Abstract
The description and evaluation of a PCR-based assay for the detection and species identification of the eight known human herpesviruses are presented. Two primer pairs targeting well-conserved regions of the genome allowed the amplification of the DNAs of all known human herpesviruses at a high level of sensitivity (10 to 100 genome copies for most viruses). Identification of the virus species was achieved through restriction enzyme digestion with BamHI and BstUI, which yielded fragment sizes that were characteristic for each herpesvirus. Furthermore, it was demonstrated that this restriction enzyme panel allowed the discrimination between human herpesvirus 6 variant A and variant B. This assay format was validated over the course of 1 year in a clinical virology laboratory setting, where it was shown that it readily detected human herpesviruses, including occasional multiple infections, in a variety of clinical samples. The PCR assay was compared to isolation and electron microscopy for the detection of herpes simplex (HSV) and varicella-zoster virus (VZV) in clinical samples. All specimens positive by conventional methods were also positive by PCR. However, in a number of clinical specimens in which HSV or VZV could not be detected by conventional methods, PCR was able to demonstrate the presence of the virus.
To date, eight human viruses of the family Herpesviridae have been identified, namely, herpes simplex virus (HSV) type 1 (HSV-1), herpes simplex virus type 2 (HSV-2), varicella-zoster virus (VZV), Epstein-Barr virus (EBV), cytomegalovirus (CMV), human herpesvirus 6 (HHV-6), human herpesvirus 7 (HHV-7), and human herpesvirus 8 (HHV-8) (1, 19). Whereas some of these agents have been known for decades and are well characterized, less is known about the pathogenic potentials of the more recently described members of this family. The eventual understanding of the full spectrum of disease caused by these viruses relies on the availability of appropriate diagnostic approaches for their detection.
While classical diagnostic laboratory methods such as electron microscopy, immunofluorescence microscopy, or viral isolation are well established for HSV-1, HSV-2, VZV, and CMV, the meaningful detection of viruses such as EBV, HHV-6, HHV-7, and HHV-8 remains more challenging in the diagnostic setting. Tests based on PCR have therefore assumed an important role for the laboratory detection of these agents. Even for viruses such as HSV which can be readily isolated, diagnosis by PCR has become the “gold standard” for some diseases such as herpetic encephalitis (3, 22, 29, 31). Because infection with different or multiple species of herpesviruses can cause similar symptoms, PCR tests have been designed to detect more than one herpesvirus at a time. Methods used to achieve this have included parallel PCRs, multiplex PCRs with several primer pairs, and tests with a consensus primer pair targeting conserved genomic regions. Methods for subsequent confirmation of the species of the virus detected have included hybridization with a species-specific probe, nested PCR with species-specific primers, and restriction enzyme analysis (4, 9, 21, 25–27, 29).
In this study we present the first comprehensive PCR assay for the detection and species identification of all eight known human herpesviruses, including differentiation between HHV-6 variant A (HHV-6A) and variant B (HHV-6B). The detection of herpesviruses in this assay is based on PCR with two primer pairs, followed by restriction enzyme analysis with BamHI and BstUI.
MATERIALS AND METHODS
Herpesvirus DNAs.
Template DNAs of HSV-1, HSV-2, and CMV were obtained from Sigma (catalog nos. D9416, D9541, and D9166, respectively). Template DNA of EBV was extracted from a defined number of Namalwa cells, a line which contains two EBV genome copies per cell (24). Template DNA for VZV PCR was synthesized by long PCR (see below) from a clinical isolate of VZV from a patient with varicella obtained from the Diagnostic Virology Laboratory of our institution. Template DNA of HHV-6A was synthesized by long PCR from DNA extracted from the supernatant of a culture of the cell line HSB2 infected with HHV-6A, provided by Carla Osiowy, Laboratory Centre for Disease Control, Winnipeg, Manitoba, Canada. Kaposi's sarcoma biopsy specimens and a non-Hodgkin's lymphoma specimen positive for HHV-8 were provided by Giorgio Inghirami, Department of Pathology, New York University Medical Center.
DNA sequence analyses.
DNA sequence analyses were performed with Gene Runner for Windows, version 3.04 (Hastings Software). The GenBank accession numbers for the DNA sequences used were as follows: HSV-1, X14112; HSV-2, Z86099; EBV, V01555; CMV, M14709; HHV-8, U93872; VZV, X04370; HHV-6A, X83413; HHV-6B, AF157706; and HHV-7, U43400.
Primers.
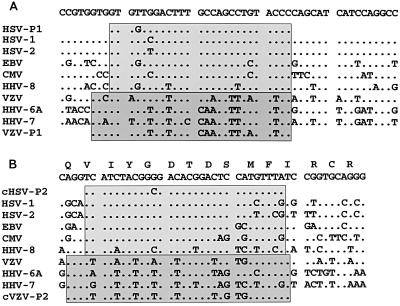
Primers were designed to bracket a well-conserved region in the DNA polymerase gene, based on an alignment of the DNA sequences of the eight known human herpesviruses (Fig. 1A and B).
FIG. 1.
Alignments of herpesvirus DNA polymerase gene sequences. Dots indicate similarities with the consensus sequence on top. (A) Homology in the region from positions 64941 to 64990 in the HSV-1 sequence, along with the sense primers HSV-P1 and VZV-P1. (B) Homology in the region from positions 65447 to 65492 in the HSV-1 region which codes for the YGDTDS motif, along with the complement of the antisense primers HSV-P2 and VZV-P2 (cHSV-P2 and cVZV-P2, respectively).
Primer pair HSV-P1 (5′-GTGGTGGACTTTGCCAGCCTGTACCC-3′) and HSV-P2 (5′-TAAACATGGAGTCCGTGTCGCCGTAGATGA-3′) was used to amplify HSV-1, HSV-2, EBV, CMV, and HHV-8. Primer pair VZV-P1 (5′-GTCGTGTTTGATTTTCAAAGTTTATATCC-3′) and VZV-P2 (5′-ATAAACACACAATCCGTATCACCATAAATAACCT-3′) was used to amplify VZV, HHV-6 (variant A or B), and HHV-7. The characteristics of the expected amplicons for each virus are listed in Table 1.
TABLE 1.
Characteristics of amplicons as predicted by DNA sequencing
| Virus | Length (bp) | % G+C content | BamHI site (position)a | BstUI site(s) [position(s)]a |
|---|---|---|---|---|
| HSV-1 | 532 | 66.2 | No site | 228, 361, 383, 397, 404, 418, 420, 476 |
| HSV-2 | 532 | 67.3 | 230 | 88, 167, 194, 228, 383, 397, 402, 404, 406, 418, 420, 476 |
| EBV | 538 | 62.8 | 252 | 258 |
| CMV | 604 | 59.4 | No site | 164, 221, 233, 372, 383, 405, 483, 511, 570 |
| HHV-8 | 526 | 54.6 | No site | 208 |
| VZV | 536 | 40.3 | No site | 228, 283 |
| HHV-6 variant A | 533 | 44.5 | 246 | No site |
| HHV-6 variant B | 533 | 44.7 | 246 | 167 |
| HHV-7 | 533 | 37.9 | No site | 470 |
The positions of restriction sites refer to the first base pair of the restriction site.
PCR.
Each reaction was performed in a 0.6-ml tube (PRE 050; Diamed) in a total volume of 50 μl overlaid with 50 μl of mineral oil. Each reaction contained 5 μl of 10× Cetus buffer II (Perkin-Elmer), 5 μl of 25 mM MgCl2, 5 μl of a deoxynucleoside triphosphate mixture (each deoxynucleoside triphosphate at a concentration of 2 mM; Pharmacia), 2.5 μl of dimethyl sulfoxide (DMSO; Sigma), 37.5 pmol of each primer, 0.5 μl (2.5 U) of Amplitaq Gold (Perkin-Elmer), and molecular-grade double-distilled water to a volume of 40 μl. The master mixture was then divided into aliquots and placed in tubes, to which 10 μl of template DNA, dissolved in molecular-grade water, was added. PCR was performed on a Stratagene Robocycler 40 instrument.
With the HSV-P1 and HSV-P2 primer pair the cycling parameters were initial preincubation at 95°C for 12 min; then 3 cycles consisting of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min; then 37 cycles of 95°C for 1 min, 55°C for 45 s, and 72°C for 1 min; and then a final incubation at 72°C for 3 min. With the VZV-P1 and VZV-P2 primer pair the cycling parameters were initial incubation at 95°C for 10 min and then 40 cycles consisting of 95°C for 1 min, 47°C for 1 min, and 72°C for 1 min, followed by a final incubation at 72°C for 3 min. A 10-μl volume of each reaction mixture was subjected to electrophoresis on 1.5% agarose gels containing ethidium bromide. The gels were visualized on a UV transilluminator and photographed.
Precautions against PCR contamination.
PCR reagents were prepared before each assay in a master mixture that was then aliquoted. The preparation of the master mixture, the extraction of the DNA and the addition of the template to the PCR mixture, and the thermal cycling were performed in three different, well-separated rooms, each with its own dedicated set of micropipettes and gowns. General precautions against contamination, including systematic use of aerosol-barrier-protected pipette tips, frequent changes of gloves, and frequent decontamination of surfaces with UV light and sodium hypochlorite, were strictly adhered to.
PCR controls.
For each clinical sample, extracted DNA was recovered in a defined volume of double-distilled H2O as described below. One aliquot of 10 μl was tested in a PCR mixture “as is,” and a second aliquot of 10 μl was tested in a PCR mixture spiked with a defined amount of template to control the integrity of the reaction mixture and to rule out the presence of PCR inhibitors originating from the sample. For the PCR with the HSV-P1 and HSV-P2 primer pair the spike consisted of approximately 1,500 genome copies of CMV DNA. For PCR with the VZV-P1 and VZV-P2 primer pair, the spike consisted of 1.2 μl of a dilution of DNA from an HHV-6A culture supernatant, containing approximately 10 to 100 genome copies. In addition, for each PCR run of up to 10 samples, an aliquot of phosphate-buffered saline was also submitted to DNA extraction as a negative control. The extracts, along with a positive control (same template used for the spike) and a water specimen as an additional negative control to rule out contamination of the reagents or by aerosols, were then subjected to PCR as described above.
Restriction enzyme digestion.
Each reaction mixture in which amplicons were detected was subjected to digestion with the restriction enzymes BamHI (Life Technologies) and BstUI (New England Biolabs). The digestion mixture consisted of 10 μl of PCR mixture, 1.5 μl of the appropriate enzyme buffer, 1 μl of enzyme, and 2.5 μl of double-distilled water for a total volume of 15 μl. The reaction mixtures were incubated for 1 h at 37°C (BamHI) or 60°C (BstUI).
Extraction of DNA from clinical samples. (i) CSF.
DNA was extracted from cerebrospinal fluid (CSF) by a guanidine thiocyanate (GTC) method (23), as follows. To 100 μl of CSF were added 400 μl of extraction buffer (5.75 M guanidine thiocyanate [G9277; Sigma], 50 mM Tris [pH 7.4], 50 μg of glycogen [901393; Boehringer-Mannheim] per ml, 10 μl of β-mercaptoethanol [Sigma] per ml). The preparation was agitated on a vortex mixer and incubated for 10 min at room temperature. After the addition of 500 μl of isopropanol, the preparation was then further agitated and centrifuged at 12,000 × g for 20 min at room temperature. The supernatant was removed and the pellet was washed with 80% ethanol and centrifuged further for 2 min at 12,000 × g. The pellet was air dried and resuspended in 25 μl of molecular-grade water.
(ii) Blood, serum, and tissue.
The Blood and Tissue QIAamp extraction kit (Qiagen) was used to extract DNA from blood, serum, and tissue according to the manufacturer's recommendations.
Templates for quantitation of PCR sensitivity for VZV and HHV-6.
DNA was extracted from the designated cells or culture supernatant by the GTC method, as outlined above. Amplicons which included the region targeted by the VZV-P1 and VZV-P2 primer pair were generated by long PCR by a previously described method (30).
For VZV, the primers used were VZVLA-1 (5′-GTTGCTACTTGGAATGTAGACGAGCGTACAAATTGTG-3′) and VZVLA-2 (5′-GCATCAAAGGTAATAAAGCCAAATCAGAGTCCG-3′), which bracketed a 5.5-kb region of the genome. For HHV-6, the primer pair HHV6LA-1 (5′-ACAGATGCTTCGCAAATACAGTCAACGATCAC-3′) and HHV6LA-2 (5′-TAGGCACTGGATTGGGCTCGGTTATAAGTG-3′), which bracketed a 5.4-kb region, was used. For both long PCRs, the cycling parameters were as follows: 35 cycles consisting of 99°C for 35 s, 67°C for 30 s, and 68°C for 8 min. The PCR mixtures were then submitted to agarose gel electrophoresis. After visualization of the gels by ethidium bromide staining, bands of the expected size were excised and purified with the Geneclean II kit (Bio 101) according to the manufacturer's recommendations. The DNA concentrations of the purified amplicons were determined by agarose gel electrophoresis and ethidium bromide staining, using the High DNA Mass Ladder (Life Technologies) as a concentration standard. Templates with a range of copies of DNA were then prepared by serial 10-fold dilutions in molecular-grade water, aliquoted, and frozen.
Sequencing.
The amplicons to be sequenced were excised from agarose gels, purified with the Jetsorb kit (Genomed, Frederick, Md.), and subjected to sequencing reactions in both directions with Thermosequenase (Amersham Canada Ltd., Oakville, Ontario, Canada). This was performed by the DNA Sequencing Facility, Center for Applied Genomics, Hospital for Sick Children.
Isolation in cell culture.
Isolation of HSV-1, HSV-2, VZV, and CMV was performed by standard techniques (2, 15, 13).
Electron microscopy.
Vesicular fluid samples dried on a glass slide were resuspended in 1% ammonium acetate and applied to Formvar- and carbon-coated electron microscopy grids and stained with 2% phosphotungstic acid. The grids were examined with a Philips EM 300 electron microscope at a magnification of ×50,000.
RESULTS
Sensitivity.
The sensitivity of the PCR assay with the HSV-P1 and HSV-P2 primer pair was assessed with serial dilutions of DNA from well-characterized templates of defined concentration. The limiting sensitivities measured were 10 to 100 genome copies for HSV-1, HSV-2, and EBV and 400 genome copies for CMV. Similarly, the sensitivity of the PCR assay with the VZV-P1 and VZV-P2 primer pair was shown to be 10 to 100 genome copies for VZV and 1 to 10 genome copies for HHV-6A.
Species identification by restriction enzyme digestion.
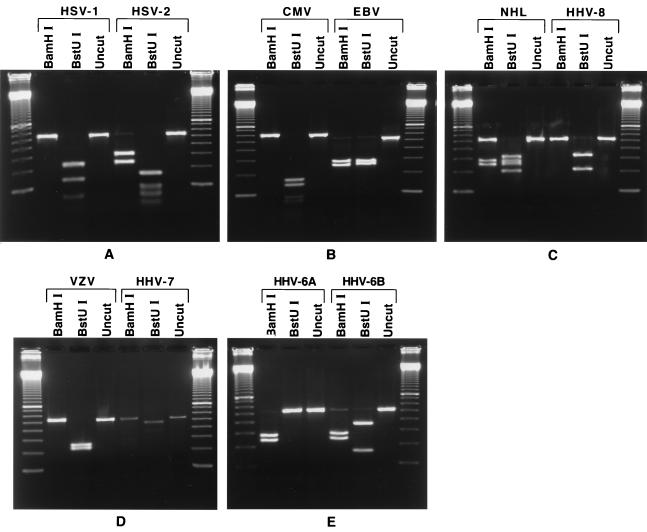
Amplicons obtained by PCR from the template DNAs described above were subjected to restriction endonuclease digestion with BamHI and BstUI. After digestion and electrophoresis, each viral DNA yielded a characteristic fragment pattern that corresponded to that predicted by DNA sequence analysis. Even though HSV-1, HSV-2, and CMV are cut at multiple sites by BstUI (8, 12, and 9 times, respectively), the larger fragments are clearly distinguishable and form a pattern unique for each virus. In addition, the undigested CMV amplicon of 604 bp migrated noticeably slower by agarose gel electrophoresis than the 526- to 536-bp amplicons obtained from the other herpesviruses (Table 1 and Fig. 2).
FIG. 2.
Restriction enzyme digestion pattern for each human herpesvirus. Digestion reaction mixtures were electrophoresed on 1.5% agarose gels. On each panel the gel is flanked by a 100-bp ladder (Life Technologies). (A) HSV-1 and HSV-2. Only the larger fragments of the BstUI digestion can be seen, but they are readily distinguishable. (B) CMV and EBV. The CMV amplicon migrates noticeably more slowly than the other amplicons. For CMV, only the larger fragments of the BstUI digestion can be seen, but they are readily distinguishable. (C) HHV-8. On the right-hand side, the expected HHV-8 pattern was obtained after PCR with a Kaposi's sarcoma sample. On the left-hand side, PCR with a non-Hodgkin's lymphoma (NHL) sample showed a mixed infection with EBV and HHV-8. (D) VZV and HHV-7. (E) HHV-6A and HHV-6B have the same BamHI patterns but have distinct BstUI patterns.
At the start of this study, the sequence of the DNA polymerase gene of HHV-6B had not been determined and the restriction enzyme digestion pattern for HHV-6B shown in Fig. 1 could not have been predicted. As clinical samples were being analyzed by PCR, several instances of this new pattern were encountered. Amplicons obtained from four different samples that displayed this new pattern were sequenced directly. The sequences determined from both strands of a 370-bp segment from amplicons from all four samples were identical and displayed 14 nucleotide differences from the corresponding segment of HHV-6A (with one of these differences accounting for the BstUI site). This new sequence clearly identifies an HHV-6 variant. Since these observations were made, two different strains of HHV-6B have been sequenced (11, 17) and they showed the same 14 nucleotide differences from the sequence of HHV-6A that we found in our variants. This demonstrated that whereas amplicons from HHV-6A and HHV-6B have the same BamHI site, they can be differentiated by the restriction pattern obtained by BstUI digestion.
In several instances we obtained amplicons from a number of clinical samples which, on digestion with restriction enzymes, yielded the pattern predicted for HHV-7 (Fig. 2). Finally, when DNA extracted from two Kaposi's sarcoma biopsy specimens and two samples of non-Hodgkin's lymphoma known to be positive for HHV-8 were subjected to the PCR assay with the HSV-P1 and HSV-P2 primer pair, the presence of an amplicon with the restriction pattern predicted for HHV-8 (Fig. 2) was demonstrated. Furthermore, the presence of EBV was demonstrated in three of these samples. By contrast, by use of the primers of Rozenberg and Lebon (27) with DNA from the dually infected Kaposi's sarcoma biopsy specimen, it was possible to amplify only the EBV DNA (data not shown).
For all the samples analyzed that gave rise to amplicons of the predicted size, the restriction patterns were always those predicted by DNA sequence analysis or the pattern we now know to be predicted for HHV-6B. In all cases in which a virus was also isolated by cell culture (HSV-1, HSV-2, VZV, CMV), the identification obtained by typing by immunofluorescence microscopy with reference monoclonal antibodies was in agreement with that determined by the PCR assay followed by restriction fragment analysis. In rare cases involving large amounts of HSV-1 DNA, very faint bands of the expected size for HSV-2 digestion products were detected in the digestion reaction with BamHI, but the fragments predicted from digestion of HSV-2 with BstUI were lacking. In these rare cases the presence of HSV-2 was ruled out by performing an additional digestion with MscI, which cuts the HSV-2 amplicon (the MscI site is at nucleotide 205) but does not cut the HSV-1 amplicon.
Detailed review of clinical samples.
A brief survey of the results obtained over a 1-year period for the samples submitted from patients in our institution, which include immunocompromised and cancer patients, as well as organ transplant recipients, is presented below.
(i) CSF.
A total of 226 CSF samples were tested. Of these, eight were positive for HSV-1, one was positive for HSV-2, five were positive for VZV, seven were positive for EBV, one was positive for CMV, two were positive for HHV-6A, one was positive for HHV-6B, and two were positive for HHV-7.
(ii) Vesicular cutaneous lesions and mucocutaneous ulcers.
Over a 1-year period, 156 lesion scrapings, aspirates, or swabs from suspected herpetic lesions were submitted for detection of HSV by PCR (with primers HSV-P1 and HSV-P2). Of these, 50 were positive for HSV-1; EBV was detected in 19 samples and CMV was detected in 2 samples. Similarly, 110 samples from suspected chicken pox or shingles lesions were submitted for detection of VZV by PCR (with the VZV-P1 and VZV-P2 primer pair). Of these, VZV was detected in 42 samples, HHV-6B was detected in 1 sample, and HHV-7 was detected in 5 samples.
Fluid from vesicular lesions caused by HSV or VZV usually contains a high concentration of viruses, which can readily be demonstrated by electron microscopy. A subset of samples from vesicles were prospectively tested by PCR, electron microscopy, and virus isolation. Ten samples were positive for HSV-1; all 10 samples were positive by PCR, 7 were positive by all three tests, 2 were positive by PCR and isolation, and 1 was positive by PCR and electron microscopy. As well, 29 samples were positive for VZV; all 29 samples were positive by PCR, 14 were positive by all three tests, and 13 were positive by PCR and electron microscopy.
In contrast to vesicular lesions, the viruses in herpetic ulcerative lesions are more difficult to detect by either electron microscopy or viral isolation. Swabs from ulcerative lesions were prospectively tested by viral isolation (for HSV) and PCR. Of 22 specimens positive for HSV-1, all were positive by PCR but only eight were also positive by viral isolation. Thus, the use of PCR allowed detection of a virus that would otherwise have been missed. In addition, a number of other viruses were detected by PCR in such lesions, including CMV in one sample, EBV in 14 samples (2 samples also contained HSV-1), HHV-6 in 1 sample (along with HSV-1), and HHV-7 in 2 samples.
(iii) Tissues and bodily fluids (other than CSF or blood).
Whenever possible (as the quantity of clinical material was often limiting) the samples were analyzed by the PCR assays with both pairs of primers. Over a 1-year period, 159 samples were analyzed with the HSV-P1 and HSV-P2 primer pair; 116 of these were also analyzed with the VZV-P1 and VZV-P2 primer pair. In all, 72 samples were positive for at least one virus (12 were positive for two viruses).
In the PCR assay with primers HSV-P1 and HSV-P2, nine samples were positive for HSV-1, including two brain biopsy specimens, two gastrointestinal biopsy specimens (one was also positive for HHV-7), one oral cavity biopsy specimen, one pleural fluid aspirate, one bronchoalveolar lavage specimen, one liver biopsy specimen (which was also positive for HHV-6B), and one bone marrow aspirate. The last two samples were from a bone marrow transplant recipient with disseminated HSV-1 disease. Forty-eight samples were positive for EBV, including 8 bone marrow aspirates, 8 lymphoid tissue biopsy specimens (1 was also positive for HHV-6B), 12 gastrointestinal biopsy specimens (3 were also positive for HHV-7), 8 lung biopsy specimens (2 were also positive for CMV and 2 others were also positive for HHV-6B), 4 bronchoalveolar lavage specimens (1 was also positive for HHV-6B), 2 brain biopsy specimens, 2 liver biopsy specimens, 1 heart biopsy specimen, 1 mediastinal mass biopsy specimen, and 2 peritoneal fluid samples.
Five samples were positive for CMV, including one bone marrow aspirate, two lung biopsy specimens (that were also positive for EBV), and two bronchoalveolar lavage specimens.
In the PCR assay with primers VZV-P1 and VZV-P2, five samples were positive for VZV. One was a lung biopsy specimen. One was a skin biopsy specimen from a newborn with congenital VZV infection. Also positive were a liver biopsy specimen and a bone marrow aspirate from a bone marrow transplant recipient with disseminated VZV infection. Finally, a gastric biopsy specimen from another bone marrow transplant recipient with disseminated VZV infection was also positive. Nine samples were positive for HHV-6. Among these, HHV-6B was found in three lung biopsy specimens (two were also positive for EBV), one bronchoalveolar lavage specimen (along with EBV), one lymph node biopsy specimen (along with EBV), one liver biopsy specimen (along with HSV-1), one gastrointestinal biopsy specimen (along with HHV-7), and one sputum sample (along with HHV-7). As well, HHV-6A was found in one brain biopsy specimen. Nine samples were positive for HHV-7, including eight gastrointestinal biopsy specimens (one was also positive for HSV-1, two were positive for EBV, and one was positive for HHV-6B) and one sputum sample (which was also positive for HHV-6B).
(iv) Blood and serum samples.
In all, 35 blood or serum samples were tested by the PCR assay with the HSV-P1 and HSV-P2 primer pair and 26 were also tested with the VZV-P1 and VZV-P2 primer pair. Whenever possible, a sample that tested positive by use of DNA extracted from a whole-blood sample was also tested by use of DNA extracted from plasma (with the blood cells having been removed by centrifugation) to attempt to differentiate between latent and actively replicating viruses (10, 28). A total of eight samples were found to be positive by the PCR assays. Five samples were positive for EBV with whole blood only (plasma samples were negative). Two samples were positive for VZV, one with both whole blood and plasma and the other with serum (whole blood was not tested). These were obtained from the two patients with disseminated VZV infection mentioned above. One sample was positive for HHV-6B with both whole blood and plasma; this patient had just received a bone marrow transplant, which then failed to engraft.
Toward an integrated multiplex herpesvirus group PCR.
Amplicons generated with the HSV-P1 and HSV-P2 primer pair have high G+C contents, particularly amplicons from HSV-1 (66.2%) and HSV-2 (67.3%), and their successful synthesis requires the presence of DMSO in the PCR mixture. In contrast, the amplicons generated with the VZV-P1 and VZV-P2 primer pair have much lower G+C contents (from 44.7 to 37.9%) and their synthesis does not require DMSO, although it was found that the presence of DMSO did not significantly affect the sensitivity of the PCR assay for VZV and HHV-6A. Consequently, all the PCR assays in this study were performed in the presence of DMSO. Because of the lower melting temperature of the VZV-P1 and VZV-P2 primer pair, the annealing temperature for the PCR with this pair had to be lowered from 55 to 47°C. It was found that PCR with the HSV-P1 and HSV-P2 primer pair could be done at an annealing temperature of 47°C, although with some lots of primers HSV-P1 and HSV-P2 artifactual bands were generated, in addition to the predicted amplicons. This problem is currently being investigated. Samples containing two viruses constituted a minority of the samples and usually involved at least one of the lymphotropic viruses (EBV, HHV-6, or HHV-7). We found that the more complicated restriction pattern that resulted from the presence of DNA from two viruses could still be readily interpreted.
DISCUSSION
A PCR test for herpesviruses with the primers and restriction enzymes originally described by Rozenberg and Lebon (27) was introduced into our laboratory in 1992. As the sequences of the other human herpesvirus genomes became available, the PCR was redesigned and a comprehensive test for all known human herpesviruses was developed. The primers designed by Rozenberg and Lebon (27) are anchored in highly conserved regions of the DNA polymerase gene; one of these codes for the YGDTDS motif present in many viral DNA polymerases (18). As the alignments in Fig. 1 show, primers akin to those of Rozenberg and Lebon (27) should be able to amplify HHV-8, in addition to HSV-1, HSV-2, EBV, and CMV. It was indeed shown that with primers HSV-P1 and HSV-P2 the DNAs of these five viruses could be amplified. Of note, of the four samples in which HHV-8 was detected, EBV was also present in three samples. The frequent association between the presence of HHV-8 and EBV has been noted previously and may play a role in pathogenesis (16). Figure 1 also suggests that the remaining herpesviruses have too many differences to be accommodated with primers that amplify the five viruses mentioned above. However, a different pair of primers could be used to amplify VZV, HHV-6, and HHV-7 DNAs, and it was shown that by use of the VZV-P1 and VZV-P2 primer pair, this could be accomplished with a high degree of sensitivity. It is surprising that the segregation of the human herpesviruses into two groups by these primer pairs also corresponded to their G+C content (Table 1) but not to the phylogenetic grouping based on the complete genome (19).
The BamHI enzyme originally recommended by Rozenberg and Lebon (27) for differentiation of viruses was retained, but BstUI was used as a second enzyme. Both enzymes work well without having to purify the amplicons from the PCR mixture, and the restriction fragments unequivocally identify all eight viruses since all viral DNAs are cut by at least one enzyme, enabling verification of restriction fragment sizes. Even for amplicons cut several times by BstUI, the larger fragments are readily identifiable. The sequence of the DNA polymerase gene of HHV-6B was determined while our study was under way, but this helped us to show that these restriction enzymes can further distinguish between HHV-6A and HHV-6B (11, 14, 17). Although the nucleotide sequences of HHV-6A and HHV-6B are approximately 90% identical, there is no genetic gradient between the two, and to date there have been no reports of recombinant virus (11). It has been proposed that HHV-6A and HHV-6B are in fact distinct virus species (8, 11). The use of these PCR assays with clinical samples in our institution illustrated that, with the exception of HHV-8, each virus could be detected repeatedly and reproducibly. Only a small number of samples were positive for HSV-2, which would be expected for a pediatric patient population. Indeed, HSV-2 was commonly detected in CSF samples from adult patients referred by other institutions (data not shown). Similarly, HHV-8, a virus encountered less frequently than other herpesviruses (19), is, furthermore, expected to be very rarely encountered in a pediatric population in North America (7). In each case in which a virus was also isolated, the type obtained with reference monoclonal antibodies agreed with that obtained by PCR. The only problem identified with the restriction enzymes chosen is that occasionally, when a large amount of the HSV-1 DNA amplicon was digested, faint bands corresponding to those expected with a BamHI digest of HSV-2 were noted. It is believed that samples with these results cannot be reported as having a mixed infection unless the BstUI bands of HSV-2 or the HSV-2 bands predicted from digestion with MscI on a supplementary digestion are apparent. Of note, HSV-1 is only 1 bp away from having the same BamHI site as HSV-2. It is hypothesized that these faint bands that correspond to the pattern of HSV-2 may be attributable to a PCR mistake, the presence of a minority of HSV-1 mutants in a large population, or a star effect (although the star effect documented for BamHI is reported to be quite different [12]).
For vesicular lesions caused by HSV and VZV, even though we found that PCR was the most sensitive detection modality, as others have reported (5, 20), it was remarkable that detection by electron microscopy had a comparable sensitivity. Electron microscopy constitutes the most rapid diagnostic modality (10 to 20 min for staining and examination) and therefore remains a worthwhile procedure if an appropriate instrument is available (6). In contrast, isolation of VZV from vesicular fluid or even of HSV-1 from ulcers had a substantially lower sensitivity. With the very high sensitivity seen by this PCR assay, it may be possible to consider this an acceptable test to actually rule out herpesvirus infection. Although vesicular and ulcerative lesions caused by either HSV or VZV are commonly seen in a pediatric setting, they also have serious implications for immunocompromised patients and for maintenance of infection control.
Difficult diagnostic problems occur with the lymphotropic viruses (EBV, HHV-6, HHV-7, HHV-8), since they can remain latent in lymphocytes without actively replicating. Their detection by PCR in various tissues (or bodily fluids) may therefore merely reflect a lymphocytic infiltration for reasons unrelated to these viruses. Alternatively, they may also contribute to disease. In some cases it may be useful to demonstrate the presence of virions in the plasma as an indication of occurrence of the lytic cycle (10, 28). In one such occurrence we found actively replicating HHV-6 in a patient whose bone marrow transplant failed to engraft, a problem that has been previously associated with HHV-6 (8, 10). The full spectrum of diseases caused by the herpesviruses remains to be established, and it is hoped that the simplicity and sensitivity of the method that we have described here will contribute to the realization of this goal.
In summary, we have described and validated two PCR tests which, when coupled with digestion with a panel of two restriction enzymes, can amplify and identify all eight human herpesviruses and distinguish between HHV-6A and HHV-6B. In their current state of development, these procedures are simple, streamlined, and cost-effective. They are well-suited for the clinical laboratory and can be performed within a day.
ACKNOWLEDGMENTS
This work was supported by the Department of Paediatric Laboratory Medicine, Hospital for Sick Children, Toronto.
REFERENCES
- 1.Arao Y, Schmid D S, Pellett P E, Inoue N. Herpesviruses beyond HSV-1 and HSV-2. Clin Microbiol Newsl. 1999;21:153–159. [Google Scholar]
- 2.Arvin A M, Prober C G. Herpes simplex viruses. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 876–883. [Google Scholar]
- 3.Atkins J T. HSV PCR for CNS infections: pearls and pitfalls. Pediatr Infect Dis. 1999;18:823–824. doi: 10.1097/00006454-199909000-00015. [DOI] [PubMed] [Google Scholar]
- 4.Baron J M, Rübben A, Grußendorf-Conen E-I. Evaluation of a new general primer pair for rapid detection and differentiation of HSV-1, HSV-2, and VZV by polymerase chain reaction. J Med Virol. 1996;49:279–282. doi: 10.1002/(SICI)1096-9071(199608)49:4<279::AID-JMV4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Beards G, Graham C, Pillay D. Investigation of vesicular rashes for HSV and VZV by PCR. J Med Virol. 1998;54:155–157. doi: 10.1002/(sici)1096-9071(199803)54:3<155::aid-jmv1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Biel S S, Gelderblom H R. Diagnostic electron microscopy is still a timely and rewarding method. J Clin Virol. 1999;13:105–119. doi: 10.1016/S1386-6532(99)00027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blauvelt A, Sei S, Cook P M, Schulz T F, Jeang K-T. Human herpesvirus 8 infection occurs following adolescence in the United States. J Infect Dis. 1997;176:771–774. doi: 10.1086/517298. [DOI] [PubMed] [Google Scholar]
- 8.Braun D K, Dominguez G, Pellett P E. Human herpesvirus 6. Clin Microbiol Rev. 1997;10:521–567. doi: 10.1128/cmr.10.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casas I, Tenorio A, de Ory F, Lozano A, Echevarría J M. Detection of both herpes simplex and varicella-zoster viruses in cerebrospinal fluid from patients with encephalitis. J Med Virol. 1996;50:82–92. doi: 10.1002/(SICI)1096-9071(199609)50:1<82::AID-JMV14>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 10.Chan P K S, Peiris J S M, Yuen K Y, Liang R H S, Lau Y L, Chen F E, Lo S K F, Cheung C Y, Chan T K, Ng M H. Human herpesvirus-6 and human herpesvirus-7 infections in bone marrow transplant recipients. J Med Virol. 1997;53:295–305. doi: 10.1002/(sici)1096-9071(199711)53:3<295::aid-jmv20>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez G, Dambaugh T R, Stamey F R, Dewhurst S, Inoue N, Pellett P E. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George J, Blakesley R W, Chirikjan J G. Sequence-specific endonuclease Bam HI. J Biol Chem. 1980;255:6521–6524. [PubMed] [Google Scholar]
- 13.Gershon A A, LaRussa P, Steinberg S P. Varicella-zoster virus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 895–904. [Google Scholar]
- 14.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 15.Hodinka R L, Friedman H R. Human cytomegalovirus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 884–894. [Google Scholar]
- 16.Horenstein M, Nador R G, Chadburn A, Hyjek E M, Inghirami G, Knowles D M, Cesarman E. Epstein-Barr virus latent gene expression in primary effusion-lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. Blood. 1997;90:1186–1191. [PubMed] [Google Scholar]
- 17.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. Comparison of the complete DNA sequences of the human herpesvirus 6 variants A and B. J Virol. 1999;73:8053–8063. doi: 10.1128/jvi.73.10.8053-8063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouzarides T, Bankier A T, Satchwell S C, Weston K, Tomlinson P, Barrell B G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987;61:125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy J A. Three new human herpesviruses (HHV6, 7 and 8) Lancet. 1997;349:558–562. doi: 10.1016/S0140-6736(97)80119-5. [DOI] [PubMed] [Google Scholar]
- 20.Madhavan H H, Priya K, Anand A R, Therese K L. Detection of herpes simplex virus (HSV) genome using polymerase chain reaction (PCR) in clinical samples. Comparison of PCR with standard laboratory methods for the detection of HSV. J Clin Virol. 1999;14:145–151. doi: 10.1016/s1386-6532(99)00047-5. [DOI] [PubMed] [Google Scholar]
- 21.Minjolle S, Michelet C, Jusselin I, Joannes M, Cartier F, Colimon R. Amplification of the six major human herpesvirus from cerebrospinal fluid by a single PCR. J Clin Microbiol. 1999;37:950–953. doi: 10.1128/jcm.37.4.950-953.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell P S, Espy M J, Smith T F, Toal D R, Rys P N, Berbari E T, Osmon D R, Persing D H. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimen. J Clin Microbiol. 1997;35:2873–2877. doi: 10.1128/jcm.35.11.2873-2877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson S, Matlow A, McDowell C, Roscoe M, Karmali M, Penn L, Dyster L. Detection of Bordetella pertussis in clinical specimens by PCR and a microtiter plate-based DNA hybridization assay. J Clin Microbiol. 1997;35:117–120. doi: 10.1128/jcm.35.1.117-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchett R, Pedersen M, Kieff E. Complexity of EBV homologous DNA in continuous lymphoblastoid cell lines. Virology. 1976;74:227–231. [PubMed] [Google Scholar]
- 25.Read S J, Jeffery K J M, Bangham C R M. Aseptic meningitis and encephalitis: the role of PCR in the diagnostic laboratory. J Clin Microbiol. 1997;35:691–696. doi: 10.1128/jcm.35.3.691-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Read S J, Kurtz J B. Laboratory diagnosis of common viral infections of the central nervous system by using a single multiplex PCR screening assay. J Clin Microbiol. 1999;37:1352–1355. doi: 10.1128/jcm.37.5.1352-1355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozenberg F, Lebon P. Amplification and characterization of herpes virus DNA in cerebrospinal fluid from patients with acute encephalitis. J Clin Microbiol. 1991;29:2412–2417. doi: 10.1128/jcm.29.11.2412-2417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Secchiero P, Carrigan D R, Asano Y, Benedetti J, Crowley R W, Komaroff A L, Gallo R C, Lusso P. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J Infect Dis. 1995;171:273–280. doi: 10.1093/infdis/171.2.273. [DOI] [PubMed] [Google Scholar]
- 29.Tang Y-W, Espy M J, Persing D H, Smith T F. Molecular evidence and clinical significance of herpes virus coinfection in the central nervous system. J Clin Microbiol. 1997;35:2869–2872. doi: 10.1128/jcm.35.11.2869-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tellier R, Bukh J, Emerson S U, Miller R H, Purcell R H. Long PCR and its application to hepatitis viruses: amplification of hepatitis A, hepatitis B, and hepatitis C virus genomes. J Clin Microbiol. 1996;34:3085–3091. doi: 10.1128/jcm.34.12.3085-3091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber T, Frye S, Bodemer M, Otto M, M, Lüke W. Clinical implications of nucleic acid amplification methods for the diagnosis of viral infection of the nervous system. J Neurovirol. 1996;2:175–190. doi: 10.3109/13550289609146880. [DOI] [PubMed] [Google Scholar]