Abstract
Positive results by cytomegalovirus (CMV) PCR of plasma are considered predictive of active CMV infection in kidney allograft recipients. To assess whether contamination with leukocyte-derived CMV DNA can distort the results, aliquots of whole-blood samples from 60 CMV immunoglobulin G-positive patients with leukocyte CMV DNAemia were stored for up to 24 h at room temperature (RT) and at 4°C before plasma preparation. Native and ultrafiltered plasma samples were tested by CMV and β-globin PCRs. Among 30 latently infected patients (negative for CMV pp65 antigens), low baseline rates (10%) and levels (median number of copies, 10 [per 10 μl]) of CMV plasma DNAemia in native plasma samples increased significantly over time (after 4 h at RT, 37% [P < 0.001]; median number of copies, 45 [P < 0.001]). Similar effects were found during storage at 4°C. Ultrafiltration reduced the levels of CMV plasma DNAemia, but by 6 h of storage the levels were significantly elevated as well. CMV and β-globin DNA kinetics in plasma were parallel. In contrast, 30 actively infected patients (pp65 positive) had high baseline rates (87% in native samples) and levels (median number of copies, 75) of CMV plasma DNAemia. No significant effects of storage or ultrafiltration and no concordance with β-globin DNA kinetics were seen. In conclusion, delayed preparation of plasma samples bears a significant risk of false-positive CMV PCR results, probably due to leukocyte lysis. This has important implications in the clinical setting and for PCR standardization.
The presence of cytomegalovirus (CMV) DNA in blood as detected by PCR is used to monitor transplant patients at risk of active CMV infection (for reviews, see references 1 and 16). Both peripheral blood leukocyte (PBL) and plasma fractions are used for CMV PCR, but the clinical significance of results varies and is dependent on the PCR approach. By quantitative PCR techniques, demonstration of high-level CMV leukocyte DNAemia and plasma DNAemia usually precedes the development of clinical features. Although levels of leukocyte DNAemia and plasma DNAemia are usually well correlated, CMV DNA load is considerably higher in PBLs, which can reasonably be explained by the strong cell association of the virus (2, 6, 18, 22). As the performance of quantitative PCR is technically demanding and cost-intensive, which limits its high-throughput use, qualitative CMV PCR assays with reliable significance remain desirable. Leukocyte DNAemia, however, is often detected in CMV immunoglobulin G (IgG)-positive patients with no further evidence of active CMV infection, whereas a positive CMV PCR with plasma has been reported to be more specifically associated with clinical manifestations (1, 16).
Despite the important diagnostic role of CMV plasma DNAemia, its biological properties are not fully understood. One notion is that it reflects active virus replication (20–22). On the other hand, the high abundance of CMV leukocyte DNAemia in actively CMV-infected patients has led to the hypothesis that plasma DNAemia may be, at least partly, a result of PBL lysis (6, 22). If cell turnover was a major mechanism that elicits CMV plasma DNAemia, this would suggest that in patients with CMV leukocyte DNAemia long-term storage of whole blood samples before separation may cause release of viral DNA into the plasma fraction, thus distorting PCR results. This question is of great importance not only in the clinical setting but also for test reproducibility and standardization, especially since commercial kits for CMV PCR of plasma samples are available (3, 10). Some investigators argue that contamination of freshly prepared plasma fractions with cellular DNA can be overcome by ultrafiltration, whereas others have reported that PCR for cellular target sequences is regularly positive even with ultrafiltered samples (6, 20, 21). None of these investigations, however, tested the effect of delays in sample processing under controlled conditions. This problem has been addressed in the present study.
MATERIALS AND METHODS
Patients.
The blood samples from kidney allograft recipients investigated in this study were obtained between days 18 and 66 posttransplantation. Sixty CMV IgG-positive recipients with CMV leukocyte DNAemia were enrolled in the study. Of these, 30 patients were latently infected with CMV and 30 experienced active CMV infection (see below for definitions). Of the latently infected patients, 19 received a graft from a CMV-seronegative donor; 21 of the donors among the recipients who became actively infected with CMV were seronegative. Ten CMV IgG-positive patients without leukocyte DNAemia (seven donors were seronegative) and five CMV IgG-negative patients (all five donors were seronegative) served as controls. To rule out factors apart from the storage conditions which might influence CMV PCR results, patients receiving antiviral therapy at the time of the investigation were excluded.
Active CMV infection was confirmed by detection of pp65 antigens in PBLs. Latent infection was assumed when leukocyte samples remained pp65 antigen negative, no virus shedding from body fluids (urine, throat wash) was detectable by culture methods, and no evidence of symptomatic CMV infection was present throughout the stay at the hospital. CMV leukocyte DNAemia alone was not rated as active infection. Symptomatic CMV infection was defined by two CMV-associated symptoms (unexplained temperature of >38°C for >3 days, arthralgia, hyperhidrosis, graft dysfunction without histological evidence of rejection, leukopenia, thrombocytopenia, or liver enzyme level elevation), and/or confirmed organ involvement (pneumonitis or gastrointestinal ulceration with virus isolation from biopsy specimens) according to an international agreement (12).
Blood sample processing.
Twenty milliliters of whole blood was freshly collected from each participant and placed into tubes that contained EDTA. One-milliliter aliquots were prepared immediately postdrawing. One aliquot was processed right away for baseline PCR of PBLs and plasma, and one was used for the antigenemia assay (see below). The remaining unprocessed sample aliquots were stored at room temperature (RT) and at 4°C. From these, PBLs and plasma were prepared for PCR analysis after storage for 2, 4, 6, 12, and 24 h.
For each aliquot plasma was separated from blood cells by centrifugation at 700 × g for 10 min. Supernatants were centrifuged for a second time to pellet the cell debris. Half of each supernatant (approximately 500 μl) was then sterile filtered through a 0.2-μm-pore-size filter (Millipore, Eschborn, Germany). The supernatants of the ultrafiltered and native plasma samples were extracted with 1 volume of phenol-chloroform and subsequently with 1 volume of chloroform-isoamyl alcohol (24:1). DNA from 100 μl of the aqueous phase was precipitated with 0.1 volume of sodium acetate (pH 5.2) and 2.5 volumes of absolute ethanol. After centrifugation at 15,000 × g for 30 min, the pellets were washed with 70% ethanol and resuspended in 100 μl of H2O. A 10-μl sample was subjected to PCR.
PBLs were obtained by treating the pelleted cells with 0.8% ammonium chloride for erythrocyte lysis. The PBLs were washed with phosphate-buffered saline (pH 7.4) and quantitated with a hematological cell counter. DNA was extracted from 5 × 105 PBLs by digestion with 100 μg of proteinase K per ml (11, 14) in a 50-μl reaction volume. After being boiled for 10 min, a 10-μl sample of the supernatant was subjected to PCR.
PCR assays.
CMV DNA was quantitated in all leukocyte and plasma extracts. To assess the role of cell lysis, samples were additionally tested for the single-copy human β-globin gene. Target sequences were quantitated by competitive PCR with cloned standard (ST) sequences as described previously (11, 14), with slight modifications. The CMV ST sequence was generated in a PCR by site-directed mutagenesis and was subcloned into the vector pSPT19 (Boehringer Mannheim, Mannheim, Germany) (14). It contained three successive point mutations within the amplified coding region of CMV glycoprotein B compared to the sequence derived from laboratory strain AD169 (4). The cloning of the β-globin ST sequence was described previously (9).
Ten-microliter samples of DNA extracts from leukocyte or plasma preparations were added to the PCR mixtures (11, 14) to a total volume of 100 μl. The reaction mixtures were spiked with either 2 × 105 copies (leukocyte PCR) or 2 × 103 copies (plasma PCR) of the β-globin ST sequence and either 1 × 103 copies (high-ST) or 50 copies (low-ST) of the CMV ST sequence. CMV target sequences were amplified for 20 cycles with the external CMV-specific primers E1 (5′-TCCAACACCCACTAGACCGGT-3′) and E2 (5′-CGGAAACGATGGTGTAGTTGG-3′). Ten microliters of each of the external reaction mixtures was reamplified in a second round of PCR for 30 cycles with the internal CMV-specific primers TGGE1B (5′-CCGGATCCCGCCGCCCGCCCCGCGCCCGCCGCGGCAGCACCTGGCT-3′) and TGGE2E (5′-GCGAATTCGTAAACCACATCACC GTGGA-3′) and the β-globin-specific primers 1aB (5′-CCGGATCCCGCCGCCCGCCCCGCGCCCCTGCCGTTACTGCCCTGT-3′) and 1bE (5′-GCGAATCCTATTGGTCTCCTTAAACCTG-3′). The ST sequence and wild-type CMV and β-globin PCR amplimers were quantitated by hybridization to a strand-specifically labeled ST sequence, separation by temperature gradient gel electrophoresis, and densitometric analysis of autoradiographs (14). For samples with ≥500 CMV DNA copies in 10 μl, the figures from the high-ST reaction were used, and for samples with <500 CMV DNA copies, those from the low-ST reaction were used. Results were expressed as the average value of two measurements, which differed by ≤15% (data not shown). Exact quantification was possible within the ranges of 5 to 1 × 104 CMV wild-type genome equivalents and 2 × 103 to 2 × 105 β-globin copies per PCR mixture. In PBLs, CMV DNA/cellular DNA ratios were expressed as the number of CMV DNA copies/2 × 105 copies of β-globin DNA (the theoretical maximum DNA yield from 105 cells). Plasma CMV and β-globin copy numbers were expressed as the genome equivalents present in 10 μl of plasma (approximately equivalent to the volume of whole blood containing 105 PBLs).
Antigenemia assay.
Aliquots of 105 PBLs obtained from freshly prepared sample aliquots were centrifuged in duplicate onto glass slides, fixed, and permeabilized with 5% paraformaldehyde–0.5% Nonidet P-40, and pp65 antigens were detected in polymorphonuclear cells by indirect immunofluorescence as described previously (5) by using Clonab CMV (Biotest, Dreieich, Germany) according to the manufacturer's instructions.
Statistical analyses.
The frequencies of positive results of the CMV PCR and the β-globin PCR for the plasma samples were compared between different groups by the χ2 test. The frequencies of positive β-globin PCRs were compared between CMV PCR-positive and CMV PCR-negative samples of the same groups by the McNemar test. Quantities of CMV and β-globin target sequences were compared (i) in PBLs and in plasma over the storage period by the Friedman test combined with the Wilcoxon rank test, (ii) in PBLs and in plasma between actively and latently CMV-infected patients by the Mann-Whitney U test, and (iii) between native and ultrafiltered plasma samples also by the Mann-Whitney U test. The correlation of the number of CMV DNA copies in PBLs with those in plasma fractions was determined by Spearman regression analysis.
RESULTS
The effects of storage time and ultrafiltration on PCR results presented here were found to be independent of the storage temperature. Therefore, detailed data are shown only for sample aliquots stored at RT.
Qualitative CMV PCR of plasma.
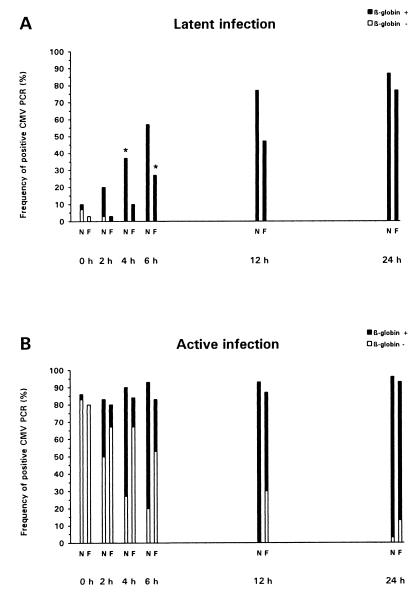
The influence of storage time and ultrafiltration on plasma CMV PCR results for the samples from 60 patients with leukocyte DNAemia (30 were latently CMV infected, and were 30 actively infected) is depicted in Fig. 1. At the baseline, among the native plasma samples only 10% from the latently CMV-infected group but 87% from the actively infected patients were CMV PCR positive (P < 0.001). Similar observations were made with the ultrafiltered plasma samples (3% positive for latently infected patients and 80% positive for actively infected individuals [P < 0.001]). In actively CMV-infected patients, the total rates of positive CMV PCRs did not increase significantly over the storage period, and no differences in the overall frequencies of positive CMV PCRs were evident between native and ultrafiltered samples. In contrast, among patients with latent CMV infection, the frequency of positive CMV PCR results was significantly higher than the baseline value by 4 h for native plasma samples (37% [P = 0.033]) and by 6 h for ultrafiltered samples (27% [P = 0.03]). The differences between actively and latently infected patients in the frequencies of positive CMV PCRs lost statistical significance by 12 and 24 h of storage regarding native and ultrafiltered plasma specimens, respectively.
FIG. 1.
Positivity rates for CMV PCR of plasma samples obtained from latently (A) and actively (B) CMV-infected patients, dependent on storage time (at RT) and ultrafiltration (N, native samples; F, ultrafiltered samples). Black bars, positivity both by CMV PCR and by β-globin PCR; white bars, positivity by CMV PCR but negativity by β-globin PCR. The storage times at which the frequencies of positivity of the CMV PCR became significantly different from the frequencies at the baseline are marked by asterisks.
To assess the influence of cell-associated DNA on the rates of positive plasma PCRs, the β-globin PCR results were additionally analyzed (Fig. 1). Different observations were made for latently and actively CMV-infected individuals. First, for latently infected patients, all CMV PCR-positive samples (native as well as ultrafiltered) were also β-globin PCR positive by 2 h postdrawing. In contrast, for patients with active CMV infection, the vast majority of CMV PCR-positive samples were β-globin PCR negative at the baseline, and for the ultrafiltered CMV PCR-positive aliquots, the β-globin PCR was negative significantly more frequently until at least 6 h of storage (P < 0.001). Second, for latently infected patients ultrafiltration significantly decreased the rates of positivity of the CMV PCR compared with those for native samples at 4 h (P = 0.033), 6 h (P = 0.036), and 12 h (P = 0.034) of storage. In contrast, for the actively CMV-infected patients, ultrafiltration did not reduce the overall rates of CMV PCR positivity but decreased only the proportion that was also β-globin PCR positive (4 h, P < 0.001; 6 h, P = 0.004; 12 h, P = 0.002). The overall rates of positivity of the β-globin PCR for native and ultrafiltered plasma sample aliquots did not differ significantly between latently and actively CMV-infected patients over the storage period (data not shown).
Only 2 of the 29 native plasma samples (3 from patients latently infected with CMV and 26 from patients actively infected with CMV) which were CMV PCR positive at the baseline became PCR negative in the course of storage. All plasma sample aliquots derived from the control patients without CMV leukocyte DNAemia remained CMV PCR negative throughout the storage period. For the samples from these patients, no differences in the overall rates of positivity of the β-globin PCR were seen compared with those for the corresponding samples from the patients who had CMV leukocyte DNAemia (data not shown).
Quantitative CMV PCR of plasma.
To further analyze the data obtained by qualitative PCR, the kinetics of CMV DNA and β-globin DNA were determined in all PBL and plasma sample aliquots over the storage period.
The CMV DNA load in PBLs at the baseline was significantly higher in actively CMV-infected patients (median, 950 copies/2 × 105 copies of β-globin DNA) than in patients with latent CMV infection (median, 45 copies [P < 0.001]). In actively CMV-infected patients, the ratio of CMV DNA levels in PBLs/CMV DNA levels in plasma (related to equivalent sample volumes) at the baseline ranged from 7.9 to 12.8 and from 8.2 to 13.1 for aliquots of native and ultrafiltered plasma, respectively. A significant correlation was found between the number of CMV DNA copies in PBLs and the number in the corresponding plasma samples (baseline values for native samples, Spearman regression coefficient [rs] = 0.77 [P = 0.025]; baseline values for ultrafiltered samples, rs = 0.82 [P = 0.022]). These values remained virtually constant over the storage period. For latently CMV-infected patients, the ratios of the CMV DNA numbers in PBLs/CMV DNA copy numbers in plasma did not exceed 2.5 (native plasma samples) and 1.9 (ultrafiltered aliquots) at the baseline. Furthermore, they dropped to nearly 1.0 during storage (data not shown).
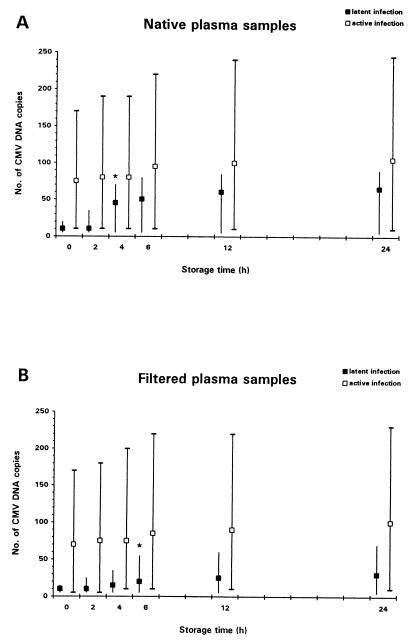
The kinetics of CMV DNA copies in plasma over the storage period are illustrated in Fig. 2. The numbers of CMV DNA copies detected in latently CMV-infected patients were significantly lower than the numbers detected in actively infected patients throughout the storage period. During storage, no relevant changes in CMV DNA copy numbers occurred in samples from patients with active CMV infection. In contrast, among the latently infected patients, the CMV copy numbers strongly increased during storage, and the differences from the baseline values became statistically significant after 4 h for native samples (P < 0.001) and 6 h for ultrafiltered samples (P < 0.01), respectively. Ultrafiltration of plasma samples only marginally decreased the CMV DNA copy numbers in actively infected patients, whereas in the latently CMV-infected individuals, the CMV DNA copy numbers were significantly lower in ultrafiltered samples than in native samples after storage times of ≥4 h (4 h, P < 0.001; 6 h, P < 0.001; 12 h, P < 0.01; 24 h, P < 0.01).
FIG. 2.
Effect of storage time (at RT) and of ultrafiltration on quantitative CMV PCR of plasma samples. The copy numbers are for 10 μl of plasma. Median values are shown for latently (■) and actively (□) CMV-infected patients. Error bars span the maximum and minimum values. The storage times at which differences from the baseline results became statistically significant are marked by asterisks.
The β-globin DNA copy numbers in native plasma samples reached about 5%, on average (mean, 9 × 103 copies in 10 μl), of the values determined in the corresponding PBL aliquots during the storage period. By storage times of ≥4 h the β-globin copy numbers measured after ultrafiltration were significantly lower compared with the numbers in aliquots of native plasma. No significant differences in β-globin DNA copy numbers were observed between the corresponding samples from patients with CMV leukocyte DNAemia and control patients at any time during storage (data not shown).
In PBL fractions of actively infected patients, CMV DNA and β-globin DNA copy numbers decreased slightly over the storage period, but the differences from the baseline values did not reach statistical significance. However, a significant decrease in CMV DNA copy numbers was noted for samples from latently CMV-infected patients (P < 0.01 after 6 h). No significant differences in β-globin DNA copy numbers were observed between the corresponding samples from patients with CMV leukocyte DNAemia and control patients (data not shown).
DISCUSSION
In latently CMV-infected renal allograft recipients CMV PCR performed with plasma yields false-positive results with significant frequency when specimens are prepared with delay, regardless of the storage temperature. In contrast, storage factors do not seem to have any relevant effect on the results of CMV PCR for patients with active CMV infection. These findings have important implications for the diagnostic significance of plasma PCR.
The presence of CMV leukocyte DNAemia proved to be necessary for positive plasma PCR results. As isolated CMV leukocyte DNAemia in transplant recipients is rated as a pathological condition per se by some investigators (1), it must be noted that in this study the definition of latent CMV infection in leukocyte DNAemia-positive patients ruled out any episodes of pp65 antigenemia, virus shedding from body fluids, or CMV-related symptoms during the stay at hospital. In contrast, truly active CMV infection was assumed only when pp65 antigenemia was confirmed, because the prognostic significance of CMV DNAemia is dubious in pp65-negative patients with other signs of virus activity, e.g., virus isolation from urine or throat swabs (11, 19).
Fundamentally different effects of storage on plasma CMV PCR were observed between plasma from latently and actively CMV-infected patients. The data suggest that lysis of latently infected PBLs is the driving force for CMV plasma DNAemia in patients with latent infection but that this does not play a key role in active CMV infection.
In latently infected leukocytes, CMV DNA has been localized to the cell nucleus (7). Preanalytical turnover of these cells, with nucleus-associated DNA entering the plasma fraction, provides a reasonable explanation for the finding that the plasma CMV DNAemia that was low grade at early storage times continuously rose over time and strongly diminished with ultrafiltration, in parallel with β-globin DNA levels in plasma. Accordingly, low absolute CMV copy numbers in PBLs and low ratios of CMV DNA in PBLs/CMV DNA in plasma gradually diminished over time.
Interpretation of the data for the actively infected patients is more complex. Plasma CMV DNA representing cell-free target sequences (6, 20, 21) is the most obvious explanation for the high-grade plasma CMV DNAemia at the baseline which remained stable over time; therefore, it did not correlate with β-globin DNA kinetics and was largely uninfluenced by ultrafiltration. Moreover, CMV PCR-positive but β-globin PCR-negative samples were present throughout storage. The notion of cell-free virus would fit reports on the recovery of infectious virus from the plasma fraction (6, 20). The release of virus DNA into the plasma fraction from other surrounding sites of infection (e.g., endothelial cells) is conceivable.
However, cell-free virus is not the only possible explanation. The significant correlation of CMV DNA copies between PBLs and plasma and the similar ratios of CMV DNA in PBLs/CMV DNA in plasma throughout storage suggests that plasma DNAemia in actively infected patients is also, to some extent, leukocyte associated (6, 18, 22). This is not necessarily contradictory to the statements made above because the CMV DNA circulating in PBLs during active infection has been localized to the cytoplasm (7) rather than the nucleus, most likely as a result of virus uptake from surrounding sites of replication. Accordingly, in actively infected patients CMV DNA and β-globin DNA behaved consistently different in terms of ultrafiltrability. During CMV dissemination there may be a steady state in the bloodstream between the viral components present in the cytoplasms of cells and in the plasma fraction, leading to a comparatively high plasma CMV DNA load. Such a hypothesis could explain why, despite the high CMV DNA load in PBLs, the plasma CMV levels found at the baseline increased only marginally during storage, although PBL lysis must have resulted in the release of cytoplasmic CMV DNA.
Although samples of actively CMV-infected patients probably also contained CMV DNA derived from latently infected PBLs which entered the plasma over time, this was unlikely to have a great influence on the interpretation of the results because the levels of latent CMV DNA derived from PBLs (predominantly mononuclear cells) are very low in both actively and latently CMV-infected patients (18).
The β-globin PCR data may reconcile some discrepant previous statements. Gerna et al. (6) regularly detected β-globin DNA by qualitative PCR in plasma samples after ultrafiltration using a highly efficient DNA extraction method. Others have reported the loss of β-globin signals by ultrafiltration, but the investigators used a proteinase K-based extraction protocol which refrained from DNA precipitation (20, 21). Therefore, the most likely reason for these discrepancies is the different sensitivities of the methods. In agreement with this interpretation, quantitation of β-globin DNA in our study revealed that only a limited amount of cellular material was effectively ultrafiltered.
The preanalytical pitfalls of plasma CMV PCR may be overcome by alternative PCR approaches that are less error prone. First, qualitative CMV PCR of polymorphonuclear leukocyte fractions depleted of the mononuclear cells by Ficoll density centrifugation is promising for the monitoring of renal allograft recipients (18). Its specificity is comparable to that plasma PCR and it is even more sensitive than plasma PCR (15), and storage of unprocessed samples for at least 72 h has no effect on the results (17). Second, serum CMV PCR is supposed to be equivalent to plasma PCR for prediction of active infections (8, 13) and therefore should also be investigated for the adverse effects of delayed sample preparation.
In conclusion, it has been demonstrated that a mere positive CMV PCR result obtained with the plasma of a renal allograft patient with CMV leukocyte DNAemia should be interpreted with extreme caution. For quantitative CMV PCR delays in specimen processing are probably less crucial. It is urgently necessary to take notice of this matter in the clinical setting and when any measures for standardization of plasma CMV PCR procedures are devised.
REFERENCES
- 1.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin G, Handfield J, Toma E, Murray G, Lalonde R, Bergeron M G. Comparative evaluation of the cytomegalovirus load in polymorphonuclear leukocytes and plasma of human immunodeficiency virus-infected subjects. J Infect Dis. 1998;177:355–360. doi: 10.1086/514190. [DOI] [PubMed] [Google Scholar]
- 3.Boivin G, Handfield J, Toma E, Murray G, Lalonde R, Tevere V J, Sun R, Bergeron M G. Evaluation of the AMPLICOR cytomegalovirus test with specimens from human immunodeficiency virus-infected subjects. J Clin Microbiol. 1998;36:2509–2513. doi: 10.1128/jcm.36.9.2509-2513.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cranage M P, Kouzarides T, Bankier A T, Satchwell S, Weston K, Tomlinson P, Barrell B, Hart H, Bell S E, Minson A C, Smith G L. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerna G, Furione M, Baldanti F, Sarasini A. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J Clin Microbiol. 1994;32:2709–2717. doi: 10.1128/jcm.32.11.2709-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackstein H, Kirchner H, Jahn G, Bein G. The intracellular localization of human cytomegalovirus DNA in peripheral blood leukocytes during active infections by high-resolution fluorescence in situ hybridization. Arch Virol. 1996;141:1293–1305. doi: 10.1007/BF01718831. [DOI] [PubMed] [Google Scholar]
- 8.Hamprecht K, Mikeler E, Jahn G. Semi-quantitative detection of cytomegalovirus DNA from native serum and plasma by nested PCR: influence of DNA extraction procedures. J Virol Methods. 1997;69:125–135. doi: 10.1016/s0166-0934(97)00148-1. [DOI] [PubMed] [Google Scholar]
- 9.Henco K, Heibey M. Quantitative PCR: the determination of template copy numbers by temperature gradient gel electrophoresis (TGGE) Nucleic Acids Res. 1990;18:6733–6734. doi: 10.1093/nar/18.22.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiyoshi M, Tagawa S, Takubo T, Tanaka K, Nakao T, Higeno Y, Tamura K, Shimaoka M, Fujii A, Higashihata M, Yasui Y, Kim T, Hiraoka A, Tatsumi N. Evaluation of the AMPLICOR CMV test for direct detection of cytomegalovirus in plasma specimens. J Clin Microbiol. 1997;35:2692–2694. doi: 10.1128/jcm.35.10.2692-2694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kühn J E, Wendland T, Schäfer P, Möhring K, Wieland U, Elgas M, Eggers H J. Monitoring of renal allograft recipients by quantitation of human cytomegalovirus genomes in peripheral blood leukocytes. J Med Virol. 1994;44:398–405. doi: 10.1002/jmv.1890440416. [DOI] [PubMed] [Google Scholar]
- 12.Ljungman P, Plotkin S A. Workshop on CMV disease; definitions, clinical severity scores, and new syndromes. Scand J Infect Dis Suppl. 1995;99:87–89. [Google Scholar]
- 13.Patel R, Smith T F, Espy M, Wiesner R H, Krom R A F, Portela D, Paya C V. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J Clin Microbiol. 1994;32:1431–1434. doi: 10.1128/jcm.32.6.1431-1434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schäfer P, Braun R W, Möhring K, Henco K, Kang J, Wendland T, Kühn J E. Quantitative determination of human cytomegalovirus target sequences in peripheral blood leukocytes by nested polymerase chain reaction and temperature gradient gel electrophoresis. J Gen Virol. 1993;74:2699–2707. doi: 10.1099/0022-1317-74-12-2699. [DOI] [PubMed] [Google Scholar]
- 15.Schäfer P, Kühn J E, Tenschert W, Eing B, Schröter M, Laufs R. Polymerase chain reaction (PCR) from Ficoll-purified polymorphonuclear leukocytes for monitoring cytomegalovirus infections in renal allograft recipients: superior sensitivity and similar specificity compared with plasma PCR. J Infect Dis. 1998;178:1544–1545. doi: 10.1086/314468. [DOI] [PubMed] [Google Scholar]
- 16.Schäfer P, Laufs R. Experience with quantitative PCR for the management of HCMV disease. Intervirology. 1996;39:204–212. doi: 10.1159/000150496. [DOI] [PubMed] [Google Scholar]
- 17.Schäfer P, Tenschert W, Gutensohn K, Laufs R. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J Clin Microbiol. 1997;35:741–744. doi: 10.1128/jcm.35.3.741-744.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schäfer P, Tenschert W, Cremaschi L, Gutensohn K, Laufs R. Utility of major leukocyte subpopulations for monitoring secondary cytomegalovirus infections in renal-allograft recipients by PCR. J Clin Microbiol. 1998;36:1008–1014. doi: 10.1128/jcm.36.4.1008-1014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinkai M, Bozzette S A, Powderly W, Frame P, Spector S A. Utility of urine and leukocyte cultures and plasma DNA polymerase chain reaction for identification of AIDS patients at risk for developing human cytomegalovirus disease. J Infect Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- 20.Spector S A, Merrill R, Wolf D, Dankner W M. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J Clin Microbiol. 1992;30:2359–2365. doi: 10.1128/jcm.30.9.2359-2365.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf D G, Spector S A. Early diagnosis of human cytomegalovirus disease in transplant recipients by DNA amplification in plasma. Transplantation. 1993;56:330–334. doi: 10.1097/00007890-199308000-00014. [DOI] [PubMed] [Google Scholar]
- 22.Zipeto D, Morris S, Hong C, Dowling A, Wolitz R, Merigan T C, Rasmussen L. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J Clin Microbiol. 1995;33:2607–2611. doi: 10.1128/jcm.33.10.2607-2611.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]