition to AH_0303-v1, other Ayurvedic herbs enriched with PEPs are AH_3088-v1: Zingiber officinale, AH_0879-v1: Curcuma longa with 24 and 20 PEPs, respectively. Both of these are well-known Ayurvedic herbs for their immune-boosting capacity and have also been studied for their efficacies against exposed asymptomatic cases associated with COVID-19 [36].
The data suggest that the targeted action of these herbs against COVID-19 may be attributed to the constituting PEPs, which hold the potential to regulate SARS-CoV-2 proteins (as may be seen in the analysis of PEPcov2-PTcov2 network, detailed in the later sections of this study). Detailed examination of herbs may also put light on their respective phytochemicals for their target specificity against SARS-CoV-2 proteins. The chemical organization of 292 PEPs was found to be distributed among six broad chemical classes (Fig. 3).
Fig. 3.
Clustering and chemical distribution of PEPs: The 292 potentially effective phytochemicals (PEPs) are clustered in a hierarchical manner using Tanimoto coefficient and atom pair descriptors using ChemMine tools. Clustering of the PEPs is represented in the form of a tree layout where outer circles represent the detailed chemical class of PEP molecule and inner circle represents the PEP identifier assigned to each phytochemical considered in this study. The 292 PEPs are found to be broadly classified into 6 chemical classes and each class is represented by a unique color code
Chemical mapping of the PEPs reveals that chemical classes of terpenoids, especially “triterpenoids” and “bicyclic monoterpenoids,” were highly abundant in the dataset. This suggests that the PEPs dataset constitutes pharmaceutically relevant molecules as the class of terpenoids is of high importance in terms of pharmaceutical value due to their broad-spectrum medical application since prehistoric times [37]. Thus, future attention toward the detailed investigation of these PEPs could be of considerable importance in drug discovery. The chemical class of each of the PEPs can be checked in Supplementary Table 5.
Phytochemical–antiviral drugs similarity network
To select potentially active phytochemicals based on compound–compound similarity with existing antiviral drugs, the Tanimoto coefficients were calculated for each Ayurvedic phytochemical (APC)–antiviral drug (AVD) pair. The similarity is depicted in the form of a bipartite network, in which nodes in either set correspond to compounds from the lists of APCs or AVDs and edges are drawn between the nodes belonging to these two sets if the Tc value between them follows the criterion mentioned in Material and Methods Section “Anti-viral drug dataset and similarity index calculation” (Fig. 4). Hence, only the APCs earlier passing the Tc-based selection criterion (referred as PEPs) were considered at this step. Satisfying this criterion, 292 PEPs were screened-in against 16 of 125 AVDs. In this manner, a Tc-based similarity network between 292 PEPs and 16 AVDs, with network size of 307 nodes and 302 edges, was constructed (referred as PEP-AVD similarity network; Fig. 4). Detailed examination of the network returned that 160 PEPs share similarity with AV_DB00632 in the PEP-AVD network. AV_DB00632 corresponds to Docosanol, a class of approved drug effective against broad-spectrum lipid-enveloped viruses [38]. Among the list of 160 PEPs, C_00323 (2-Isopropyl-5-methylcyclohexanol) shares the maximum similarity with this AVD with Tc score of 0.92. C_00323 (2-Isopropyl-5-methylcyclohexanol) is a cyclohexanol molecule that has gained massive attention for its isoprenylated forms and is reported to be effective against viral infections as caused by HIV-1 and H1N1 [39, 40]. According to the phytochemical dataset prepared in this study, C_00323 (2-isopropyl-5-methylcyclohexanol) is found to be present in 39 Ayurvedic herbs (including varieties); the abundance of this phytochemical in various Ayurvedic herbs strengthens the therapeutic relevance of Ayurveda against viral infections.
Fig. 4.
PEP-AVD similarity network: The PEP-AVD network represents the Tanimoto coefficient (Tc)-based similarities between the PEPs and antiviral drugs listed in DrugBank. 292 PEPs (blue-colored diamonds) are found to be associated with 16 (pink-colored arrows) of total 125 AVDs considered in this study via 302 PEP-AVD pairs. Only PEP-AVD pairs following the selection criterion detailed in Materials and Methods section are considered for constructing the network. The edge widths of 302 pairs in the network are plotted in proportion to their Tc values. Red-colored circular outlined sub-networks represent the multi-similarity APCs against more than one AVD class, highlighting 8 APCs (C_01204, C_04300, C_01145, C_02130, C_01197, C_07863, C_04774 and C_01979) having multi-level similarity. The size of the nodes varies according to its degree centrality value in this network
It is interesting to note that while the majority of PEPs share one-to-one connection, i.e., showing similarity with only one AVD, few of them have one-to-many similarity-based connections. Out of 292 PEPS, 8 (C_01204: octylamine, C_04300: primene, C_01145: hexahydroaniline, C_02130: myristylamine, C_01197: 1-aminoheptane, C_07863: 2'-deoxycytidine B-D-threo, C_04774: 2-aminononadecane, and C_01979: 2'-deoxycytidine) were found to have similarity with more than one AVD, suggesting the importance of detailed examination of these compounds to be examined in detail for their molecular features thereby aiding in future pharmacophore-based antiviral drug design approaches.
Phytochemicals–SARS-CoV-2 protein target association
As per the approach mentioned in Material and Methods Section “In-silico molecular docking and interaction analysis,” each SARS-CoV-2 protein was associated with their screened-in PEPs and their association was represented in form of PEPcov2-PTcov2 network (Fig. 5). As already stated, the cutoff criterion resulted in selecting 129 of 292 PEPs against 24 SARS-CoV-2 proteins; therefore, PEPcov2-PTcov2 network was limited to 153 nodes (129 PEPcov2 & 24 SARS-CoV-2 proteins) having 1179 edges between them. The information of PEPcov2-PTcov2 network can be checked in Supplementary Table-3.
Fig. 5.
Phytochemicals–SARS-CoV-2 protein target association (PEPcov2-PTcov2 network): The PEPcov2-PTcov2 network represents the association of 129 PEPcov2 with SARS-CoV-2 proteins leading to the network size of 153 nodes (129 PEPcov2 and 24 SARS-CoV-2 proteins) and 1,179 edges. The PEPcov2 is represented using blue-colored diamond-shaped nodes and SARS-CoV-2 proteins (PTcov2) as yellow-colored octagons. The size of the nodes varies according to its degree centrality and width of the edges varies according to their binding energy values, where the pairs having lower value of binding energy (which represents the most suitable protein–ligand interaction pair) are given more weight and are ranked higher
For QHD43415_6, a non-structural protein nsp6 of SARS-CoV-2, 62 PEPcov2 were screened-in, where the least-binding energy was observed as −8.3 kcal/mol for C_04396 (Epi-Friedelanol) and C_16048 (hexadecahydropicen-3-ol derivative). Studies suggest that nsp6 is linked to the virulence of the virus as it is involved in cellular DNA synthesis [41, 42]. Similarly, for the main protease protein, QHD43415_3, 44 PEPcov2 were screened-in of which 4 (C_32090: tohogenol, C_11130: octahydronaphthalene-2,6-diol derivative, C_17085: taraxastane-3beta,20alpha-diol, and C_22189: inophyllolide chromanol) show very good binding affinities, with the lowest one being −8.5 kcal/mol. QHD43415_3 is a coronavirus 3 chymotrypsin-like protease (3CLpro) which is often termed as “the Achilles” heel of coronaviruses and is a validated target for identification of novel leads against corona virus [43]. Thus, the relevance of above mentioned 4 compounds in the regulation of QHD43415_3 is highly noticeable and requires special attention for in vitro and in vivo evaluation of their activity as potential anti-coronavirus inhibitors.
During the detailed analysis of local network structures, it was found that C_03212 (inophyllum B) possesses the multitargeting ability against 20 of 24 SARS-CoV-2 proteins. The shift from single-target to multi-target drugs has made rapid and remarkable progress and has emerged as an evolving paradigm of drug discovery [44], and as highlighted in a recent study network pharmacology acts as a powerful tool in identifying effective combination therapies in drug development [45]. Hence, other protein targets may also be looked for their potential regulators from the PEPcov2-PTcov2 network and may be ranked on the basis of their binding energy values, thereby giving an overall idea about the protein-specific regulatory role of Ayurvedic herbs against COVID-19 disease.
Phytochemicals–human protein target association
To detect the poly-pharmacological action of PEPs on the human system, the association of PEPs with their human PTs was represented in the form of potentially effective phytochemicals–human protein target (PEPs-PThs) network. For a phytochemical having ID C_31134, no protein target could be screened-in against the selection criterion adopted for protein target identification. Therefore, the network was constructed by associating 291 PEPs with their 621 protein targets identified using three target prediction algorithms as mentioned in Material and Methods Section “Protein Target identification of phytochemicals.” This resulted in the construction of PEPs-PThs network with network size of 912 nodes (291 PEPs + 621 PThs) and 6299 edges (Supplementary Fig. 1). Each of the 6299 PEP-PThs interaction pairs was prioritized based on their prediction supported from three target prediction algorithms. This led to the identification of 1265 high-confidence pairs, as predicted by at least two of the three target prediction algorithms (HCI pairs). The 6299 PEP-PThs pairs and the pairs corresponding to HCI data are detailed in Supplementary Table 6.
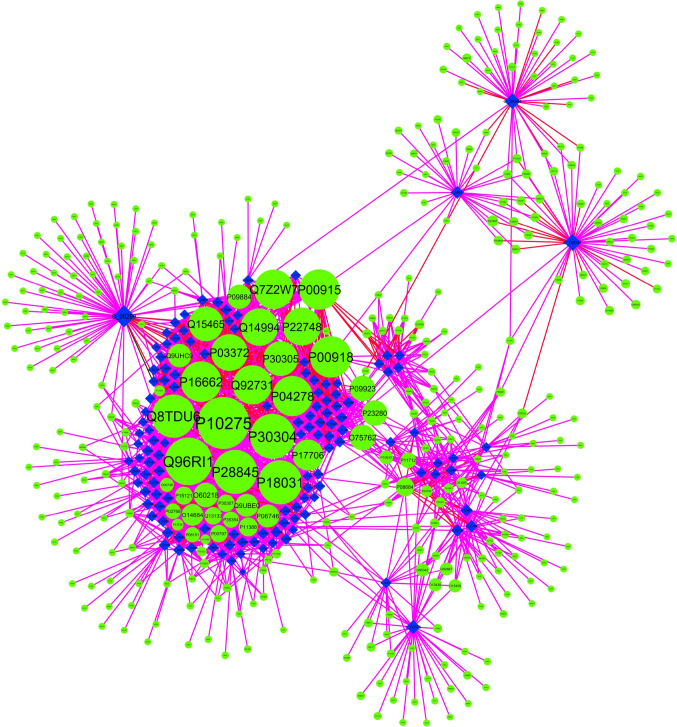
A sub-network of the PEP-PThs network consisting of 502 nodes and 2690 edges, specific to 129 PEPcov2 and their 373 PTs (referred to as PEPcov2-PThs network), was derived to focus on human proteins being targeted by them (Fig. 6). In the PEPcov2-PThs network, C_00289 and C_02937 hold the maximum targeting capacity among other PEPcov2, as these can target 74 and 49 proteins, respectively. Their high degree centrality value represents the importance of these phytochemicals in the overall PEPcov2-PThs network. It was interesting to note that all the 129 PEPcov2 were of multi-targeting nature with the capability to regulate several human proteins simultaneously. Among the protein targets, maximum number of regulators could be identified for Q96RI1, P28845 and P10275 with 102, 100 and 95 PEPcov2, respectively. P10275 is an androgen receptor encoded by AR gene, and the relevance of the androgens has been associated with increased viral load and dissemination as observed in case of COVID [46]. Androgen-mediated induction of COVID-19 suggests that the role of these 95 PEPcov2 in regulating the AR gene is noticeable for the management of COVID-19.
Fig. 6.
PEPcov2-PThs network: PEPcov2-PThs network represents a sub-network of PEPs-PThs network, specific to the association of 129 PEPcov2 and their 373 human protein targets. The network consists of 502 nodes and 2690 edges, with the size of nodes varying as per their degree values in the PEPs-PThs network
Case-study I: PEPcov2 as bi-directional regulators effective against COVID comorbidities
Since the focus of the study is to identify phytochemicals with a regulatory role in both the pathogen and its host, bi-directional regulators were searched among the PEPcov2 list. Such compounds have an added advantage as they work in dual scale mode, where at one end they can target pathogen proteins that may be crucial for its survival while at another end they tend to regulate the human proteins required to strengthen its defense mechanism against the pathogen. To identify the desired PEPcov2 with bi-directional regulation ability, 129 PEPcov2 were examined against SARS-CoV-2 and human protein targets in the PEPcov2-PTcov2 and PEPcov2-PThs networks, respectively.
In PEPcov2-PThs network, 129 PEPcov2 were found to be associated with 373 human protein targets. Using all these data, a tripartite network consisting of 129 PEPcov2, their 373 human targets and 24 SARS-CoV-2 protein targets referred to as “Bi-regulatory PEPcov2 network” was developed. The obtained network of size 526 nodes and 3869 edges, along with the list of 373 human targets, is given in Supplementary Fig. 2; Supplementary Table 7.
It is well known that not all proteins of the human system are suitable for drug interactions, only a fraction of the total human proteome can bind to drug molecules with high affinity and are potential drug targets, i.e., they have an association with a disease or disorder. Therefore, a “druggable bi-regulatory PEPcov2 network” was extracted from the “bi-regulatory PEPcov2 network” by considering only those proteins that are considered to be targets of FDA-approved drugs. While “Bi-regulatory PEPcov2 network” gives an overall idea of the dual regulatory mode of PEPcov2, the sub-network may provide valuable help in protein-specific drug designing of PEPcov2 with multi-targeting action. Confidence was also added at this level by considering only those PEPcov2-PThs pairs that belong to the HCI data. In this manner, there is a high-confidence druggable subnetwork of size 179 nodes and 1588 edges, consisting of 24 SARS-CoV-2 proteins, 115 PEPcov2 and 40 human protein-approved targets (Fig. 7; Supplementary Table 7).
Fig. 7.
Druggable bi-regulatory PEPcov2 network: The network represents the dual-regulatory mode of 115 PEPcov2 (middle layer, blue diamond shaped nodes) against 24 SARS-CoV-2 proteins (top-layer, yellow-colored octagon nodes) and 40 approved protein targets of Homo sapiens (bottom-layer, green-colored circular nodes). For the differentiation, the edges between PEPcov2 and SARS-CoV-2 proteins are represented using violet color while edges between PEPcov2 and human proteins using orange color. The size of the nodes among the network varies according to its degree in this network
Recent studies on the treatment procedure given to COVID-19 patients address the need of special attention toward the problem of cardiovascular system [47]. Since most of the current antiviral drugs cause cardiac complications, alternative therapeutic strategies effective to combat the cardiac toxicity should be given consideration. Therefore, we searched for PEPcov2 that can target COVID-19 proteins without imposing a load on the cardiac system. To achieve the desired list of PEPcov2, their protein targets in the human system were checked for the participation in cardiovascular diseases. To extract high-confidence disease association data, Gene–disease association (GDA) score (S) of 0.05 was chosen as the threshold [48] so as to have a nonzero contribution from either of the C (curated data), M (animal model data) or I (inferred data), or a support of at least 5 publications. Thirty-six proteins among the 373 human targets of 129 PEPcov2 were found to be involved in cardiovascular diseases within the desired cutoff score. The interactors specific to these 36 proteins were extracted from the bi-regulatory PEPcov2 network, where they were found to have an association with 123 PEPcov2 and all the 24 SARS-CoV-2 proteins and presented as a sub-network specific to cardiovascular diseases with network size of 183 nodes and 1471 edges (Supplementary Fig. 3). In the network, the most multitargeting PEPcov2, C_03212 (Inophyllum B) shows its targeting action against 20 SARS-CoV-2 proteins and also supports the cardiac system by regulating 3 cardiovascular diseases-associated proteins, i.e., O00206 (TLR4), Q13093 (PLA2G7) & P42336 (PIK3CA). C_03212 (Inophyllum B) corresponds to the most active component of Calophyllum inophyllum, an important component of Ayurvedic drug therapy. Besides regulating an important therapeutic target TLR4 [49], PLA2G7 [50] and PIK3CA [51] against various cardiac-related diseases, literature data are plenteous for the antiviral activity of C_03212 [52, 53]. These findings suggest that future research endeavors toward exploring the anti-COVID-19 activity of C_03212 must be given a proper consideration and examined in detail as per the in vivo and in vitro studies.
In the context of disease comorbidities associated with COVID, a study by Roth and group suggests that the severity of COVID infection is higher for the patients suffering from diabetes mellitus and hypertension [54]. Therefore, using a similar strategy as applied for cardiovascular diseases, drug targets involved in diabetes and hypertension were also checked and a separate network for each disease was constructed and investigated. This is essential to analyze the underlying disease comorbidity pattern and the compounds from Ayurvedic herbs that may regulate them while dealing against the COVID-19 infection. The proteins associated with each disease considered can be checked in Supplementary Table 8. For the identification of PEPcov2 that may act on multiple scales, the protein targets of PEPcov2 were checked for their multi-disease association, considering the above 3 comorbid diseases. Detailed examination could help us to identify that multi-disease associations of a protein were observed at this point, where multiple proteins overlap between the 3 classes of diseases discussed here (Fig. 8A).
Fig. 8.
Regulatory role of phytochemicals in dealing COVID-19-associated comorbidities. A Distribution of proteins among COVID-19 comorbid diseases: The Venn diagram shows the distribution of human protein targets of PEPcov2 in 3 COVID-19-associated comorbid diseases, namely, cardiovascular diseases, hypertension and diabetes. Of total 373 human targets of 129 PEPcov2 considered in this study, 36 were associated with cardiovascular diseases, 40 with hypertension and 100 with diabetes mellitus. 14 overlapping proteins common to all the 3 comorbid diseases were identified and considered for detailed analysis. B Bi-regulatory PEPcov network specific to 14 common proteins: The network is a subnetwork of bi-regulatory PEPcov2 network specific to the PEPcov2 effective in dealing COVID-19-associated comorbidity diseases; cardiovascular diseases hypertension and diabetes mellitus. The network contains 14 human proteins (green-colored circular nodes) being regulated by 73 PEPcov2. The information of SARS-CoV-2 proteins targeted by these 73 PEPcov2 is also added to the network. For the differentiation, the edges between PEPcov2 and SARS-CoV-2 proteins are represented using violet color while edges between PEPcov2 and human proteins using orange color. The size of the nodes among the network varies according to their degree value, representing the high number of regulators for P04150 and P35354 (as depicted by their large size among all the nodes). C Multi-targeting role of a bi-regulatory phytochemical C_17006: A phytochemical having ID C_17006 shows a dual action mode in both the human and SARS-CoV-2 systems. The multi-targeting nature of this compound against two human FDA-approved protein targets P04150 and P35354 is shown in the left side of the panel. In the virus system, the compound can target 4 viral proteins within the binding energy range of −9.4 to −6.7 kcal/mol where best of −9.4 kcal/mol was obtained for nsp2 protein QHD43415_2 (shown in the right side of the panel). The binding energy values of the compound with each viral protein are represented along the edges of the network
The 14 human proteins common to all these 3 comorbid diseases (Table 1) were selected, and their regulatory PEPcov2 was checked. Mapping of these 14 proteins onto bi-regulatory PEPcov2 network could help us in deriving a sub-network specific to them with size of 111 nodes (73 PEPcov2 + 14 Human proteins + 24 SARS-CoV-2 proteins) and 749 edges (Fig. 8B). To identify high-confidence regulatory PEPcov2, only pairs corresponding to HCI-pair data were considered, leading to the selection of 12 PEPcov2 against 2 proteins (P04150 and P35354). Both these proteins, i.e., P04150 and P35354, also belong to the FDA-approved protein target list, thereby suggesting the key relevance of these proteins targets and phytochemicals against COVID-19.
Table 1.
Distribution of the proteins among different classes of comorbid diseases
| Serial no. | Class | No. of proteins | Proteins |
|---|---|---|---|
| 1 | Cardiovascular and Hypertension | 4 | P09917, P37023, P07949, P19320 |
| 2 | Cardiovascular and Diabetes | 12 | P03372, O00206, P05362, P01130, P02647, P02768, O95477, Q07869, P04114, Q8NBP7, P27169, P11473 |
| 3 | Hypertension and Diabetes | 6 | P24530, P10145, P25101, P80365, P42345, P03956 |
| 4 | Associated with all three (Cardiovascular, Hypertension, and Diabetes) | 14 | P35354, P02649, P00797, P37231, P04035, P08253, P16581, P30556, P04150, P35228, P42336, P12821, P06858, P29474 |
Detailed association of these interactions helped us to highlight the multitargeting role of C_17006 (CHEMBL141117), as it targets both these protein targets (Fig. 8C). When checked for its SARS-CoV-2 targeting capacity, C_17006 was found to have its binding affinity against 4 SARS-CoV-2 proteins within the binding energy range of −9.4 to −6.7 kcal/mol where best of −9.4 kcal/mol was noted for QHD43415_2, a non-structural protein 2 (nsp2), shown in Fig. 8C. This suggests the role of C_17006 is highly noteworthy in dealing with the co-morbidities associated with COVID-19. In this manner, other regulatory molecules can also be checked for their multi-targeting capacity and can be prioritized based on their binding affinity with SARS-CoV-2 proteins.
Case study II: Immunoregulatory potential of PEPcov2
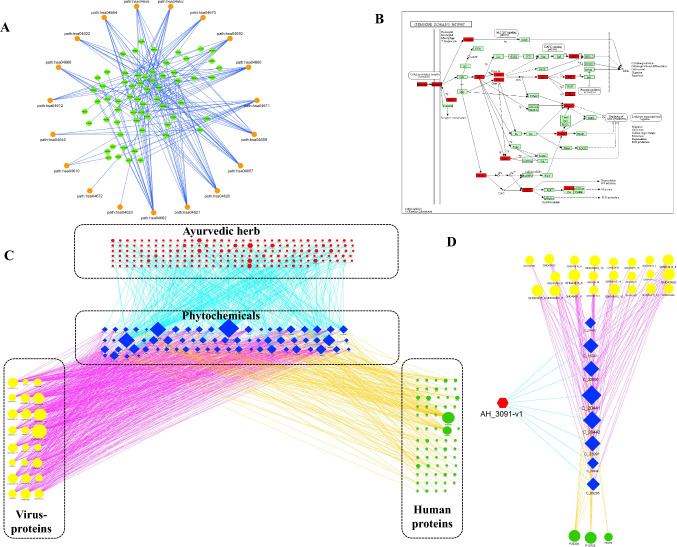
To explore the underlying mechanisms of Ayurvedic herbs being studied toward promoting the human immune system, a sub-network of immune pathways being regulated by PEPcov2 was constructed. It has been studied that in the early stages of infection or during incubation period, host needs a specific adaptive immune response to exterminate virus from the system [55]. This is necessary to inhibit the progression of the disease to its chronic form or more severe stages. At this stage, immune system of the host body plays an important role to promote a state of good health. A strong immune system in addition to the genetic background (e.g., HLA) is essential to elicit a strong antiviral immunity at initial stages [55]. Therefore, immunomodulatory potential of PEPcov2 was investigated by characterizing potential PEPcov2-PThs interactions potentially responsible for immune system pathways. For this, 21 pathways specific to immune-system as described by KEGG database (i.e., hsa04062, hsa04610, hsa04611, hsa04612, hsa04620, hsa04621, hsa04622, hsa04623, hsa04624, hsa04625, hsa04640, hsa04650, hsa04657, hsa04658, hsa04659, hsa04660, hsa04662, hsa04664, hsa04666, hsa04670 & hsa04672) and their protein targets in human were selected for further analysis [56]. Among 373 human protein targets of 129 PEPcov2, 63 were involved in the above-mentioned 19 of 21 immune pathways via 163 interactions (as presented in the immunoregulatory network; Fig. 9A; Supplementary Table 9). For two immune pathways hsa04624 and hsa04625, no protein target shows their involvement, and therefore, network is restricted to 19 immune pathways. The network suggests that the immunomodulatory potential of PEPcov2 is largely via regulating chemokine and NOD-like receptor signaling pathways. The high regulatory potential of 21 PEPcov2 against chemokine signaling pathways may also give an added advantage in managing cardiovascular diseases like atherosclerosis, as such patients are at high risk in developing COVID-19 infection [47, 57]. The chemokine regulation is mainly via these 21 PEPcov2 which target 18 proteins involved in this pathway. The location of these 18 proteins onto the pathway is shown in red rectangles in Fig. 9B.
Fig. 9.
Immune regulatory potential of Ayurvedic herbs. A Immunomodulatory network: the immunoregulatory network represents the participation of protein targets of 129 PEPcov2 in regulating the immune system of Homo sapiens. The outer layer representing the circular nodes depicts 19 of 21 immune system-related pathways in humans as described by KEGG database. Among 373 human protein targets of 129 PEPcov2, 63 were found to be involved in 19 immune pathways, arranged inside the circular layout as shown by green color circular nodes. No protein target was associated with hsa04624 and hsa04625, thereby restricting the number to 19. B Chemokine signaling pathway (path:hsa04062) obtained from KEGG database: The location of the mapped genes corresponding to protein targets of PEPcov2 is highlighted in red-colored boxes in the pathway. C Herb-specific immune regulatory network (HSIR-Network): HSIR network is the 4-component network of size 352 nodes and 1,128 edges containing association of 198 AHs, 67 PEPcov2, 24 SARS-CoV-2 proteins and 63 human proteins. The network is limited to the protein targets involved in immune system-related pathways in humans as described by KEGG database. D Subnetwork of HSIR network specific to AH_3091-v1 (Ziziphus jujube): The dual regulatory role of Ziziphus jujube in targeting SARS-CoV-2 and human immune system-related proteins through 8 of its phytochemicals (C_23991, C_28934, C_22690, C_05205, C_16341, C_28442, 51,025,490, C_2044 and C_24442) represented by blue-colored diamonds
The immune regulatory network suggests that immunoregulatory effect may be conferred by carefully designed combination of phytochemicals. The combined effect of these PEPs may be associated with the molecular-scale rationale behind the immune-boosting capacity of Ayurvedic herbs and formulations. Among 67 PEPcov2 involved in immunoregulation (via targeting 63 human proteins of immunoregulatory network), C_34364 (guanosine-5'-triphosphate) and C_02937 (crotonoside) are the top immunoregulators with 13 and 9 protein targets, respectively (Table 2).
Table 2.
Information of the top-5 immunomodulators
| S. No. | Phytochemical information | No. of protein targets | Herbal-association | |||
|---|---|---|---|---|---|---|
| Phytochemical ID | Phytochemical name | Human immune system-associated proteins | SARS-Cov-2 proteins | No. of herbs | Herb details | |
| 1 | C_34364 | Guanosine 5'-triphosphoric acid | 13 | 5 | 1 | AH_0264 |
| 2 | C_02937 | Crotonoside | 9 | 1 | 1 | AH_0858 |
| 3 | C_09617 | 5-Methoxypodophyllotoxin | 7 | 5 | 3 | AH_2275, AH_2276, AH_17301 |
| 4 | C_00378 | Arteannuin | 6 | 1 | 1 | AH_0303 |
| 5 | C_12746 | Benzofuran-1-one derivative | 6 | 1 | 2 | AH_2274, AH_0318 |
Additional details of these phytochemicals may be checked by studying the phytochemical and their protein–ligand complexes for their structural and analytical properties. To derive the information of herbs these 67 PEPcov2 (involved in immunoregulation) belongs to, the AH-PEPs network was checked which led to the identification of association of 198 AHs with these 67 PEPcov2. The information was added to Bi-regulatory PEPcov2 network to construct a 4-component network consisting of 198AHs, 67 PEPcov2 and their regulators from both human and SARS-CoV-2 proteins. This led to the construction of a herb-specific immune regulatory network (HSIR network), with network-size of 352 nodes and 1128 edges containing association of 198 AHs, 67 PEPcov2, 24 SARS-CoV-2 proteins and 63 proteins from humans (Fig. 9C). Each immune regulatory herb from the 198 AHs was prioritized based on their both human and virus targeting capability. When checked for the AHs having at least 5 virus targets, 149 AHs got selected, and among those the AHs who contribute maximally to the protein targets are Zea mays (AH_3081-v1) with 32 proteins being targeted by its 9 PCs, Cucurbita maxima (AH_0865-v1) with 29 proteins being targeted by its 10 PCs, Pisum sativum (AH_2237-v1) with 29 proteins being targeted by its 9 PCs, Thlaspi arvense (AH_2874-v1) with 28 protein being targeted by its 8 PCs, Calophyllum inophyllum (AH_0504-v1) with 28 protein being targeted by its 5 PCs and AH_3091-v1/v3 (Ziziphus jujube) with 27 proteins being targeted by its 2 PCs (for AH_3091-v3) and with 26 proteins being targeted by its 8 PCs (for AH_3091-v1).
Among these, Ziziphus jujube was found to target maximum number of viral proteins. It is interesting to note that the decoction of Ziziphus jujube has also been suggested in the advisory issued by Ministry of AYUSH (Ayurveda, Yoga, Unani, Siddha and Homeopathy), Government of India, toward the management of COVID at its preventive and prophylactic stage [9]. The observation strengthens the credibility of the network toward suggesting potential herbs and their phytochemicals for dealing COVID-19 pandemic. The network also sheds light on the phytochemical-specific targeted action of herbs; for example, the targeted action of Ziziphus jujube is shown in Fig. 9D, where the mechanism of its management against COVID-19 can be attributed to its 8 phytochemicals (C_23991: colubrinic acid, C_28934: CHEMBL1782597, C_22690: zizyberanalic acid, C_05205: ceanothic acid, C_16341: di-carboxylic acid derivative, C_28442: carboxylic acid derivative, C_2044: alphitolic acid, and C_24442: epiceanothic Acid) that have targeting potential against 23 SARS-CoV-2 proteins. The plant also aids in regulation the host immune system through these 8 phytochemicals by targeting three human proteins P20292, P13726 and P35354; among these P13726 and P35354 belong to the class of FDA-approved targets. The complete interaction data used for constructing HSIR-network are given in Supplementary Table 10. The data may be checked for other herbs to decipher their phytochemical specific targeted action in the management of COVID-19. Although the network is limited to the immune regulatory potential of those phytochemicals having the ability to target SARS-CoV-2 proteins, the approach holds the potential to give a mechanistic understanding of therapeutic relevance of traditional herbs.
The information of the data used in the present study and the results generated is summarized in Table 3.
Table 3.
Summary of the data used and results generated in the present study
| Serial No. | Particulars of the data used and results generated | Associated numbers | Reference |
|---|---|---|---|
| 1 | Ayurvedic herbs available at IMPD | 7258 | Supplementary Table-1 |
| 2 | Ayurvedic herbs used in the study | 3038 | Supplementary Table-1 |
| 3 | Ayurvedic Phytochemicals (APCs) | 34,472 | Supplementary Table-1 |
| 4 | Anti-Viral Drugs (AVDs) | 125 | Supplementary Table-2 |
| 5 | Potentially Effective Phytochemicals (PEPs) | 292 | Supplementary Table-2 |
| 6 | SARS-CoV-2 proteins from I-TASSER | 24 | Supplementary Table-3 |
| 7 | Potentially effective phytochemicals against SARS-CoV-2 proteins (PEPcov2) | 129 | Supplementary Table-3 |
| 8 | PEPcov2-PTcov2 network |
153 (nodes) 1179 (edges) |
Supplementary Table-3 |
| 9 | Herbs containing 292 PEPs | 558 | Supplementary Table-4 |
| 10 | Chemical class of PEPs | 6 (Broad classes) | Supplementary Table-5 |
| 11 | PEP-PThs pairs | 6299 | Supplementary Table-6 |
| 12 | HCI pairs | 1265 | Supplementary Table-6 |
| 13 | Bi-regulatory PEPcov2 network |
526 (nodes) 3869 (edges) |
Supplementary Table-7 |
| 14 | Human protein targets of 129 PEPcov2 | 373 | Supplementary Table-7 |
| 15 | Approved drug targets in Druggable bi-regulatory PEPcov2 network | 40 | Supplementary Table-7 |
| 16 | Human protein targets of PEPcov2 involved in comorbid diseases |
36 (cardiovascular diseases) 40 (hypertension) 100 (diabetes mellitus) |
Supplementary Table-8 |
| 17 | Immunomodulatory network | 63 protein targets associated with 19 immune pathways | Supplementary Table-9 |
| 18 | HSIR network |
352 (nodes) 1,128 (edges) |
Supplementary Table-10 |
Summary
The exceptional state of health crisis emerged due to the novel SARS-CoV-2 virus has forced the researchers across the globe to constantly work toward searching the preventive measures as well as developing its possible cure. Ayurveda, considered as the oldest healing schema on Earth, describes thousands of herbs and their formulations for the well-being of mankind. It has always remained a great source of drugs and other lead-like molecules. To explore the therapeutic relevance of Ayurveda for combating the current situation, the network pharmacological evaluation of Ayurvedic herbs was carried out in this study. An extensive collection of the phytochemicals present in Ayurvedic herbs and the study of their regulatory prospects form the basis of present work. To decipher the phytochemical-specific targeted action of herbs, a collection of 34,472 Ayurvedic phytochemicals (APCs) was developed from 7258 botanical names. 292 (referred to as PEPs) of these phytochemicals were found to be similar (based on Tc value) with 16 of 125 currently available antiviral drugs considered in the study. Herb-wise distribution of PEPs was found to be maximally concentrated to Artemisia annua with 35 of its phytochemicals in PEPs category. When checked for the association of these 292 PEPs with SARS-CoV-2 proteins based on their binding energy value distribution, 129 (referred to as PEPcov2) were screened-in against 24 SARS-CoV-2 proteins, thereby restricting the further analysis to PEPcov2. The therapeutic relevance of PEPs was assessed using the information of their 621 human protein targets and 24 SARS-CoV-2 protein targets, where targeting capabilities of 62 PEPcov2 were identified against non-structural protein nsp6 of SARS-CoV-2. Among the list of 292 PEPcov2, 115 were identified with dual regulatory mode having targeting capability in both virus and its host system, thereby indicating their future implications in pharmacophore-based drug design approaches. For example, a phytochemical C_03212 (Inophyllum B from Calophyllum inophyllum) was found to support the cardiovascular system by targeting genes involved in cardiovascular diseases, like TLR4, PLA2G7 & PIK3CA. The ability of this compound to target 20 SARS-CoV-2 proteins further strengthens its role in managing COVID-19. The multi-regulatory role of 73 phytochemicals was highlighted for their ability to manage the complication of COVID-19-associated comorbidity; among them the effect of C_17006 (CHEMBL141117) is highly noticeable for its multi-targeting strategy. In addition to this, the high binding affinity of the compound for nsp2 protein of SARS-CoV-2 attracts attention for its ability to act as a potential lead moiety. Immunoregulatory ability of the Ayurvedic herbs was also explored and presented as a special case study. The analysis helps to decipher the role of 63 PEPcov2 for their regulatory role on the immune system of host body where the effect is mainly via regulating chemokine and NOD-like receptor signaling pathways. C_34364 (Guanosine 5'-triphosphoric acid) and C_02937 (Crotonoside) were found as top immunoregulators with ability to regulate 13 and 9 proteins of the immune system, respectively. In our study, Ziziphus jujube appeared as a potential candidate with dual regulatory effects in SARS-CoV-2 targeting and immune-supportive role; the detailed phytochemical special protein targeting ability of the plant has been deciphered and presented as an example where the effect is found to be mainly through its 8 phytochemicals. Other potential herbs may also be explored for their systems-level effects, and the role of multi-targeting phytochemicals can be identified via analyzing the interaction networks generated in the study. The developed protocol provides novel insights about the complex regulatory role of traditional medicines and their target specificity in a much deeper and simpler context for managing the current global situation. This study can be considered a major attempt toward integrating the wealth of traditional practices with modern scientific approaches to meet the therapeutic demands in the current scenario.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
N.C. is grateful to the Indian Council of Medical research (ICMR) for support provided through ICMR-SRF. Authors would like to thank Dr. Arun Kumar for useful discussions.
Abbreviations
- TIM system
Traditional Indian Medicinal System
- AHs
Ayurvedic herbs
- APCs
Ayurvedic phytochemicals
- AVDs
Antiviral drugs
- HCI
High-confidence interactions
- IMPD
Indian Medicinal Plants Database
- IMPPAT
Indian Medicinal Plants, Phytochemistry and Therapeutics
- NPASS
Natural Product Activity and Species Source Database
- PCIDB
Phytochemical Interactions DB
- PEPs
Potentially effective phytochemicals
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- Tc
Tanimoto coefficient
- TCM
Traditional Chinese Medicine
Authors’ contribution
VS conceptualized the study and designed the research framework. NC contributed to data-collection, data integration, computational analyses. NC and VS investigated and analyzed the results, and prepared the manuscript.
Data availability
The complete data used and generated in this study are given in the form of Supplementary material available at https://chemrxiv.org/articles/preprint/Extensive_Exploration_of_Ayurvedic_Herbs_to_Prioritize_Anti-Viral_Drugs_Alike_Phytochemicals_Against_SARS-CoV-2_Using_Network_Pharmacology/13385480/1.
Declarations
Conflict of interest
Authors declare that there is no conflict of interest regarding the publication of this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xioing Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh V and Sigh V (2020) C19-TraNet: an empirical, global index-case transmission network of SARS-CoV-2. https://arxiv.org/abs/2006.15162
- 3.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 4.Dobson J, Whitley RJ, Pocock S, Monto AS. Oseltamivir treatment for influenza in adults: a meta-analysis of randomised controlled trials. Lancet. 2015;385:1729–1737. doi: 10.1016/S0140-6736(14)62449-1. [DOI] [PubMed] [Google Scholar]
- 5.Mercorelli B, Palù G, Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren J, Zhang A-H, Wang X-J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res. 2020;155:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Zhang Y. Traditional Chinese medicine treatment of COVID-19. Compl Ther Clin Pract. 2020;39:101165. doi: 10.1016/j.ctcp.2020.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ministry of AYUSH Gov. of India. Annexure-I advisory from ministry of ayush For meeting the challenge arising out of spread of corona virus (COVID-19) In India. 1–9. https://www.ayush.gov.in/docs/125.pdf
- 10.Jadhav P, Kapoor N, Thomas B, Lal H, Kshirsagar N. Antiviral potential of selected Indian medicinal (ayurvedic) plants against herpes simplex virus 1 and 2. N Am J Med Sci. 2012;4:64. doi: 10.4103/1947-2714.104316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 12.Choudhary N, Singh V. A census of P. longum’s phytochemicals and their network pharmacological evaluation for identifying novel drug-like molecules against various diseases, with a special focus on neurological disorders. PLoS ONE. 2018;13:0191006. doi: 10.1371/journal.pone.0191006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi: 10.3389/fphar.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanraj K, Karthikeyan BS, Vivek-Ananth RP, Chand RB, Aparna SR, Mangalapandi P, Samal A. IMPPAT: a curated database of Indian medicinal plants. Phytochem Therapeut Sci Rep. 2018;8:4329. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Yu S, Bai H, Ning K. TCM-Mesh: the database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-03039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng X, Zhang P, He W, Qin C, Chen S, Tao L, Wang Y, Tan Y, Gao D, Wang B, Chen Z. NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucl Acids Res. 2018;46:D1217–D1222. doi: 10.1093/nar/gkx1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng X, Zhang P, Wang Y, Qin C, Chen S, He W, Tao L, Tan Y, Gao D, Wang B, Chen Z. CMAUP: a database of collective molecular activities of useful plants. Nucl Acids Res. 2019;47:D1118–D1127. doi: 10.1093/nar/gky965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Ann Rep Comput Chem. 2008;4:217–241. doi: 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- 19.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucl Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feunang YD, Eisner R, Knox C, Chepelev L, Hastings J, Owen G, Fahy E, Steinbeck C, Subramanian S, Bolton E, Greiner R. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J Cheminform. 2016;8:61. doi: 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backman TWH, Cao Y, Girke T. ChemMine tools: An online service for analyzing and clustering small molecules. Nucl Acids Res. 2011;39:486–491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Santos A, Von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucl Acids Res. 2016;44:380–384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucl Acids Res. 2019;47:W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucl Acids Res. 2007;35:198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J Chem Inf Comput Sci. 1998;38:983–996. doi: 10.1021/ci9800211. [DOI] [Google Scholar]
- 28.Choudhary N, Singh V. Insights about multi-targeting and synergistic neuromodulators in Ayurvedic herbs against epilepsy: integrated computational studies on drug-target and protein-protein interaction networks. Sci Rep. 2019;9:10565. doi: 10.1038/s41598-019-46715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucl Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris G, Huey R. AutoDock4 and AutoDockTools4: Automated docking with selective receptor felxibility. J Comput Chem. 2009;30:2785–3279. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhary N, Singh V (2021) Neuromodulators in food ingredients: insights from network pharmacological evaluation of Ayurvedic herbs. https://arxiv.org/abs/2108.09747
- 34.Li SY, Chen C, Zhang HQ, Guo HY, Wang H, Wang L, Zhang X, Hua SN, Yu J, Xiao PG, Li RS. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haq FU, Roman M, Ahmad K, Rahman SU, Shah SM, Suleman N, Ullah S, Ahmad I, Ullah W. Artemisia annua: trials are needed for COVID-19. Phyther Res. 2020;2020:1–2. doi: 10.1002/ptr.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rastogi S, Pandey DN, Singh RH. COVID-19 pandemic: a pragmatic plan for Ayurveda Intervention. J Ayurveda Integr Med. 2020 doi: 10.1016/j.jaim.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Tang W, Bidigare RR (2005) Terpenoids as therapeutic drugs and pharmaceutical agents bt - natural products: drug discovery and therapeutic medicine. Humana Press, Totowa, NJ, pp 197–227. 10.1007/978-1-59259-976-9_9
- 38.De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Liu D, Proksch P, Zhou D, Lin W. Truncateols OV, further isoprenylated cyclohexanols from the sponge-associated fungus Truncatella angustata with antiviral activities. Phytochemistry. 2018;155:61–68. doi: 10.1016/j.phytochem.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y, Si L, Liu D, Proksch P, Zhou D, Lin W. Truncateols A-N, new isoprenylated cyclohexanols from the sponge-associated fungus Truncatella angustata with anti-H1N1 virus activities. Tetrahedron. 2015;71:2708–2718. doi: 10.1016/j.tet.2015.03.033. [DOI] [Google Scholar]
- 41.Geng H, Liu YM, Chan WS, Lo AW, Au DM, Waye MM, Ho YY. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 2005;579:6763–6768. doi: 10.1016/j.febslet.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tangudu C, Olivares H, Netland J, Perlman S, Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J Virol. 2007;81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Bartlam M, Rao Z. Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Curr Pharm Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 44.Ramsay RR, Popovic-Nikolic MR, Nikolic K, Uliassi E, Bolognesi ML. A perspective on multi-target drug discovery and design for complex diseases. Clin Transl Med. 2018;7:3. doi: 10.1186/s40169-017-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng F, Kovács IA, Barabási A-L. Network-based prediction of drug combinations. Nat Commun. 2019;10:1197. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wambier CG, Goren A. SARS-COV-2 infection is likely to be androgen mediated. J Am Acad Dermatol. 2020;83:308–309. doi: 10.1016/j.jaad.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y-Y, Ma Y-T, Zhang J-Y, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choudhary N, Choudhary S, Kumar A, Singh V. Deciphering the multi-scale mechanisms of Tephrosia purpurea against polycystic ovarian syndrome (PCOS) and its major psychiatric comorbidities: studies from network pharmacological perspective. Gene. 2021;773:145385. doi: 10.1016/j.gene.2020.145385. [DOI] [PubMed] [Google Scholar]
- 49.Jia SJ, Niu PP, Cong JZ, Zhang BK, Zhao M. TLR4 signaling: a potential therapeutic target in ischemic coronary artery disease. Int Immunopharmacol. 2014;23:54–59. doi: 10.1016/j.intimp.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 50.Zalewski A, Macphee C. Role of lipoprotein-associated phospholipase A2 in atherosclerosis: biology, epidemiology, and possible therapeutic target. Arterioscler Thromb Vasc Biol. 2005;25:923–931. doi: 10.1161/01.ATV.0000160551.21962.a7. [DOI] [PubMed] [Google Scholar]
- 51.Durrant TN, Hers I. PI3K inhibitors in thrombosis and cardiovascular disease. Clin Transl Med. 2020;9:8. doi: 10.1186/s40169-020-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yimdjo MC, Azebaze AG, Nkengfack AE, Meyer AM, Bodo B, Fomum ZT. Antimicrobial and cytotoxic agents from Calophyllum inophyllum. Phytochemistry. 2004;65:2789–2795. doi: 10.1016/j.phytochem.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 53.Patil AD, Freyer AJ, Eggleston DS, Haltiwanger RC, Bean MF, Taylor PB, Caranfa MJ, Breen AL, Bartus HR. The inophyllums, novel inhibitors of HIV-1 reverse transcriptase isolated from the Malaysian tree, Calophyllum inophyllum Linn. J Med Chem. 1993;36:4131–4138. doi: 10.1021/jm00078a001. [DOI] [PubMed] [Google Scholar]
- 54.Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucl Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel J, Channon KM, McNeill E. The downstream regulation of chemokine receptor signalling: implications for atherosclerosis. Mediators Inflamm. 2013;2013:459520. doi: 10.1155/2013/459520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete data used and generated in this study are given in the form of Supplementary material available at https://chemrxiv.org/articles/preprint/Extensive_Exploration_of_Ayurvedic_Herbs_to_Prioritize_Anti-Viral_Drugs_Alike_Phytochemicals_Against_SARS-CoV-2_Using_Network_Pharmacology/13385480/1.