Abstract
Staphylococcus saprophyticus is one of the most frequently encountered microorganisms associated with acute urinary tract infections (UTIs) in young, sexually active female outpatients. Conventional identification methods based on biochemical characteristics can efficiently identify S. saprophyticus, but the rapidities of these methods need to be improved. Rapid and direct identification of this bacterium from urine samples would be useful to improve time required for the diagnosis of S. saprophyticus infections in the clinical microbiology laboratory. We have developed a PCR-based assay for the specific detection of S. saprophyticus. An arbitrarily primed PCR amplification product of 380 bp specific for S. saprophyticus was sequenced and used to design a set of S. saprophyticus-specific PCR amplification primers. The PCR assay was specific for S. saprophyticus when tested with DNA from 49 gram-positive and 31 gram-negative bacterial species. This assay was also able to amplify efficiently DNA from all 60 strains of S. saprophyticus from various origins tested. This assay was adapted for direct detection from urine samples. The sensitivity levels achieved with urine samples was 19 CFU with 30 cycles of amplification and 0.5 CFU with 40 cycles of amplification. This PCR assay for the specific detection of S. saprophyticus is simple and rapid (approximately 90 min, including the time for urine specimen preparation).
Coagulase-negative staphylococci are commensal organisms of human skin flora but have become major etiological agents of nosocomial bacteremia and can colonize a variety of medical devices (18). These nosocomial infections are usually (>80%) caused by Staphylococcus epidermidis (18, 24). However, Staphylococcus saprophyticus is the second most frequently encountered agent of acute urinary tract infections (UTIs) after Escherichia coli (10, 11). S. saprophyticus is often isolated from the urine of young, sexually active female outpatients presenting with symptoms of acute UTI (1, 15, 25) indistinguishable from the symptoms of UTIs caused by Escherichia coli. These coagulase-negative staphylococci are rarely found as a cause of UTIs in hospitalized patients or as a contaminant of urine cultures (15) and are characterized by the low bacterial counts (less than 105 CFU per ml) required to elicit a UTI (24). S. saprophyticus could be the cause of chronic bacterial prostatitis in men (4), and there is evidence that suggests that this staphylococcal species could be the etiological agent of sexually transmitted urethritis (9). The use of spermicide-coated condoms has now been associated with an increased risk of UTIs caused by S. saprophyticus (6).
The classical phenotypic identification of staphylococci by Kloos and Schleifer (19) remains the “gold standard” for reference laboratories, but it is too lengthy and cumbersome for routine use in hospital microbiology laboratories. S. saprophyticus is differentiated from other urinary coagulase-negative staphylococci (i.e., S. epidermidis) by its uniform resistance to novobiocin, aerobic growth requirements, urease production, and carbohydrate utilization (15). Several culture-based commercially available systems including API Staph strips and the RapiDEC system have been evaluated for the identification of S. saprophyticus (12, 26). However, these systems require at least 20 h for staphylococcal species identification and occasionally misidentify S. saprophyticus. A few DNA-based assays that target variable regions of the 16S rRNA gene of S. saprophyticus have been developed (7, 8). However, these assays have not been evaluated for direct detection of S. saprophyticus from clinical specimens.
Although S. saprophyticus is easy to cultivate, phenotypic analysis requires overnight growth of the microorganism. A rapid and sensitive DNA-based assay which is specific for S. saprophyticus and which is suitable for direct detection of the organism from urine specimens would allow a significant reduction in the time required for the diagnosis of S. saprophyticus infections. In this study, we present the development of an S. saprophyticus-specific DNA-based assay. An arbitrarily primed PCR (AP-PCR) protocol was used to find a prominent fingerprinting product of 380 bp shared by a panel of clinical strains of S. saprophyticus but not encountered in other closely related bacterial species. This DNA fragment was sequenced and used to design a pair of PCR primers suitable for the specific and ubiquitous detection of S. saprophyticus. This S. saprophyticus-specific PCR assay was adapted for direct detection of the organism from urine specimens. This assay will be combined in multiplex with other PCR assays currently under development in our laboratory to allow the concomitant detection of other bacteria frequently associated with UTIs.
MATERIALS AND METHODS
Bacterial strains.
The bacterial isolates used in this study were selected from the culture collection of the Microbiology Laboratory of the Centre Hospitalier Universitaire de Québec (Pavillon Centre Hospitalier de l'Université Laval [CHUL], Ste-Foy, Québec, Canada). Three S. saprophyticus strains obtained from the American Type Culture Collection (ATCC; strains ATCC 15305, ATCC 35552, and ATCC 43867) were also used for this study. The strains were cultured on sheep blood agar or in brain heart infusion (BHI) medium. Bacterial cultures were stored frozen (−80°C) in BHI broth containing 10% glycerol.
Forty-nine gram-positive, 31 gram-negative, and 60 clinical isolates were used to establish the performance of the S. saprophyticus PCR assay. This battery of bacterial strains includes isolates obtained from both ATCC and the Microbiology Laboratory of CHUL.
Clinical specimens.
A total of five culture-negative urine specimens received at the Microbiology Laboratory of CHUL, all collected from different patients, were used in this study. Urine samples (stored at 4°C) were tested by PCR less than 48 h after reception at the laboratory.
DNA isolation.
Genomic DNA was purified with the G NOME kit (Bio 101, Inc., Vista, Calif.) according to the manufacturer's instructions, except that the bacterial cells were initially resuspended in 250 μl of a lysis solution containing 200 μg of lysostaphin (Sigma Chemical Co., St. Louis, Mo.) per ml, 20 mM Tris, 2 mM EDTA, and 1.2% Triton X-100 and were incubated for 30 min at 37°C. Purified genomic DNA was diluted at a concentration of 1 ng/μl in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA).
Urine specimens were prepared for PCR amplification by using the IDI DNA extraction kit (Infectio Diagnostic [IDI] Inc., Sainte-Foy, Québec, Canada) according to the manufacturer's instructions.
AP-PCR amplification.
Twenty 10-nucleotide primers (kit AD; Operon Technologies Inc., Alameda, Calif.) were used for AP-PCR to search for a specific amplicon shared exclusively by the species of interest, S. saprophyticus (5, 27, 28). Amplifications were performed directly from 1 μl (0.1 ng/μl) of purified genomic DNA from 5 S. saprophyticus strains and 27 other staphylococcal (non-S. saprophyticus) strains. The 25-μl AP-PCR mixture contained 50 μM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 2.5 mM MgCl2, 1 of the 20 10-nucleotide primers at a concentration of 1.5 μM, 200 μM (each) the four deoxynucleoside triphosphates, and 0.5 U of Taq DNA polymerase (Promega Corp., Madison, Wis.) combined with the TaqStart antibody (Clontech Laboratories Inc., Palo Alto, Calif.). The TaqStart antibody, which is a neutralizing monoclonal antibody of Taq DNA polymerase, was added to all PCR mixtures to enhance the efficiency of the amplifications (17). The PCR mixtures were subjected to thermal cycling (3 min at 96°C and then 42 cycles of 1 min at 94°C for the denaturation step, 1 min at 31°C for the annealing step, and 2 min at 72°C for the extension step) with a PTC-200 thermal cycler (MJ Research Inc., Watertown, Mass.). A final extension step of 7 min at 72°C was performed to allow completion of amplicons. Random amplified polymorphic DNA (RAPD) fragment fingerprints were obtained by electrophoresis in 1.5% agarose gels containing 0.5 μg of ethidium bromide per ml in Tris-borate-EDTA buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 4 V/cm for 90 min. The gels were visualized under 254-nm UV light. The sizes of the amplification products were estimated by comparison with a 50-bp-molecular-size standard ladder.
Subsequently, AP-PCR products of the predicted size were recovered from the gel by using the QIAquick gel extraction kit (QIAGEN Inc., Mississauga, Ontario, Canada). The purified DNA fragments were then cloned into the pCR 2.1 T/A cloning vector (Invitrogen Corp., Carlsbad, Calif.). Plasmids were isolated from transformed E. coli strains by using the QIAGEN plasmid mini kit (QIAGEN Inc.). The presence of a DNA insert in the recombinant plasmids was confirmed by digesting the purified plasmid DNA with EcoRI (New England Biolabs Ltd., Mississauga, Ontario, Canada), which allowed the inserted fragment to be cut out. Both strands of the DNA inserts for each of the selected recombinant plasmids were sequenced with the PRISM Ready Reaction DyeDeoxy Terminator cycle sequencing kit with an Applied Biosystems 373A sequencer (Perkin-Elmer Corp., Foster City, Calif.). From the 380-bp sequence, we designed a pair of PCR primers suitable for the specific and ubiquitous detection of S. saprophyticus. The selected primer pair was verified by using the primer analysis software Oligo, version 5.0 (National Bioscience, Plymouth, Minn.).
Conventional PCR amplification.
Amplifications were performed either from 1 μl of a purified genomic DNA preparation or from a standardized bacterial suspension whose turbidity was adjusted to equal that of a 0.5 McFarland standard, which corresponds to approximately 1.5 × 108 bacteria per ml. The 20-μl PCR mixture contained 0.4 μM (each) the two S. saprophyticus-specific primers 5′-TCA AAA AGT TTT CTA AAA AAT TTA C-3′ (annealing positions 169 to 193) and 5′-ACG GGC GTC CAC AAA ATC AAT AGG A-3′ (annealing positions 355 to 379), 200 μM (each) the four deoxyribonucleoside triphosphates (Pharmacia Biotech Inc., Baie d'Urfé, Québec, Canada), 10 μM Tris-HCl (pH 9.0), 50 μM KCl, 0.1% Triton X-100, 2.5 mM MgCl2, 3.3 μg of bovine serum albumin (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) per ml, 10 copies of linearized plasmid pSL1138, which served as a target for the internal control, and 0.5 U of Taq DNA polymerase (Promega Corp.) combined with the TaqStart antibody (Clontech Laboratories Inc.) (16, 22). An internal control was integrated into every PCR mixture (16). Use of this control allowed verification of the efficiency of the amplification and ensured that significant PCR inhibition was absent. The PCR mixtures were subjected to thermal cycling (3 min at 96°C and then 30 or 40 cycles of 1 s at 95°C for the denaturation step and 30 s at 55°C for the annealing-extension step with a PTC-200 thermal cycler). Analysis by agarose gel electrophoresis was performed as described previously (22).
The specificities of the DNA-based tests were verified by using a panel of clinical isolates consisting of 49 gram-positive and 31 gram-negative bacterial species (Table 1). The ubiquity (i.e., the ability to detect all strains of S. saprophyticus) of the DNA-based tests was verified by using a panel of 60 clinical isolates identified as S. saprophyticus by using the MicroScan Autoscan-4 system equipped with the Positive BP Combo Panel Type 6 (Dade Diagnostics, Mississauga, Ontario, Canada).
TABLE 1.
Bacterial strains used to test specificity of S. saprophyticus-specific PCR assay
| Gram-positive bacteria (n = 49) | Gram-negative bacteria (n = 31) |
|---|---|
| Staphylococcus arlettae ATCC 43957 | Acinetobacter baumannii ATCC 19606 |
| Staphylococcus aureus subsp. anaerobius ATCC 35844 | Bacteroides fragilis ATCC 25285 |
| Staphylococcus aureus subsp. aureus ATCC 43300 | Bordetella pertussis ATCC 9797 |
| Staphylococcus auricularis ATCC 33803 | Bulkholderia cepacia ATCC 25416 |
| Staphylococcus capitis subsp. capitis ATCC 27840 | Citrobacter diversus ATCC 27028 |
| Staphylococcus caprae ATCC 35538 | Citrobacter freundii ATCC 8090 |
| Staphylococcus carnosus ATCC 51365 | Enterobacter aerogenes ATCC 13048 |
| Staphylococcus chromogenes ATCC 43764 | Enterobacter cloacae ATCC 13047 |
| Staphylococcus caprae ATCC 35538 | Escherichia coli ATCC 25922 |
| Staphylococcus cohnii subsp. urealyticum ATCC 20260 | Gardnerella vaginalis ATCC 14018 |
| Staphylococcus delphini ATCC 49171 | Haemophilus ducreyi ATCC 33940 |
| Staphylococcus epidermidis ATCC 14990 | Haemophilus influenzae ATCC 9007 |
| Staphylococcus equorum ATCC 43958 | Hafnia alvei ATCC 13337 |
| Staphylococcus felis ATCC 49168 | Kingella indologenes ATCC 25869 |
| Staphylococcus gallinarum ATCC 35539 | Klebsiella oxytoca ATCC 13182 |
| Staphylococcus haemolyticus ATCC 29970 | Klebsiella pneumoniae ATCC 13883 |
| Staphylococcus hominis ATCC 27844 | Moraxella catarrhalis ATCC 25240 |
| Staphylococcus hyicus ATCC 11249 | Morganella morganii ATCC 25830 |
| Staphylococcus intermedius ATCC 29663 | Neisseria gonorrhoeae ATCC 35201 |
| Staphylococcus kloosi ATCC 43959 | Neisseria meningitidis ATCC 13077 |
| Staphylococcus lentus ATCC 29070 | Pasteurella aerogenes ATCC 27883 |
| Staphylococcus lugdunensis ATCC 43809 | Proteus mirabilis ATCC 25933 |
| Staphylococcus saprophyticus ATCC 15305 | Proteus vulgaris ATCC 13315 |
| Staphylococcus saprophyticus ATCC 35552 | Providencia rettgeri ATCC 9250 |
| Staphylococcus saprophyticus ATCC 43867 | Pseudomonas aeruginosa ATCC 27853 |
| Staphylococcus schleiferi subsp. coagulans ATCC 49545 | Salmonella typhimurium ATCC 14028 |
| Staphylococcus sciuri subsp. sciuri ATCC 29060 | Serratia marcescens ATCC 8100 |
| Staphylococcus simulans ATCC 27848 | Shigella flexneri ATCC 12022 |
| Staphylococcus warneri ATCC 27836 | Shigella sonnei ATCC 29930 |
| Staphylococcus xylosus ATCC 29971 | Stenotrophomonas maltophilia ATCC 13843 |
| Bacillus subtilis ATCC 27370 | Yersinia enterocolitica ATCC 9610 |
| Enterococcus avium ATCC 14025 | |
| Enterococcus durans ATCC 19432 | |
| Enterococcus faecalis ATCC 29212 | |
| Enterococcus faecium ATCC 19434 | |
| Enterococcus flavescens ATCC 49996 | |
| Enterococcus gallinarum ATCC 49573 | |
| Lactococcus lactis ATCC 11454 | |
| Lactobacillus acidophilus ATCC 4356 | |
| Listeria monocytogenes ATCC 15313 | |
| Macrococcus caseolyticus ATCC 13548 | |
| Mobiluncus curtissi ATCC 35242 | |
| Streptococcus agalactiae ATCC 27591 | |
| Streptococcus anginosus ATCC 33397 | |
| Streptococcus bovis ATCC 33317 | |
| Streptococcus dysgalactiae ATCC 43078 | |
| Streptococcus pneumoniae ATCC 27336 | |
| Streptococcus pyogenes ATCC 19615 | |
| Streptococcus salivarius ATCC 7073 |
For determination of the sensitivities of the 30- and 40-cycle PCR assays, cultures of three strains of S. saprophyticus (strains ATCC 15305, ATCC 35552, and ATCC 43867) in the logarithmic phase of growth (optical density at 600 nm, ≈0.7 to 0.8) were diluted in phosphate-buffered saline (PBS). Each dilution (1 μl) was tested in PCR assays to determine the minimal number of CFU which could be detected. The number of CFU was estimated by standard plating procedures. A similar approach was applied to determine the minimal number of genome copies which could be detected.
To assess the sensitivity of the PCR assay for detection of S. saprophyticus directly from urine specimens, five bacterium-free urine specimens were spiked with various amounts of S. saprophyticus cells in the mid-logarithmic phase of growth in order to determine the minimal number of CFU which could be detected. The sensitivity was determined with 40 cycles of amplification.
Nucleotide sequence accession number.
The nucleotide sequence of the S. saprophyticus-specific AP-PCR amplicon is available from GenBank as accession no. AF144088.
RESULTS
Isolation of an S. saprophyticus-specific DNA fragment by AP-PCR.
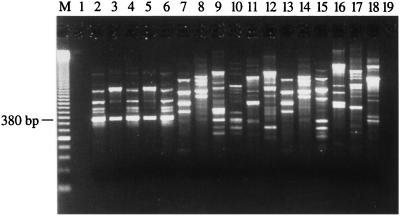
The generation of RAPD fingerprints for 5 S. saprophyticus strains (including the 3 ATCC strains) and 29 other staphylococcal species with the 20 different AP-PCR primers (10-mer) allowed determination of which primer produced amplification patterns specific for the 5 S. saprophyticus strains. Primer OPAD-16 (5′-AACGGGCGTC-3′) allowed isolation of a DNA fragment of 380 bp found in all RAPD patterns for the 5 S. saprophyticus strains tested but absent from the RAPD patterns for the other bacterial species tested (Fig. 1). Subsequently, we confirmed that this 380-bp amplification product was also absent from a wider array of bacterial species consisting of 19 other genetically related gram-positive species (Table 1). This S. saprophyticus-specific amplification product was gel purified and then cloned into the T/A cloning vector pCR 2.1.
FIG. 1.
AP-PCR amplification with the OPAD-16 primer performed with 100 pg of purified genomic DNA from reference and clinical strains of S. saprophyticus, various staphylococcal species, and gram-positive bacteria genetically related to S. saprophyticus. The content of each lane is as follows: 2, S. saprophyticus ATCC 15305; 3, S. saprophyticus ATCC 33552; 4, S. saprophyticus ATCC 43867; 5, S. saprophyticus Cssa-18; 6, S. saprophyticus Ssa-165; 7, S. aureus ATCC 43300; 8, S. capitis subsp. capitis ATCC 27840; 9, S. epidermidis ATCC 14990; 10, S. haemolyticus ATCC 29970; 11, S. hominis ATCC 27844; 12, S. simulans ATCC 27848; 13, S. warneri ATCC 27836; 14, Bacillus subtilis ATCC 27370; 15, Enterococcus faecalis ATCC 29212; 16, Lactobacillus acidophilus ATCC 4356; 17, Listeria monocytogenes ATCC 15313; 18, Streptococcus pneumoniae ATCC 27336; 1 and 19, controls to which no DNA was added; M, 50-bp ladder (molecular size standard).
Subsequently, the sequences of both strands of the S. saprophyticus 380-bp genomic DNA insert were determined for the five strains. We performed a multiple sequence alignment of these sequences and found homology of over 99%, indicating that this genomic target is well conserved in S. saprophyticus and, consequently, is promising for diagnostic purposes. Searches for this sequence in various data banks did not reveal any significant homologies with known sequences. A pair of PCR primers for the specific detection of S. saprophyticus was derived from conserved regions of this DNA fragment with the help of the Oligo software.
PCR assays.
Specificity tests performed with the panel of gram-positive and gram-negative bacterial species (Table 1) with 30 and 40 cycles of amplification showed that the selected PCR primer pair amplified only DNA from S. saprophyticus strains. In order to ensure that the negative PCR results obtained with the bacterial species other than the target species were not attributable to PCR inhibitors or to the inadequacy of the PCR assay, all reactions included an internal control simultaneously amplified. This control was always efficiently amplified when no target DNA was present, thereby showing the absence of PCR inhibitors. It is important that the S. saprophyticus-specific PCR assay did not yield any specific amplification product with 27 staphylococcal species other than S. saprophyticus (Table 1). No false-positive results with the set of 49 gram-positive bacteria comprising 30 staphylococcal species and 19 other genetically related gram-positive species was observed, indicating that the targeted genomic sequences are unique to S. saprophyticus. Increasing the number of amplification cycles from 30 to 40 did not appear to affect the specificity of the S. saprophyticus-specific PCR assay because all staphylococcal species other than S. saprophyticus as well as closely related species (Table 1) could not be amplified by the 40-cycle PCR assay (data not shown).
The S. saprophyticus-specific PCR assay was further validated by testing DNA from 60 clinical isolates of S. saprophyticus from the region of Quebec City, Quebec, Canada. These ubiquity tests showed that DNAs from all isolates were efficiently amplified by this PCR assay, thereby showing a perfect correlation with standard bacterial identification methods. DNAs from all reference strains tested were also shown to be efficiently amplified, thereby demonstrating a 100% ubiquity.
We determined the sensitivity of the 30-cycle PCR assay by using genomic DNA purified from the three S. saprophyticus strains from ATCC. These results indicated a detection limit of 100 copies of the S. saprophyticus genome for the three S. saprophyticus strains. In order to enhance the sensitivity of the assay, we increased the number of cycles. For PCR assays with 40 cycles, the sensitivity was increased to about six copies of the S. saprophyticus genome, while the length of time for completion of the assay was increased by approximately 10 min.
Sensitivity assays were also performed to determine the minimal number of S. saprophyticus cells which can be detected in urine specimens spiked with various amounts of cells in the mid-logarithmic phase of growth (Table 2). The detection limits in terms of the numbers of CFU determined with the IDI DNA extraction kit with five different urine specimens spiked with various amounts of S. saprophyticus cells were in the range of 300 to 1,400 CFU/ml of urine for the 40-cycle PCR. Furthermore, there was no significant PCR inhibition because the internal control was always efficiently amplified. For comparison, we have determined the sensitivity levels achieved with the same spiked urine specimens added directly to the PCR mixture without pretreatment. We found much lower detection limits (i.e., in the range of 700,000 to 800,000 CFU/ml of urine). Moreover, there was partial inhibition of the PCR on the basis of the amplification of the internal control. The sensitivity levels achieved with S. saprophyticus cells diluted in PBS were 500 CFU/ml with the IDI extraction kit, as opposed to 650,000 CFU/ml for samples added directly to the PCR mixture without pretreatment (Table 2). As expected, no significant PCR inhibition was observed for any experiment with PBS.
TABLE 2.
Sensitivity levels achieved by the 40-cycle PCR assay with PBS or urine specimens spiked with various amount of S. saprophyticus ATCC 15305 cells
| Sample preparation | Detection limit
|
|
|---|---|---|
| CFU/PCR mixture | CFU/ml of PBS or urine | |
| Spiked PBS | ||
| IDI DNA extraction kit | 0.5 | 500 |
| No treatment | 650 | 650,000 |
| Spiked urine samplesa | ||
| IDI DNA extraction kit | 0.3–1.4 | 300–1,400 |
| No treatment | 700–800 | 700,000–800,000 |
The given ranges of detection limits are for five urine samples.
DISCUSSION
S. saprophyticus is relatively easy to culture and identify by phenotypic methods. However, there is a need for the development of rapid and sensitive DNA-based assays which are suitable for the direct detection of S. saprophyticus from clinical specimens, especially urine specimens, to improve the rapidity and the accuracy of the diagnosis of S. saprophyticus infections. At present, resistance to novobiocin is still used in most laboratories to presumptively identify S. saprophyticus, but other coagulase-negative species, including S. epidermidis, are occasionally resistant to novobiocin. We have previously demonstrated the usefulness of nucleic acid amplification by PCR for detection and identification of S. aureus, S. epidermidis, and their clinically relevant antibiotic resistance genes (21–23). In the present study, we have developed a rapid PCR-based assay suitable for specific detection of S. saprophyticus in urine specimens. Initially, a set of 20 10-mer AP-PCR primers was tested with five different strains of S. saprophyticus in order to find a prominent shared amplicon. By this strategy, we were able to obtain such an amplicon of 380 bp that was consistently found in all S. saprophyticus strains tested but that was absent from other staphylococcal species and genetically related gram-positive bacteria. This S. saprophyticus-specific AP-PCR amplicon was sequenced and used to derive optimal PCR primers for the detection of S. saprophyticus. The S. saprophyticus-specific PCR assay developed in this study was specific because it did not amplify DNAs from a variety of gram-positive and gram-negative bacterial species including 26 staphylococcal species other than S. saprophyticus. Furthermore, this assay was shown to be 100% ubiquitous on the basis of testing of 60 S. saprophyticus clinical isolates from various patients, of which 92% were etiologic agents of UTIs. All 60 of these clinical isolates from CHUL as well as the three strains from ATCC were initially reconfirmed to be S. saprophyticus with the MicroScan Autoscan-4 system, thereby showing a perfect correlation with the identification obtained by the S. saprophyticus-specific PCR assay. Therefore, the S. saprophyticus genomic target of unknown coding potential selected for the PCR assay appears to be present in all S. saprophyticus strains and also appears to be well conserved in this species at the nucleotide level but either absent from or distinct in other closely related bacterial species including other staphylococcal species. The PCR assay, which was performed directly from standardized bacterial suspensions or urine specimens spiked with a known number of S. saprophyticus cells, was designed and optimized to be simple and performed in approximately an hour and a half.
Others have developed S. saprophyticus-specific PCR amplification assays targeting variable regions V3 and V6 of the 16S rRNA gene (7, 8). However, these assays have not been applied for direct detection of S. saprophyticus from clinical specimens. In our study, we have used a different approach to develop S. saprophyticus-specific DNA-based diagnostic tests. Our goal was to develop a simple and rapid PCR assay which is specific and ubiquitous for S. saprophyticus and which can be applied to detection directly from bacterial cultures or urine specimens in about 1 h. The 30-cycle PCR protocol showed sensitivity levels of about 100 copies of the S. saprophyticus genome. This sensitivity level is sufficient for culture confirmation assays from urine specimens or blood cultures. Increased levels of sensitivity of the PCR are required for detection of S. saprophyticus directly from urine specimens, in which the number of target cells can be much lower. The 40-cycle PCR protocol, which showed sensitivity levels of about six copies of the S. saprophyticus genome per PCR mixture or 300 to 1,400 CFU/ml of urine, appears to be suitable for that purpose. Such a high sensitivity level will be particularly critical in patients with UTIs with low S. saprophyticus cell counts (i.e., less than 104 CFU/ml), found in approximately one-third of women with acute lower UTIs caused by S. saprophyticus (14, 20). Similarly, acute pyelonephritis caused by UTIs have been reported in association with low bacterial counts in voided urine (13).
The sensitivity assays performed with S. saprophyticus cells diluted in PBS or urine samples demonstrated the efficacy of the IDI DNA extraction kit for (i) control of the PCR inhibitors present in urine samples and (ii) lysis of S. saprophyticus cells. On the basis of the minimum number of CFU detected in PBS, this extraction kit allows a cell lysis which is about 1,300 times more efficient than that from the direct addition of diluted cells to the PCR mixture without pretreatment. Furthermore, the sensitivity levels achieved with spiked urine samples and cells diluted in PBS prepared with the IDI DNA extraction kit were both very similar, thereby suggesting that it eliminated PCR inhibition completely. For comparison, there was partial to complete PCR inhibition with spiked urine samples added directly to the PCR mixture without pretreatment.
Preliminary studies performed with the IDI DNA extraction kit, which requires about 10 min for urine sample preparation, indicate that it is also suitable for efficient recovery of DNA from gram-negative bacilli including E. coli, enterococci, and streptococci, which are also frequently encountered in urine specimens. However, this application needs to be confirmed in a clinical study to validate the procedure for the diagnosis of UTIs. We have previously developed PCR assays for the specific detection of other staphylococcal species as well as associated antibiotic resistance genes (21–23). The S. saprophyticus PCR assay reported in this study will be combined in multiplex with these PCR assays as well as with others which are under development, especially for the identification and detection from urine specimens of E. coli and other bacteria frequently associated with UTIs. A direct impact of such diagnostic tests is that they should allow the faster establishment of effective antibiotic therapy and a reduction of empirical treatments with broad-spectrum antibiotics which are associated with high costs and toxicity (2, 3). The consequent reduction of antibiotic use should reduce the emergence of resistance.
ACKNOWLEDGMENTS
We thank Louise Côté, who is the director of the Microbiology Laboratory of CHUL, for free access to the laboratory and for providing the S. saprophyticus clinical isolates. We thank Maurice Boissinot for critical comments regarding the manuscript.
Francis Martineau has a scholarship from the Fonds de la Recherche en Santé du Québec. Marc Ouellette is a Medical Research Council Scientist. This research project was supported by grant PA-15586 from the Medical Research Council of Canada and by IDI.
REFERENCES
- 1.Abrahamsson K, Hansson S, Jodal U, Lincoln K. Staphylococcus saprophyticus urinary tract infections in children. Eur J Pediatr. 1993;152:69–71. doi: 10.1007/BF02072520. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron M, Ouellette M. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J Clin Microbiol. 1998;36:2169–2172. doi: 10.1128/jcm.36.8.2169-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron M G, Ouellette M. Diagnosing bacterial infectious diseases in one hour: an essential upcoming revolution. Infection. 1995;23:69–72. doi: 10.1007/BF01833867. [DOI] [PubMed] [Google Scholar]
- 4.Bergman B, Wedren H, Holm S E. Staphylococcus saprophyticus in males with symptoms of chronic prostatitis. Urology. 1989;5:241–245. doi: 10.1016/0090-4295(89)90316-6. [DOI] [PubMed] [Google Scholar]
- 5.Fani R, Damiani G, Di Serio, Gallori E, Grifoni A, Bazzicalupo M. Use of random amplified polymorphic DNA (RAPD) for generating specific DNA probes for microorganisms. Mol Ecol. 1993;2:243–250. doi: 10.1111/j.1365-294x.1993.tb00014.x. [DOI] [PubMed] [Google Scholar]
- 6.Fihn S D, Boyko E J, Chen C L, Normand E H, Yarbro P, Scholes D. Use of spermicide-coated condoms and other risk factors for urinary tract infection caused by Staphylococcus saprophyticus. Arch Intern Med. 1998;158:281–287. doi: 10.1001/archinte.158.3.281. [DOI] [PubMed] [Google Scholar]
- 7.Gaszewska-Mastalarz A, Zakrzewska-Czerwinska J, Mordarski M. Rapid detection of Staphylococcus saprophyticus using primer specific PCR. Acta Biol Hung. 1997;48:319–322. [PubMed] [Google Scholar]
- 8.Gaszewska-Mastalarz A, Zakrzewska-Czerwinska J, Mordarski M. Rapid identification of Staphylococcus saprophyticus using an oligonucleotide probe complementary to 16S rRNA. System Appl Microbiol. 1995;18:123–126. [Google Scholar]
- 9.Goldenring J M. Urinary tract infection with Staphylococcus saprophyticus. J Adolesc Health Care. 1986;6:417–418. doi: 10.1016/s0197-0070(86)80247-9. [DOI] [PubMed] [Google Scholar]
- 10.Gupta K, Scholes D, Stamm W E. Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA. 1999;281:736–738. doi: 10.1001/jama.281.8.736. [DOI] [PubMed] [Google Scholar]
- 11.Henry D, Ellison W, Sullivan J, Mansfield D L, Magner D J, Dorr M B, Talbot G H for The Sparfloxacin Multi Center UUTI Study Group. Treatment of community-acquired acute uncomplicated urinary tract infection with sparfloxacin versus ofloxacin. Antimicrob Agents Chemother. 1998;42:2262–2266. doi: 10.1128/aac.42.9.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janda W M, Ristow K, Novak D. Evaluation of RapiDEC Staph for identification of Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus saprophyticus. J Clin Microbiol. 1994;32:2056–2059. doi: 10.1128/jcm.32.9.2056-2059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J R, Stamm W E. Diagnosis and treatment of acute urinary tract infections. Infect Dis Clin N Am. 1987;1:773–791. [PubMed] [Google Scholar]
- 14.Johnson J R, Stamm W E. Urinary tract infections in women: diagnosis and treatment. Ann Intern Med. 1989;111:906–917. doi: 10.7326/0003-4819-111-11-906. [DOI] [PubMed] [Google Scholar]
- 15.Jordan P A, Iravani A, Richard G A, Baer H. Urinary tract infection caused by Staphylococcus saprophyticus. J Infect Dis. 1980;142:510–515. doi: 10.1093/infdis/142.4.510. [DOI] [PubMed] [Google Scholar]
- 16.Ke D, Picard F J, Martineau F, Ménard C, Roy P H, Ouellette M, Bergeron M G. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellogg D E, Rybalkin I, Chen N, Mukhamedova N, Vlasik T, Siebert P D, Chenchick A. TaqStart Antibody: “hot start” PCR facilitated by a neutralizing monoclonal antibody directed against Taq DNA polymerase. BioTechniques. 1994;16:1134–1137. [PubMed] [Google Scholar]
- 18.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloos W E, Schleifer K H. Simplified scheme for routine identification of human Staphylococcus species. J Clin Microbiol. 1975;1:82–88. doi: 10.1128/jcm.1.1.82-88.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunin C M, Van Arsdale White L, Hua Hua T. A reassessment of the importance of “low-count” bacteriuria in young women with acute urinary symptoms. Ann Intern Med. 1993;119:454–460. doi: 10.7326/0003-4819-119-6-199309150-00002. [DOI] [PubMed] [Google Scholar]
- 21.Martineau F, Picard F J, Lansac N, Ménard C, Roy P H, Ouellette M, Bergeron M G. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns in Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–238. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;36:618–623. doi: 10.1128/jcm.36.3.618-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/jcm.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rupp M E, Archer G L. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19:231–245. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 25.Svanborg C, Godaly G. Bacterial virulence in urinary tract infections. Infect Dis Clin N Am. 1997;11:513–529. doi: 10.1016/s0891-5520(05)70371-8. [DOI] [PubMed] [Google Scholar]
- 26.von Baum H, Klemme F R, Geiss H K, Sonntag H G. Comparative evaluation of a commercial system for identification of gram-positive cocci. Eur J Clin Microbiol Infect Dis. 1998;17:849–852. doi: 10.1007/s100960050205. [DOI] [PubMed] [Google Scholar]
- 27.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]