Abstract
The sequences of the internal transcribed spacer (ITS) ribosomal DNA (rDNA) domain data obtained by restriction fragment length polymorphism analysis with 18S rDNA and fingerprinting (M13) for clinical and environmental strains of Pseudallescheria boydii (anamorph, Scedosporium apiospermum) were compared to those for related species of Pseudallescheria, Petriella, and Scedosporium. The infraspecific variability of P. boydii was considerable. There were five different lengths in the 18S rDNAs within P. boydii due to the occurrence of introns. In several cases, strains isolated from a single pond or ditch proved to be genetically very different. Nevertheless, some lineages had a regional distribution. The variability found is unlikely to be explained by meiotic recombination alone. Pseudallescheria fusoidea, Pseudallescheria ellipsoidea, and Pseudallescheria angusta were found to be synonyms for P. boydii. Scedosporium prolificans was found amid Petriella species in the ITS tree and showed no infraspecific variability. The type strain of Rhinocladium lesnei proved to be identical to Graphium putredinis. Acladium castellanii, which is morphologically reminiscent of S. apiospermum, was also found to be a separate species, but with an unknown affiliation.
Pseudallescheria boydii (Shear) McGinnis et al. [anamorph, Scedosporium apiospermum (Sacc.) Sacc.] is one of the emerging agents of opportunistic mycoses in humans. Twelve case reports for hospitalized patients suffering from myeloid leukemia or subjected to immunosuppressive therapy were published in 1999 alone. Infections are difficult to treat because of its resistance to common antifungal drugs such as amphotericin B. The diagnostic problems with this species are considerable. In tissue P. boydii is indistinguishable from other filamentous fungi (20), and Cryptococcus antigen testing may lead to false-positive reactions (23). Systemic infections may easily be misidentified as aspergillosis, leading to inappropriate therapy. This is one of the reasons that the mortality rate due to invasive pseudallescheriasis is high.
Judging from the number of patient isolates received by the Centraalbureau voor Schimmelcultures (CBS) Identification Department, there may still be a considerable underdetection of P. boydii (9), probably due to the species' clinical and morphological diversity. In the past, etiologic agents were frequently introduced as new taxa, as their identity with existing species was not recognized. Still, the literature contains a number of “ghost taxa” in such divergent genera as Acremonium, Actinomyces, Cephalosporium, Graphium, Madurella, and Verticillium that may be identical to P. boydii (8).
The anamorph of the species was introduced more than 100 years ago by Harz and Bezold (see references 27 and 28) as an agent of human otitis. Since the 1920s it became known as one of the major agents of mycetoma (26) and other subcutaneous infections. During the last few decades, rhinopharyngeal and pulmonary colonization was reported in leukemic, cystic fibrosis, and otherwise impaired patients (35). In disseminated cases the species is significant because of its neurotropism, with marked predilection for the cerebrospinal fluid (12). Such infections were sometimes acquired after near-drowning events in polluted waters (24).
Much of the observed morphological variability can be ascribed to variable abundance of (syn)ana- and teleomorphs. This was confirmed by the investigation of characters independent of morphology, such as nutritional physiology (10) and 18S ribosomal DNA (rDNA) sequencing (16). Wedde et al. (36), using rDNA ITS2 sequencing, were able to select primers for species recognition. However, infraspecific molecular variability of the internal transcribed spacer (ITS) rDNA of P. boydii seems to be larger than that of species such as Trichophyton rubrum (Castell.) Sab. (14), Hortaea werneckii (Horta) Nishimura et Miyaji (38), or Cladophialophora bantiana (Sacc.) de Hoog et al. (13). Previously, nuclear DNA (nDNA) homology studies had indicated the existence of three infraspecific groups in P. boydii (10, 15), and the clinical pictures and environmental sources of isolation of these three groups broadly matched. Bell (2) had found differences in virulence between a strain from patients with subcutaneous infections and one from the environment. Thus, P. boydii may comprise entities with different pathogenicities.
In the study described in the present paper, the molecular variability of P. boydii was evaluated. This should help to establish a clear species concept which is required for reliable molecular identification of this opportunist in the routine laboratory. To this aim we compared the published nDNA-DNA homology data with results of small subunit (SSU) restriction analysis and sequencing and of fingerprinting by PCR (primer M13). The strains of P. boydii compared included isolates that originated from a single source (patient or environmental), as well as a worldwide selection of strains collected over a period of 70 years (Table 1). Some closely related taxa are included for comparison.
TABLE 1.
Strains examined and sources of isolation
| Species | Strain no. | Status | Source of isolation | Origin |
|---|---|---|---|---|
| P. boydii 1 | CBS 101.22 | Type strain of Allescheria boydii Shear | Mycetoma | Texas |
| P. boydii 2 | IP 1411.82 | Mycetoma | ||
| P. boydii | CBS 100.870 | Mycetoma | ||
| P. boydii | RKI 2782/95 | Trauma and sepsis | Hamburg, Germany | |
| P. boydii | RKID 386 | Subcutaneous mycosis, leukemic patient | Hungary | |
| P. boydii | CBS 330.93 | Bronchial secretion | Alkmaar, The Netherlands | |
| P. boydii | CBS 329.93 | Bronchial secretion | Alkmaar, The Netherlands | |
| P. boydii | CBS 101718 | Encephalitis of a child after near drowning | Germany | |
| P. boydii 1 | IP 1698.87 | Lung of a leukemic patient | France | |
| P. boydii 1 | IP 1945.90 | Sputum of a cystic fibrosis patient | France | |
| P. boydii | RKI 866/94 | Sputum of a patient after heart transplantation | Berlin, Germany | |
| P. boydii | RKI 2956/93 | BAL fluida of a patient after heart transplantation | Berlin, Germany | |
| P. boydii 2 | CBS 695.70 | Type strain of Acremonium suis Bakai | Sinus of a pig | Kiev, Ukraine |
| P. boydii 2 | IP 1946.90 | Sinus | France | |
| P. boydii 2 | CBS 987.73 | Otitis | CSSR | |
| P. boydii | CBS 100.26 | Type strain of Acladium castellanii Pinoy in Castellani | Human | |
| P. boydii | CBS 591.90 | Type strain of Pseudallescheria shearii Negroni & Fischer | Mycetoma of the knee | Buenos Aires, Argentina |
| P. boydii 3 | CBS 108.54 | Soil | Zaire | |
| P. boydii 3 | IP 1742.88 | Soil | France | |
| P. boydii | CBS 101717 | Soil | Brazil | |
| P. boydii 2 | CBS 499.90 | Mud of a pond | Groningen, The Netherlands | |
| P. boydii | CBS 101719 | Mud of a pond | Groningen, The Netherlands | |
| P. boydii | CBS 101720 | Sandy soil of a polluted ditch | Site of a car accident (compare CBS 330.93 and CBS 329.93) | |
| P. boydii | CBS 101721 | Mud | s'Graveland, The Netherlands | |
| P. boydii | CBS 101722 | Mud | Gooimeer, The Netherlands | |
| P. boydii | CBS 101723 | Mud | Eempolder, The Netherlands | |
| P. boydii | CBS 101724 | Mud | Wasmeer, The Netherlands | |
| P. boydii | CBS 101725 | Mud | Lapersveld, The Netherlands | |
| P. boydii | CBS 101726 | Mud | Lapersveld, The Netherlands | |
| P. africana | CBS 311.72 | Type strain of Pseudallescheria africana (von Arx et G. Franz) McGinnis et al. | Brown sandy soil | 25 km west of Tsintsabis, Namibia |
| P. angusta | CBS 254.72 | Type strain of Pseudallescheria angusta (Malloch et Cain) McGinnis et al. | Half-digested sewage tank | Ohio |
| P. desertorum | CBS 489.72 | Type strain of Pseudallescheria desertorum (von Arx et Moustafa) McGinnis et al. | Salt marsh soil | Kuwait |
| P. ellipsoidea | CBS 418.73 | Type strain of Pseudallescheria ellipsoidea (von Arx et Fassatiová) McGinnis et al. | Soil | Tadzjikistan |
| P. fimeti | CBS 129.78 | Type strain of Pseudallescheria fimeti (von Arx et al.) McGinnis et al. | Dung of a goat | Aligarh, India |
| P. fusoidea | CBS 106.53 | Type strain of Pseudallescheria fusoidea (von Arx) McGinnis et al. | Soil | Guipo, Panama |
| G. calicioides | CBS 102084 | Decayed wood | Aomori Prefecture, Japan | |
| G. calicioides | CBS 102080 | Decayed wood | Yamanashi Prefecture, Japan | |
| G. penicillioides | CBS 320.72 | Type strain of Stilbum basitruncatum Matsushima | Forest soil | Honara, Solomon Islands |
| G. putredinis | CBS 102083 | Chrysalidiocarpus lutescens | Tokyo, Japan | |
| G. tectonae | CBS 127.84 | Type strain of Graphium tectonae C. Booth | Seed of Tectona grandis | Jamaica |
| Petriella guttulata | CBS 362.61 | Synonymous type strain of Petriella guttulata Barron et Cain | Dung of partridge | Germany |
| Petriella lindforsii | CBS 352.59 | Woodland soil | ||
| Petriella musispora | CBS 745.69 | Type strain of Petriella musispora Malloch | Decayed wood of Populus tremuloidesetriella | Ontario, Canada |
| Petriella setifera | CBS 559.80 | Ocho Rios, Jamaica | ||
| S. prolificans | CBS 114.90 | Type strain of Scedosporium inflatum Malloch et Salkin | Bone biopsy specimen of a 6-year old male | United States |
| S. prolificans | RKID 85 | Drain of the sinus maxillaris of a patient with AMLb | Rostock, Germany | |
| S. prolificans | RKI 1482/95 | Human | ||
| S. prolificans | RKI 2399/94 | Human | ||
| S. prolificans | CBS 467.74 | Type strain of Lomentospora prolificans Hennebert et Desai | Greenhouse soil, from mixed forest litter | Heverlee, Belgium |
BAL, bronchoalveolar lavage.
AML, amyelocytic leukemia.
MATERIALS AND METHODS
DNA extraction.
The methods applied for DNA extraction were described previously (13).
Restriction analysis.
Amplification of the 18S rDNA was performed with primers Oli4 and NS24 (31). The resulting amplicons were digested with the restriction endonucleases HaeIII, HinfI, DdeI, RsaI, TaqI, MspI, and HhaI (Amersham/Pharmacia Biotech) under the conditions recommended by the manufacturer. The corresponding products were electrophoresed in 1.5% agarose gels at 150 V for 2 to 3 h. The fragments were analyzed by computer-aided image analysis (ImageMaster; Pharmacia Inc.) and were verified by comparison with the expected patterns on the basis of the sequences of the same or related strains.
Sequence determination and analysis.
The ribosomal ITS region was amplified with primers V9G and LS266 (13). Both strands were sequenced with the internal primers ITS2, ITS3, ITS4, and ITS5 and the Big Dye terminator cycle sequencing kit (PE Applied Biosystems, Warrington, United Kingdom), as recommended by the manufacturer, combined with an ABI automatic DNA sequencer. Sequences were sampled with the Seqman package (DNAStar Inc., Madison, Wis.) and were aligned with BioNumerics software (Applied Maths, Kortrijk, Belgium). The distance tree was constructed with the neighbor-joining algorithm with the Kimura correction in the Treecon package (32). The robustness of the branches was assessed by bootstrap analysis with 100 replicates. The topology of the tree was verified with several algorithms including the parsimony algorithm.
PCR fingerprinting.
The core sequence of phage M13 was used as a single primer in the PCR experiments. Amplification reactions were performed as described by Weising et al. (37) in an Amplitron II thermocycler. For each reaction 10 ng of DNA was used. Forty PCR cycles were programmed as follows: 20 s at 93°C, 60 s at 50°C, and 20 s at 72°C, with elongation at 72°C for 6 min and chilling to 4°C. The PCR products were electrophoresed on 1.5% agarose gels at 100 V for 4 to 6 h. The gels were stained with ethidium bromide and photographed under UV light. The DNA fragment profiles were analyzed with the help of Gelcompar software (Applied Maths).
RESULTS
Restriction analysis.
Data obtained by restriction fragment length polymorphism (RFLP) analysis of the 18S rDNA gene are summarized in Table 2. Amplicons varied in length because of the occurrence of inserts of about 380, 470, 920, or 1,500 bp. Only restriction patterns that resulted from the digestion of 18S rDNA genes of the same length are comparable. Twenty-two strains had no inserts. Ten such strains, identified as P. boydii, yielded identical patterns when the same enzymes were used; the exception was RKI 866/94 digested with MspI. The patterns of strains of nDNA reassociation groups 1 to 3 were identical. The patterns of type strains of Pseudallescheria africana (von Arx et G. Franz) McGinnis et al., Pseudallescheria angusta (Malloch et Cain) McGinnis et al., Pseudallescheria fusoidea (von Arx et al.) McGinnis et al., and Pseudallescheria ellipsoidea (von Arx et Fassatiová) McGinnis et al. were identical to those of P. boydii. In contrast, strain CBS 100.26, maintained in the CBS culture collection as P. boydii, was deviant with all enzymes used. Graphium tectonae C. Booth and Pseudallescheria fimeti (von Arx et al.) McGinnis et al. deviated from P. boydii in HaeIII restriction patterns. Four strains of Scedosporium prolificans (Hennebert et Desai) Guého et de Hoog had the same profiles only with MspI and TaqI; they were identical to each other except for the HinfI pattern for CBS 467.74. Petriella guttulata Barron et Cain and Petriella lindforsii Curzi showed significant differences with most enzymes.
TABLE 2.
Summary of RFLP analysis of the 18S rDNA region with seven restriction enzymesa
| Species | Strain no. | Length (bp) of SSU rDNA | Pattern obtained with the following enzyme:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| HaeIII | HinfI | DdeI | RsaI | MspI | HhaI | TaqI | |||
| P. boydii 1 | IP 1698.87 | 1,800 | A | A | A | A | A | A | A |
| P. boydii 1 | CBS 101.22 | 1,800 | A | A | A | A | A | A | A |
| P. boydii 1 | IP 1945.90 | 1,800 | A | A | A | A | A | A | |
| P. boydii 2 | CBS 499.90 | 1,800 | A | A | A | A | A | A | A |
| P. boydii 3 | CBS 108.54 | 1,800 | A | A | A | A | A | A | A |
| P. boydii 3 | IP 1742.88 | 1,800 | A | A | A | A | A | A | A |
| P. boydii | CBS 591.90 | 1,800 | A | A | A | A | A | A | A |
| P. boydii | RKI 866/94 | 1,800 | A | A | A | A | B | A | A |
| P. boydii | CBS 101720 | 1,800 | A | A | A | A | A | A | A |
| P. boydii | CBS 101717 | 1,800 | A | A | A | A | A | A | A |
| P. africana | CBS 311.72 | 1,800 | A | A | A | A | A | A | A |
| P. angusta | CBS 254.72 | 1,800 | A | A | A | A | A | A | |
| P. ellipsoidea | CBS 418.73 | 1,800 | A | A | A | A | A | A | A |
| P. fusoidea | CBS 106.53 | 1,800 | A | A | A | A | A | A | A |
| P. fimeti | CBS 129.78 | 1,800 | D | A | A | A | A | A | A |
| G. tectonae | CBS 127.84 | 1,800 | E | A | A | A | A | A | A |
| S. prolificans | RKI 1482/95 | 1,800 | C | J | D | C | A | C | A |
| S. prolificans | RKI 2399/94 | 1,800 | C | J | D | C | A | C | A |
| S. prolificans | CBS 467.74 | 1,800 | C | D | D | C | A | C | A |
| S. prolificans | CBS 114.90 | 1,800 | C | J | D | A | A | ||
| P. guttulata | CBS 362.61 | 1,800 | D | A | A | L | H | E | A |
| P. lindforsii | CBS 352.59 | 1,800 | F | F | F | E | E | A | D |
| P. boydii | CBS 100.26 | 1,800 | M | K | G | H | F | K | C |
| P. boydii | CBS 330.93 | 2,180 | B | B | B | B | D | B | F |
| P. boydii | CBS 329.93 | 2,180 | B | B | B | B | D | B | F |
| P. boydii 2 | IP 1946.90 | 2,180 | B | B | B | B | D | B | B |
| P. boydii | RKI 2956/93 | 2,180 | B | B | B | B | D | B | B |
| P. boydii | RKID 386 | 2,180 | B | B | B | B | D | B | B |
| P. boydii 2 | IP 1411.82 | 2,180 | B | B | B | B | D | B | |
| P. boydii 2 | CBS 987.73 | 2,180 | H | H | H | J | M | H | F |
| P. boydii | CBS 101719 | 2,180 | H | H | J | J | M | H | F |
| P. boydii | CBS 101718 | 2,270 | I | I | I | I | I | I | I |
| P. desertorum | CBS 489.72 | 2,720 | G | E | E | D | D | D | |
| P. boydii | RKI 2782/95 | 2,720 | H | G | L | G | C | G | B |
| P. boydii 2 | CBS 695.70 | 3,300 | B | C | C | F | G | F | |
The different lengths of the corresponding SSU rDNAs are mentioned. For P. boydii the numbers following the species name indicate the nDNA-DNA reassociation groups (15). Banding patterns are characterized as letters.
Eight strains identified as P. boydii had amplicon lengths of about 2,180 bp, corresponding to the presence of an intron of about 380 bp that was demonstrated in two P. boydii strains by Issakainen et al. (16). Identical RFLP patterns were generated for four strains; however, some deviations were found with TaqI. The TaqI patterns of strains CBS 330.93 and CBS 329.93 of P. boydii deviated. Two strains (strains CBS 987.73 and CBS 101719) were different in tests with all enzymes except TaqI, which suggests that another intron of a similar length is present in P. boydii. The few strains with longer 18S rDNA amplicons yielded incomparable restriction patterns.
Sequencing.
For the distance trees presented in Fig. 1 and 2 we included ITS1 and ITS2 sequences published by Wedde et al. (36) and Lennon et al. (18). Sequences could not always be aligned with confidence, particularly the sequence of P. boydii CBS 100.26 (Acladium castellanii Pinoy), which was excluded from further analysis, and a group around Petriella setifera, which could be only partially aligned, as indicated in Fig. 1. Separate partial and complete ITS domain analyses were performed with and without this group. Table 3 summarizes the numbers of base substitutions in the ITS domains by taking P. boydii CBS 330.93 as a reference. Within the entire group compared, a maximum of 58 positions in ITS1 were variable, 79 positions in ITS2 were variable, and 1 position in the 5.8S rDNA gene was variable.
FIG. 1.
Phylogenetic tree of species of the family Microascaceae studied. The tree was constructed on the basis of confidently aligned positions of the rDNA ITS domain. The tree was generated with the Treecon package with the neighbor-joining algorithm and Kimura correction. G. calicioides was used as the outgroup. The tree was subjected to 100 bootstrap replications; only values >90 are shown. G, Graphium; P, Pseudallescheria; Pe, Petriella; R, Rhinocladium; S, Scedosporium; (T), ex type strain; 1 to 3, nDNA homology groups (15).
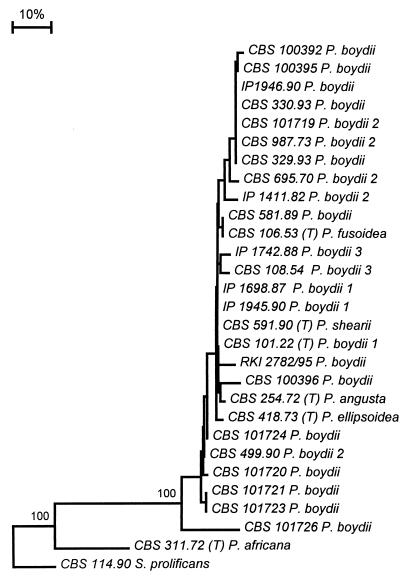
FIG. 2.
Phylogenetic tree of P. boydii, constructed as described for the tree in Fig. 1. S. prolificans CBS 114.90 was used as the outgroup.
TABLE 3.
ITS sequence diversity in Pseudallescheria and related fungi
| Strain | No. of base substitutionsa
|
||
|---|---|---|---|
| ITS1 | 5.8S | ITS2 | |
| P. boydii CBS 330.93 | 0 | 0 | 0 |
| P. boydii CBS 100396 | 7 | 3 | 8 |
| P. boydii CBS 101721 | 17 | 0 | 5 |
| P. africana CBS 311.72T | 32 | 3 | 35 |
| G. tectonae CBS 127.84T | 43 | 0 | 41 |
| R. lesnei CBS 108.10T | 45 | 0 | 51 |
| S. prolificans CBS 114.90 | 46 | 0 | 43 |
| P. fimeti CBS 129.78T | 55 | 0 | 74 |
| P. guttulata CBS 362.61T | 81 | 0 | 69 |
| G. penicillioides CBS 320.72 | 70 | 0 | 78 |
| P. lindforsii CBS 352.59 | 84 | 3 | 100 |
Numbers of base substitutions with respect to the sequence of P. boydii CBS 330.93.
In the general tree (Fig. 1), P. setifera ATCC 26490 was taken as the outgroup. All strains morphologically identified as P. boydii composed a main group. The 10 strains that had deviating 18S rDNA amplicon lengths are now confirmed to fall within the range of variability of P. boydii. Among these were the type strains of P. angusta (CBS 254.72), P. ellipsoidea (CBS 418.73), and P. fusoidea (CBS 106.53). P. africana (CBS 311.72) and Pseudallescheria desertorum (v. Arx et Moustafa) McGinnis et al. (CBS 489.72) were located outside the P. boydii clade. CBS 101721 had considerable deviations in ITS1, while the ITS2 domain was nearly identical (Table 3).
P. guttulata-P. setifera, Pseudallescheria fimeti, G. tectonae, Graphium calicioides (Fr.) Cooke et Massee, and Graphium putredinis (Corda) S. Hughes took rather isolated positions. The type strain of Rhinocladium lesnei Vuill., CBS 108.10, was nearly identical to G. putredinis. Five strains of S. prolificans were strictly identical to each other. They did not match any of the teleomorphs included in the study.
Internal branches within the P. boydii clade (Fig. 2) were found to correspond partly with the postulated infraspecific groups on the basis of nDNA-DNA reassociation data (15), but this could not be statistically confirmed. Strains CBS 329.90 and CBS 330.93, isolated from a single comatose patient after a near drowning, had identical sequences. In contrast, strain CBS 101720, isolated 2 years later from water at the site of the accident, is significantly different. Strains CBS 499.90 and CBS 101719, both isolated from mud of the same pond, had different ITS sequences. P. boydii strains CBS 101725 and CBS 101726, which were isolated from a single sample of polluted pond water, proved to be a significant distance from each other. In contrast, the ITS sequences and M13 fingerprints of pairs of separately isolated strains, strains CBS 101721 and CBS 101723 and strains IP 1698.87 and 1945.90, were (nearly) identical.
Fingerprinting with M13.
Examples of the banding patterns resulting from PCR fingerprinting are shown in Table 4. Profiles were found to be highly heterogeneous. Almost every strain had its own pattern; very few bands could be matched with confidence. All tests were repeated several times and proved to be reproducible. The fingerprints of the type strains of P. africana (CBS 311.72) and P. ellipsoidea (CBS 418.73), having somewhat deviant ITS sequences, proved to be identical to each other. Clinical strains IP 1698.87 and IP 1945.90 were also identical. The two strains from a single patient, CBS 329.93 and CBS 330.93, had the same patterns (data not shown), whereas strain CBS 101720, from the site of the accident but with another ITS sequence, proved to have different patterns.
TABLE 4.
Banding patterns from fingerprinting with M13
| Strain | Presence or absence of bands at the following positionsa:
|
||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | |
| P. boydii RKI D 386 | + | − | − | + | + | − | − | − | − | + | − | − | + | − | − | − | − | + | − | − | + | − | − | − | + | − | + | − | − | − | + | − | − | − | − | − | − | + | + |
| P. boydii RKI 2956/93 | + | − | − | + | − | − | − | − | − | + | − | − | + | − | + | − | − | + | + | − | + | − | − | − | − | + | − | − | + | − | + | − | − | + | − | + | − | − | − |
| P. boydii 3 CBS 108.54 | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | + | − | + | + | − | − | − | − | − | + | − | − | + | − | − | + | − | − | − | + | − | − | − |
| P. fimeti CBS 129.78 | − | − | − | − | + | − | − | − | − | − | + | − | − | + | − | + | − | + | − | + | − | − | − | − | − | − | − | + | − | + | − | − | − | + | − | − | − | − | − |
| P. boydii CBS 101717 | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | + | − | − | − | − | − | − | − | − | + | − | − | + | − | − | − | + | − | − |
| P. angusta CBS 254.72 | + | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | + | − | − | + | − | − | − | − | − | + | − | − | − | − | + | + | − | − | − | + | − | − |
| P. boydii CBS 101718 | − | − | + | − | + | − | − | − | − | − | + | − | − | − | + | − | − | + | − | + | − | − | − | − | + | − | + | − | − | − | + | − | − | − | + | − | − | − | − |
| P. boydii 2 CBS 695.70 | − | − | − | − | + | − | − | − | − | − | + | − | − | + | + | − | − | + | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | − |
| P. desertorum CBS 489.72 | + | − | − | − | + | − | − | − | − | + | − | − | − | + | − | − | + | + | − | − | − | + | − | − | − | + | − | − | − | − | + | − | − | − | − | − | + | − | |
| P. boydii 2 IP 1946.90 | + | − | − | + | + | − | − | − | + | − | + | − | − | + | − | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | + | − |
| P. fusoides CBS 106.53 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | + | − | − | + | − | − | − | − | − | − | − | − | − | + | − | − |
| P. boydii 1 IP 1698.87 | + | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | − | + | − | + | − | − | − | − | + | − | + | − | − | − | − | + | − | − | − | − | + | + | − |
| P. boydii 1 IP 1945.90 | + | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | − | + | − | + | − | − | − | − | + | − | + | − | − | − | − | + | − | − | − | − | + | + | − |
| P. boydii CBS 591.90 | + | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | − | + | − | + | − | − | + | − | + | − | + | − | − | − | − | − | − | − | + | − | − | + | − |
| G. tectonae CBS 127.84 | − | − | − | − | − | − | − | + | − | + | − | − | − | + | + | − | − | + | − | − | + | − | − | − | − | − | − | − | − | + | − | − | + | − | + | − | − | − | − |
| P. boydii CBS 100.26 | − | − | − | − | − | − | + | − | − | − | − | − | + | − | + | − | − | + | − | − | − | + | − | − | − | + | − | − | + | + | − | + | − | − | + | − | + | − | − |
| P. lindforsii CBS 352.59 | − | − | − | − | − | + | − | + | − | − | − | + | − | − | + | − | − | + | − | + | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − |
| P. boydii CBS 101720 | − | − | − | − | − | + | − | + | − | − | + | − | − | − | + | − | − | − | + | − | − | + | − | − | − | − | + | − | + | − | − | + | − | − | + | − | − | − | − |
| P. boydii 2 CBS 499.90 | − | − | − | − | − | + | + | − | − | − | + | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | + | − | − | + | − | + | − | − | + | − | − | − | − |
| P. guttulata CBS 362.61 | + | − | − | − | − | + | + | − | − | + | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + |
| P. boydii RKI 866/94 | − | − | − | − | + | − | − | − | − | − | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | − | − | + | |
The occurrence (+) or lack of occurrence (−) of bands with a certain molecular weight is indicated. Thirty-nine band positions were taken into account.
All S. prolificans strains except CBS 467.74 had identical banding patterns. Strain CBS 467.74 deviated in some bands. This strain also had different RFLP patterns.
DISCUSSION
P. boydii is a cleistothecial ascomycete that belongs to the family Microascaceae. This corresponds to 18S rDNA sequencing data (16), which also indicated that Microascus cirrosus Curzi is a close relative. In a similar study, Okada et al. (21) found three Graphium species among the members of the family Microascaceae. From our more detailed ITS data, G. putredinis was found to be close to G. tectonae, while Graphium penicillioides Corda took an isolated position amid the Petriella species.
Phenetically the genera Petriella and Pseudallescheria are distinct by having ostiolate versus nonostiolate ascomata, respectively (33). Our ITS1 and ITS2 distance tree (Fig. 1) does not show a clear bipartition. Most of the nonostiolate species are found in the upper part of the tree, and the ostiolate species are found in the lower branch of the tree. Issakainen et al. (16) supposed that S. prolificans is close to P. setifera, but in our study S. prolificans proved to be clearly different. No teleomorph has yet been found for S. prolificans.
von Arx et al. (33) distinguished seven species in the genus Pseudallescheria. Of these, only P. fimeti was found to have clearly different ITS sequences. P. angusta, P. ellipsoidea, and P. fusoidea were found within the P. boydii main group and must be regarded as synonymous names, as already supposed on the basis of morphological studies by McGinnis et al. (19) and on the basis of the ITS2 sequences by Wedde et al. (36). The species have been distinguished primarily on the basis of sizes of the cleistothecia and ascospores, but ranges of variability were strongly overlapping. The positions of P. desertorum and P. africana remain ambiguous, since their sequences differed from that of the type strain of P. boydii, CBS 101.22, by 12.8 and 6.8%, respectively.
The patterns of the ITSs of strain CBS 100.26, included in the CBS List of Cultures as P. boydii, obtained by RFLP analysis with SSU were markedly different from those of the remaining species; the ITSs could not be aligned. Earlier, it was found to have a clearly different mole percent G+C DNA content (15). The strain is of the Acladium castellanii type and was described from a patient with subcutaneous, suppurative mycosis (5). The fungus was described as having yellowish cultures which became blackish on some media; conidiogenous cells were aggregated in fascicles and produced apical clusters of truncate conidia (4). Butler (4) supposed that it was related to Sporothrix schenckii Hektoen et Perkins, while de Hoog (6) suggested an affinity to Raffaelea on the basis of its truncate sympodial conidia. Both suggestions were refuted on the basis of ITS (data not shown) and 18S rDNA sequence data (M. Blackwell, personal communication; compare these suggestions with those in reference 17), respectively. The taxonomic position of A. castellanii remains uncertain.
Rhinocladium lesnei (34) was found to be identical to G. putredinis and represents an anamorph taxon clearly apart from P. boydii. Morphologically and culturally, the two strains analyzed, CBS 108.10 and CBS 102083, were very different. CBS 108.10 was whitish, showing a Scedosporium anamorph only. However, in the original publication of Vuillemin (34), fasciculate conidiophores and denticulate conidiogenous cells are depicted. Thus, the identity of the two taxa is highly probable.
The genetic variability within P. boydii is considerable. Guého and de Hoog (15) found three infraspecific ecological and clinical groups on the basis of nDNA-DNA reassociation experiments. Reassociation between these groups was consistently about 50%, whereas within the groups the values were >80%. Similar reassociation groups in Galactomyces geotrichum Redhead et Malloch have recently been recognized as separate taxonomic entities (29). Contrary to these findings, two groups in Cladophialophora were judged to belong to different species, despite nDNA-DNA homology values of >80% (13). In bacteria, homology values are more precise than sequence data (30), but this is particularly due to the use of SSU sequences. In fungi, DNA homology values compared to ITS sequence data indicate a level of diversity which is similar or somewhat lower. The P. boydii reassociation groups are not supported statistically in the ITS tree (Fig. 1). It is remarkable, however, that all strains of reassociation group 2 except strain CBS 499.90 contain an intron in the SSU rDNA gene and can be found in a single branch in the ITS tree.
Judging from the data obtained by molecular fingerprinting with M13, the variability within P. boydii is considerable. Nearly all strains proved to be different from each other; the exceptions were three pairs (CBS 329.93 and CBS 330.93, CBS 101721 and CBS 101723, and IP 1698.87 and IP 1945.90). The ITS sequences of these pairs were also identical, while for two of the three pairs the sources of isolation were remote from each other. This finding indicates the existence of widespread genotypes. Due to the molecular variability of P. boydii, identification based on species-specific primers or RFLP analysis should be interpreted with care in routine diagnostics.
In the case of strains CBS 329.93, CBS 330.93, and CBS 101720, molecular fingerprinting with M13 proved to be an appropriate method for characterization of individual strains, since identical fingerprinting patterns were generated for the strains isolated from a single patient. Data obtained by molecular biological analysis for all strains from the same nonclinical isolation site (strains CBS 499.90 and CBS 101719 and strains CBS 101725 and CBS 101726) were different (Fig. 1 and 2; Tables 2 to 4). For one pair of strains (CBS 101725 and CBS 101726), the strains were different as determined by culture, and their ITS sequences deviated to such an extent that they may be two different species (Fig. 1). These findings are in agreement with those of April et al. (1), who found P. boydii strains from closely similar sites to be highly variable in cultural and physiological parameters.
Apparently, environments which are suitable for the growth of P. boydii are consistently inhabited by different populations. The small amount of correspondence between fingerprinting bands indicates that variation is continually generated, probably by meiotic recombination. However, the variability of the ITS sequences of P. boydii strains exceeds that of intermating populations. This may be a reason why such a broad spectrum of clinical pictures caused by P. boydii occurs, and a tendency to neurotropic colonization has evolved. The distance between CBS 101725 and CBS 101726 from a single sample is particularly illustrative. It is unclear why different populations have been maintained in a single environment and isolates with similar genotypes have been isolated from locations that are separated by significant geographic distances. A population genetic study is required to obtain more insight into speciation and evolution of P. boydii. M13 fingerprints, ITS sequences, and RFLP patterns proved to be more conserved in S. prolificans. Wedde et al. (36) and San Millán et al. (25) showed that variability in ITS sequences and randomly amplified polymorphic DNA analysis data, respectively, for this anamorphic taxon seem low. These results indicate that the life cycle of this species might be reduced to clonal reproduction. In contrast to P. boydii, a teleomorph name that indicates a sexual mode of reproduction is not known for S. prolificans.
Clinical strains are distributed over the entire tree. Infections caused by P. boydii-related species other than the opportunistic species P. boydii and S. prolificans are a case of lobomycosis caused by Petriella setifera in a dolphin (11) (another case of infection in a dolphin was attributed to P. boydii [see also reference 22]) and a human skin infection due to R. lesnei (34). Within P. boydii, the amount of variability of clinical strains is comparable to that of the environmental strains of that species (Fig. 1). This suggests that no particular selection by the host occurs and, thus, that all environmental strains have equal pathogenic potential. The natural ecological niche of P. boydii remains unknown, but nutrient-rich, poorly aerated environments have been described as the ecological niche of P. boydii (10). Ecological data should be taken into account in clinical practice, as they differ considerably between P. boydii and Aspergillus fumigatus. P. boydii has often been isolated from polluted ponds frequented by waterbirds; the fungus possibly occurs as a commensal organism in their intestinal tracts. It may then be expected that P. boydii has a significant potential to be an opportunistic pathogen. Given the fact that the species is particularly controlled by unspecific defense systems (3), primary pathogenicity seems unlikely.
ACKNOWLEDGMENTS
We thank P. Singer for data on strains and P. Rhein, A. H. G. Gerrits van den Ende, K. Luijsterburg, and D. Zimmerman for technical assistance.
REFERENCES
- 1.April T M, Abbott S P, Foght J M, Currah R S. Degradation of hydrocarbons in crude oil by the ascomycete Pseudallescheria boydii (Microascaceae) Can J Microbiol. 1998;44:270–278. doi: 10.1139/w97-152. [DOI] [PubMed] [Google Scholar]
- 2.Bell R G. Comparative virulence and immunodiffusion analysis of Petriellidium (Shear) Malloch strains isolated from feedlot manure and a human mycetome. Can J Microbiol. 1978;24:856–863. doi: 10.1139/m78-142. [DOI] [PubMed] [Google Scholar]
- 3.Bouza E, Muñoz P, Vega M, Rodriguez-Creixems M, Berenguer J, Escudero A. Clinical resolution of Scedosporium prolificans fungemia associated with reversal of neutropenia following administration of granulocyte colony-stimulating factor. Clin Infect Dis. 1996;23:192–193. doi: 10.1093/clinids/23.1.192. [DOI] [PubMed] [Google Scholar]
- 4.Butler E J. Acladium castellanii Pinoy and the existence of the so-called acladiosis of Castellani. Parasitology. 1937;29:259–265. [Google Scholar]
- 5.Castellani A, Jacono I. Acladiosis and paracladiosis. J Trop Med Hyg. 1934;37:360–363. [Google Scholar]
- 6.De Hoog G S. The genera Blastobotrys, Sporothrix, Calcarisporium and Calcarisporiella gen. nov. Stud Mycol. 1974;7:1–84. [Google Scholar]
- 7.De Hoog G S, Gerrits van den Ende A H G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 8.De Hoog, G. S., J. Guarro, M. J. Figueras, and J. Gené. Atlas of clinical fungi, 2nd ed., in press. Centraalbureau voor Schimmelcultures, Baarn, The Netherlands, and Universitat Rovira i Virgili, Reus, Spain.
- 9.De Hoog G S, Kuijpers A F A, van den Tweel K. Zeldzame schimmels: doodgewoon? Ned Tijdschr Med Microbiol. 1998;5:32–35. [Google Scholar]
- 10.De Hoog G S, Marvin-Sikkema F D, Lahpoor G A, Gottschall J C, Prins R A, Guého E. Ecology and physiology of Pseudallescheria boydii, an emerging opportunistic fungus. Mycoses. 1993;37:71–78. doi: 10.1111/j.1439-0507.1994.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 11.De Vries G A, Laarman J J. A case of Lobo's disease in the dolphin Sotalia guianensis. Aquatic Mammals. 1974;13:1–8. [Google Scholar]
- 12.Dworzack D L, Clark R B, Borkowski W J, et al. Pseudallescheria boydii brain abscess: association with near-drowning and efficacy of high-dose, prolonged miconazole therapy in patients with multiple abscesses. Medicine. 1989;68:218–224. [PubMed] [Google Scholar]
- 13.Gerrits van den Ende A H G, De Hoog G S. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud Mycol. 1999;43:151–162. [Google Scholar]
- 14.Gräser Y, Kühnisch J, Presber W. Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum. J Clin Microbiol. 1999;37:3713–3717. doi: 10.1128/jcm.37.11.3713-3717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guého E, De Hoog G S. Taxonomy of the medical species of Pseudallescheria and Scedosporium. J Mycol Med. 1991;1:3–9. [Google Scholar]
- 16.Issakainen J, Jalava J, Eerola E, Campbell C K. Relatedness of Pseudallescheria, Scedosporium and Graphium pro parte based on SSU rDNA sequences. J Med Vet Mycol. 1997;35:389–398. [PubMed] [Google Scholar]
- 17.Jones K G, Blackwell M. Phylogenetic analysis of ambrosial species in the genus Raffaelea based on 18S rDNA sequences. Mycol Res. 1998;102:661–665. [Google Scholar]
- 18.Lennon P A, Cooper C R, Salkin I F, Lee S B. Ribosomal DNA internal transcribed spacer analysis supports synonymy of Scedosporium inflatum and Lomentospora prolificans. J Clin Microbiol. 1994;32:2413–2416. doi: 10.1128/jcm.32.10.2413-2416.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinnis M R, Padhye A A, Ajello L. Pseudallescheria Negroni et Fischer, 1943 and its later synonym Petriellidium Malloch, 1970. Mycotaxon. 1982;14:94–102. [Google Scholar]
- 20.McGuire T W, Bullock J D, Bullock J D, Jr, Elder B L, Funkhouser J W. Fungal endophthalmitis. An experimental study with a review of 17 human ocular cases. Arch Ophthalmol. 1991;109:1289–1296. doi: 10.1001/archopht.1991.01080090115034. [DOI] [PubMed] [Google Scholar]
- 21.Okada G, Seifert K A, Takematsu A, Yamaoka Y, Miyazaki S, Tubaki K. A molecular phylogenetic reappraisal of the Graphium complex based on 18S rDNA sequences. Can J Bot. 1998;76:1495–1506. [Google Scholar]
- 22.Poelma F G, De Vries G A, Blythe-Russell E A, Luyckx M H F. Lobomycosis in an Atlantic bottle-nosed dolphin in the Dolphinarium Harderwijk. Aquatic Mammals. 1974;11:11–15. [Google Scholar]
- 23.Rüchel R. False-positive reaction of a Cryptococcus antigen test owing to Pseudallescheria mycosis. Mycoses. 1994;37:69. doi: 10.1111/j.1439-0507.1994.tb00779.x. [DOI] [PubMed] [Google Scholar]
- 24.Rüchel R, Willichowski E. Cerebral Pseudallescheria mycosis after near-drowning. Mycoses. 1995;38:473–475. doi: 10.1111/j.1439-0507.1995.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 25.San Millán R, Quindós G, Garaizar J, Salesa R, Guarro J, Pontón J. Characterization of Scedosporium prolificans clinical isolates by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1997;35:2270–2274. doi: 10.1128/jcm.35.9.2270-2274.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shear C L. Life history of an undescribed ascomycete from a granular mycetoma of man. Mycologia. 1922;14:239–243. [Google Scholar]
- 27.Siebenmann F. Die Schimmelmykosen des menschlichen Ohres. Wiesbaden, Germany: Bergmann; 1889. [Google Scholar]
- 28.Siebenmann F. Neue botanische und klinische Beiträge zur Otomykose. Z Ohrenheilk. 1889;19:7–50. [Google Scholar]
- 29.Smith M T, De Cock A W A M, Poot G, Steensma H Y. Genome comparisons in the yeast-like fungal genus Galactomyces Redhead et Malloch. Int J Syst Bacteriol. 1999;45:826–831. doi: 10.1099/00207713-45-4-826. [DOI] [PubMed] [Google Scholar]
- 30.Stackebrandt E, Goebel U. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 31.Sterflinger K, De Baere R, De Hoog G S, De Wachter R, Krumbein W E, Haase G. Coniosporium perforans and C. apollinis, two rock-inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece) Antonie Leeuwenhoek. 1997;72:349–363. doi: 10.1023/a:1000570429688. [DOI] [PubMed] [Google Scholar]
- 32.Van De Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 33.von Arx J A, Figueras M J, Guarro J. Sordariaceous Ascomycetes without ascospore ejaculation. Beih Nova Hedw. 1988;94:1–59. [Google Scholar]
- 34.Vuillemin P. Les conidiosporés. Bull Soc Sci Nancy. 1910;2:129–172. [Google Scholar]
- 35.Warnock D W, Richardson M D. Fungal infection in the compromised patient. 2nd ed. Chichester, United Kingdom: John Wiley & Sons; 1991. [Google Scholar]
- 36.Wedde M, Müller D, Tintelnot K, De Hoog G S, Stahl U. PCR-based identification of clinically relevant Pseudallescheria/Scedosporium strains. Med Mycol. 1998;36:61–67. [PubMed] [Google Scholar]
- 37.Weising K, Nybom H, Wolff K, Meyer W. DNA fingerprinting in plants and fungi. Boca Raton, Fla: CRC Press, Inc.; 1995. [Google Scholar]
- 38.Zalar P, De Hoog G S, Gunde-Cimerman N. Ecology of halotolerant dothideaceous black yeasts. Stud Mycol. 1999;43:38–48. [Google Scholar]