Abstract
From February to November 1997, 29 inpatients at Ramón y Cajal Hospital, Madrid, Spain, were determined to be either colonized or infected with imipenem- and meropenem-resistant Acinetobacter baumannii (IMRAB) strains (MICs, 128 to 256 μg/ml). A wide antibiotic multiresistance profile was observed with IMRAB strains. For typing IMRAB isolates, pulsed-field gel electrophoresis was used. For comparative purposes, 30 imipenem- and meropenem-susceptible A. baumannii (IMSAB) strains isolated before, during, and after the outbreak were included in this study. The molecular-typing results showed that the outbreak was caused by a single IMRAB strain (genotype A). By cloning experiments we identified a class D β-lactamase (OXA-24) encoded in the chromosomal DNA of this IMRAB strain which showed carbapenem hydrolysis. Moreover, the outer membrane profile of the IMRAB strain showed a reduction in the expression of two porins at 22 and 33 kDa when compared with genetically related IMSAB isolates. In addition no efflux mechanisms were identified in the IMRAB strains. In summary, we report here the molecular characterization of a nosocomial outbreak caused by one multiresistant A. baumannii epidemic strain that harbors a carbapenem-hydrolyzing enzyme. Although alterations in the penicillin-binding proteins cannot be ruled out, the reduction in the expression of two porins and the presence of this OXA-derived β-lactamase are involved in the carbapenem resistance of the epidemic nosocomial IMRAB strain.
Acinetobacter spp. are opportunistic pathogens with increasing relevance in nosocomial infections (6). They cause a wide range of clinical complications, such as pneumonia, septicemia, urinary tract infection, wound infection, and meningitis, especially in immunocompromised patients (6, 22). Antimicrobial treatment of these clinical infections, particularly those caused by Acinetobacter baumannii strains, may be compromised by the multiple-drug resistance of many isolates to β-lactams, aminoglycosides, and fluoroquinolones (4, 23).
During the last decade, hospital-acquired infections involving multiresistant A. baumannii isolates have been reported, often in association with contamination of the hospital equipment or cross-contamination by the colonized hands of patient-attending personnel (5, 6, 20, 37–39).
Regarding the resistance to β-lactam antibiotics of A. baumannii clinical strains, different mechanisms have been involved (2, 4). As in other gram-negative rods, the main mechanism of resistance to β-lactam antibiotics is the production of β-lactamases encoded either by the chromosome or by plasmids (19). In addition, a low permeability of the outer membrane of A. baumannii as well as alterations in the penicillin-binding protein (PBP) affinity has been involved in the resistance of A. baumannii to these antibiotics (2, 4, 11, 32).
In the last few years, carbapenem-resistant A. baumannii isolates have been reported worldwide (1, 12, 29). Loss of porins, PBP with reduced affinity, and different class B and D β-lactamases have been associated with resistance to carbapenems in A. baumannii clinical strains (2, 4, 7, 8, 9, 11, 12, 15, 18, 27, 29, 32).
The main objectives of this work were to characterize a nosocomial outbreak by antibiotyping and pulsed-field gel electrophoresis (PFGE) and to investigate the mechanisms of resistance to carbapenems in an epidemic multiresistant A. baumannii strain with a high level of resistance to carbapenems (i.e., the imipenem- and meropenem-resistant A. baumannii [IMRAB] strain), which caused a 10-month-long epidemic outbreak at our hospital in 1997, involving 29 patients.
(This work was presented in part at the 38th and 39th Interscience Conferences on Antimicrobial Agents and Chemotherapy, San Diego and San Francisco, Calif., respectively, 1998 and 1999, respectively. [G. Bou, G. Cerveró, D. Malpica, M. Pérez-Vázquez, L. de Rafael, and J. Martínez-Beltrán, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-120, 1998; G. Bou and J. Martinez-Beltran, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1461, 1999].)
MATERIALS AND METHODS
Description of the outbreak.
From February through November 1997, an IMRAB A. baumannii strain was isolated from 29 patients, 23 of whom were hospitalized in five different medical and surgical intensive care units (ICUs) at the Ramón y Cajal Hospital, a 1,200-bed tertiary-care teaching hospital. The original strain of the outbreak was simultaneously isolated from a bronchoaspirate and urine specimens of one patient admitted to the medical ICU. Afterwards, IMRAB isolates were obtained from 4 patients at the same ICU, 1 patient from a pediatric ICU, 17 patients from three different surgical ICUs, and 6 patients hospitalized in the internal medicine and dermatology medical departments. Criteria for infection with IMRAB were documented by infectious diseases unit physicians for 15 patients; meanwhile, the rest of the patients were considered to be colonized only. Infection control measures and the use of disposable gloves and aprons while caring for IMRAB-infected and -colonized patients were immediately implemented, the need for handwashing was reinforced, and, as far as possible, patients colonized or infected with IMRAB were isolated. Likewise, use of imipenem and meropenem, particularly in the areas involved in the outbreak, was restricted, and compliance with this antibiotic use policy was monitored by infectious diseases unit physicians.
Bacterial strains and plasmids.
A total of 226 A. baumannii clinical isolates were included in this study: 196 IMRAB isolates obtained from these 29 patients during the outbreak and 30 imipenem- and meropenem-susceptible A. baumannii (IMSAB) isolates, obtained from clinical specimens before the outbreak (November 1996 through January 1997), during the outbreak (February through November 1997), and after the outbreak (January through February 1998). Also, A. baumannii ATCC 189, ATCC 17978, ATCC 50853, and ATCC 9935 isolates were included in this study. The organisms were identified by the scheme by Kämpfer et al. (21) adapted by Dijkshoorn (14), which includes bacterial growth at 37, 41, and 44°C. The antimicrobial phenotypic susceptibility pattern was studied in 196 IMRAB and 30 IMSAB isolates. For the genotypic characterization of isolates, the PFGE method was performed with 30 IMRAB isolates, including the first isolate from 28 patients and 2 isolates from the patient who had clinical strains susceptible and resistant to tobramycin. Also 10 IMSAB isolates, obtained before, during, and after the outbreak (Table 1), and A. baumannii ATCC strains were used.
TABLE 1.
IMRAB and IMSAB A. baumannii isolates analyzed by PFGE
| Isolate no.a | Date of isolation (mo[s]/yr) | Wardb | Susceptibility patternc
|
PFGE group | ||
|---|---|---|---|---|---|---|
| CBP | CAZ | TOB | ||||
| IMRAB 1–5 | 2–4/97 | MICU | R | R | S/I | A |
| IMRAB 6–23 | 3–6/97 | SICU | R | R | S/I | A |
| IMRAB 24 | 5–6/97 | SICU | R | R | R | A |
| IMRAB 25–27 | 4–6/97 | DERM | R | R | R | A |
| IMRAB 28–30 | 7–11/97 | IMED | R | R | R | A |
| IMSAB-PRE 31 | 11/96 | MICU | S | S | S | B |
| IMSAB-PRE 32 | 12/96 | MUNI | S | I | R | C |
| IMSAB-PRE 33 | 1/97 | MUNI | S | I | S | C |
| IMSAB-AT 34 | 2/97 | SUNI | S | R | S | D-1 |
| IMSAB-AT 35 | 7/97 | SURG | S | R | S | D-1 |
| IMSAB-AT 36 | 7/97 | IMED | S | R | S | D-2 |
| IMSAB-AT 37 | 4/97 | SICU | S | S | S | E |
| IMSAB-PST 38 | 1/98 | SUNI | S | R | R | F |
| IMSAB-PST 39 | 1/98 | SICU | S | S | S | G |
| IMSAB-PST 40 | 2/98 | SICU | S | I | R | H |
PRE, Preoutbreak; AT, At outbreak; PST, postoutbreak.
MICU, medical ICU; SICU, surgical ICU; DERM, dermatology; IMED, internal medicine; SUNI, surgical unit; MUNI, medical unit.
CBP, carbapenem (imipenem and meropenem); CAZ, ceftazidime; TOB, tobramycin. R, resistant; I, intermediate; S, susceptible according to NCCLS criteria.
A. baumannii RYC 52763/97 is a carbapenem-resistant clinical strain isolated in February 1997, from a bronchial aspirate of a patient admitted to the Medical Intensive Care Unit in the Ramón y Cajal Hospital, Madrid, Spain. It was the initial isolate obtained from the first patient involved in the carbapenem-resistant A. baumannii nosocomial outbreak.
For conjugation experiments, the ampicillin-susceptible Acinetobacter junii strain MA RYC95 and Escherichia coli BM21 (a kind gift from J. M. Gomez) were used. For cloning experiments, E. coli TG1 [Δ(lac-pro) hsdD5 supE thi] was used as the host strain. Plasmid pBGS18− (36) with a kanamycin resistance marker was used for cloning the β-lactamase gene. Plasmid pUC18 with an ampicillin resistance marker was used for sequencing experiments (42).
Antimicrobial-agent susceptibility testing.
The MICs of the antimicrobial agents were determined by the agar dilution method (26), using Mueller-Hinton agar (Oxoid, United Kingdom), antibiotic-standard powders with stated potencies supplied by the drug manufacturers, and an inoculum of 104 CFU per spot. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as internal controls in each susceptibility determination.
PFGE.
Macrorestriction analysis of chromosomal DNA with SmaI and ApaI (New England Biolabs, Boston, Mass.) was carried out by PFGE following published procedures (16). PFGE was run in a CHEF-DRIII apparatus (Bio-Rad Laboratories, Richmond, Calif.), with pulses ranging from 0.5 to 15 s at a voltage of 6 V/cm at 14°C for 20 h. Products were detected after staining with ethidium bromide (50 μg/ml) and photographed with Polaroid type 665 film. Criteria for interpreting PFGE patterns according to the number of fragment differences compared with the outbreak pattern were as follows: 0 differences, isolate is part of outbreak; 2 to 3, isolate is probably part of the outbreak; 4 to 6, isolate is possibly part of the outbreak; and ≥7, isolate is not part of the outbreak (40).
REP-PCR.
Amplification reactions were performed in a final volume of 50 μl. Mg2+-free PCR buffer was purchased as a 10× concentrate consisting of 500 mM KCl, 100 mM Tris-HCl (pH 9.0), and 1% Triton X-100 (Perkin Elmer, Applied Biosystem Division). Each reaction mixture contained 5 μl of 10× PCR buffer–2 U of AmpliTaq Gold (Perkin-Elmer, Roche Molecular Systems, Inc., N.J.)–200 μM (each) dATP, dCTP, dGTP, and dTTP (Perkin-Elmer, Roche Molecular Systems, Inc.). The Mg2+ concentration was 3 mM, and the primers were used at 0.5 μM. The primer pair REP1 5′-IIIGCGCCGICATCAGGC-3′ and REP2 5′-ACGTCTTATCAGGCCTAC-3′ (41) was used to amplify putative REP-like elements in the genomic bacterial DNA. The amount of the chromosomal DNA added to the reaction was 500 ng. Amplification reactions were carried out in a Progene thermal cycler (Techne, Cambridge, United Kingdom), with an initial denaturation (10 min at 94°C) followed by 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 45°C), and extension (2 min at 72°C), with a single final extension of 16 min at 72°C. Aliquots (20 μl) of each sample were subjected to electrophoresis in 1.2% agarose gels. Amplified products were detected after staining with ethidium bromide (50 μg/ml) and photographed with Polaroid type 665 film. Strains belonging to the same DNA group showed identical profiles or highly similar profiles (up to two bands different).
Analytical isoelectric focusing.
β-Lactamases were characterized by isoelectric focusing of ultrasonic bacterial extracts prepared by sonication (25). β-Lactamases were analyzed by isoelectric focusing of cell extracts on polyacrylamide gel containing ampholytes with a pH range of 3.5 to 9.5 (Ampholine PAGplate; Pharmacia Biotech) in a Multiphor II System (Pharmacia-LKB). The focused β-lactamases were detected by overlaying the gel with nitrocefin (0.5 mg/ml) in phosphate buffer (100 mM, pH 7.0). pI values were determined by comparison with those of β-lactamases and proteins with known pIs: TEM-1 (pI 5.4), TEM-3 (pI 6.3), SHV-1 (pI 7.6), lentil lectin acid (pI 8.15), MIR-1 (pI 8.4), trypsinogen (pI 9.30), and A. baumannii RYC 52763/97 AmpC (pI 9.4).
Conjugation experiments.
Transfer of resistance by conjugation was attempted using E. coli BM21 and A. junnii MA RYC95 strains as recipients. Overnight filter mating experiments were performed at 30°C and 37°C, and the transconjugants were selected on MacConkey agar plates supplemented with ampicillin (25 μg/ml) and nalidixic acid (50 μg/ml) for E. coli and Columbia agar plates supplemented with d-glucose (2% [wt/vol]), neutral red, and ampicillin (25 μg/ml) for A. junnii.
Plasmid isolation and cloning of the TEM-1 gene.
A. baumannii RYC 52763/97 had only one plasmid, of about 22 kb, that was isolated by the alkaline lysis method (30). A blaTEM-1 gene was amplified from this plasmid by PCR using the specific blaTEM primers C1 5′-GGGAATTCTCGGGGAAATGTGCGCGGAAC and C2 5′-GGGATCCGAGTAAACTTGGTCTGACAG and cloned into the pBGS18− plasmid (pAB1).
Extraction of chromosomal DNA, cloning, and sequencing of the β-lactamase genes.
For the chromosomal-DNA purification, the strains were grown overnight on MacConkey agar plates at 37°C, and the growth from approximately one-fourth plate was resuspended in 180 μl of distilled water. Afterwards, 200 μl of buffer solution (0.01 M Tris-Cl [pH 7.8]–0.005 M EDTA–0.5% sodium dodecyl sulfate [SDS]) and 20 μl of proteinase K (1 mg/ml) were added. The mixture was incubated for 2 h at 55°C, and then 400 μl of phenol-chloroform solution was added, mixed with gentle agitation, and centrifuged at 11,000 rpm with a microcentrifuge for 5 min. The supernatant was collected, and the DNA was precipitated after the addition of 0.5 volumes of 7.5 M ammonium acetate and 2 volumes of ethanol. The DNA was washed with 70% ethanol, dried, and resuspended in 100 μl of Tris-EDTA buffer.
The restriction enzymes were purchased from Boehringer GmbH (Mannheim, Germany) and were used according to the manufacturer's directions.
Cloning procedures were performed as described by Sambrook et al. (30). Bacterial cells were made competent by the calcium chloride method. For cloning experiments, pBGS18 and pUC18 plasmids were used. Transformants were selected on Luria-Bertani (LB) plates supplemented with kanamycin (10 μg/ml) and ampicillin (50 μg/ml). Kanamycin was omitted when bacteria were transformed with the pUC18 plasmid. For the pUC18 plasmid, 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) were used for transformant selection.
Templates were sequenced on both strands by the method of Sanger et al. (31). Sequencing was carried out with the Taq DyeDeoxiTerminator cycle sequencing kit, using specific primers to the coding sequence. The sequence was analyzed in an automatic DNA sequencer (377 Abi-Prims; Perkin-Elmer).
Kinetic experiments and carbapenem hydrolysis.
For kinetic studies, cell-free lysate was obtained by sonication of the sediment of a 1-liter exponentially growing culture of A. baumannii RYC 52763/97 and E. coli harboring the OXA-24 β-lactamase (plasmid pBMB-1) gene (at 37°C in LB broth medium containing 50 μg of ampicillin per ml. The sonicated extracts were dialyzed overnight at 4°C in 0.05 M phosphate buffer (pH 7.4) and then loaded into a 300-ml (75- by 2.5-cm) Sephadex G100 column (Pharmacia Fine Chemicals AB, Uppsala, Sweden) previously equilibrated with the same buffer. The β-lactamases were eluted with 0.05 M phosphate buffer (pH 7.4), and that activity was tested by the nitrocefin method. Fractions containing β-lactamase activity were collected, concentrated with centricon (Amicon B15; W. R. Grace and Co., Danvers, Mass.), stored for a maximum of 1 week at −70°C, and used for the determination of kinetic constants. Controls for hydrolysis studies consisted of the same bacterial extract without the β-lactamase. Km, Vmax, 50% inhibitory concentration, and hydrolysis rates were monitored spectrophotometrically (UVIKON-930) and determined as previously described (8).
To study the inactivation of imipenem by A. baumannii OXA-24 β-lactamase, a microbiological disk assay was performed with a modification of the Masuda method (24). An imipenem disk (10 μg) was placed in the center of a Mueller-Hinton agar plate seeded with E. coli ATCC 25922 strain. Four filter paper disks, each containing 20, 10, or 5 μl of the enzyme preparation or 20 μl of sodium phosphate buffer (pH 7.0), were applied 15 mm from the imipenem disk. Plates were incubated at 37°C overnight, and inactivation of imipenem was shown by growth of the indicator strain within the expected inhibition zone.
OMP analysis.
Outer membrane proteins (OMPs) of the IMRAB A. baumannii RYC 52763/97 strain and the IMSAB strain 34 (Table 1) were analyzed from logarithmic cultures by previously described methods using SDS-polyacrylamide gel electrophoresis (3). All isolates were cultured in LB medium. Purified OMPs were obtained by treatment of the cell envelopes with sodium-N-lauryl sarcosinate. Samples of each preparation were applied to the gels and electrophoresed with an Electrophoresis Power Supply 500/400 (Pharmacia, Uppsala, Sweden). Densitometry analysis of the gels was performed by using a Unison Unipower PS 4.5 apparatus.
Efflux mechanism.
To determine the presence of an efflux mechanism involved in the resistance to carbapenems in the A. baumannii RYC 52763/97 strain, MIC assays were performed with Mueller-Hinton agar plates with (25 and 50 μg/ml) and without reserpine. A P. aeruginosa strain (RYC 44629/97) with a clear and defined efflux mechanism was used as a control. Meropenem and norfloxacin were used as antibiotic controls to verify that reserpine inhibits the efflux mechanism.
Detection of the OXA-24 gene in the IMRAB A. baumannii strains.
To determine the presence or absence of the OXA-24 gene in different A. baumannii strains and to study their putative role in carbapenem resistance, a PCR assay was performed. Six IMRAB and 10 IMSAB strains were used. Reactions were carried out with a 50-μl volume of a reaction mixture containing 20 mM Tris-HCl (pH 8.8), 100 mM potassium chloride, 2.0 mM magnesium chloride, 200 μM deoxynucleotide triphosphates, 50 pmol of each primer, 250 ng of the chromosomal DNA, and 2.5 U of Taq polymerase (Roche). The primers 5′-GTACTAATCAAAGTTGTGAA (P1) and 5′-TTCCCCTAACATGAATTTGT (P2) adjacent to the OXA-24-coding region were used. Amplification reactions were submitted to the following program: initial denaturation (4 min at 94°C) followed by 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 50°C), and extension (2 min at 72°C), with a single extension of 10 min at 72°C. The amplified 995-bp product was resolved by electrophoresis in a 1.5% (wt/vol) agarose gel containing ethidium bromide (50 μg/ml).
RESULTS
Antimicrobial susceptibility pattern.
A. baumannii RYC 52763/97 (Table 2) and all IMRAB isolates exhibited a similar multiresistance profile, including resistance to semisynthetic penicillins, ceftazidime, cefepime, cefpirome, aztreonam, gentamicin, netilmicin, amikacin, and ciprofloxacin (MICs of >128 μg/ml for all antibiotics) and a high level of resistance to imipenem and meropenem (MICs, 128 to 256 μg/ml). Considering the critical concentrations of susceptibility, only tobramycin, sulbactam (MIC, 16 to 32 μg/ml), and colistin (MIC, 4 to 8 μg/ml) showed activity. Tobramycin MICs for the IMRAB isolates obtained from 23 patients were 4 to 8 μg/ml, while those for isolates from 5 patients were ≥128 μg/ml. One patient simultaneously harbored tobramycin-susceptible and -resistant IMRAB isolates. Regarding IMSAB isolates, apart from carbapenem susceptibility, several antimicrobial susceptibility patterns were obtained irrespective of the isolation time (Table 1).
TABLE 2.
MICs of β-lactams for A. baumannii RYC 52763/97 and recipient strain E. coli TG1 harboring various β-lactamases
| β-Lactam | MIC (μg/ml) for strain (enzyme[s])a
|
||||
|---|---|---|---|---|---|
| A. baumannii RYC 52763/97 (AmpC, OXA-24 enzyme, and TEM-1) | E. coli TG1 | E. coli TG1 with pGER1 (AmpC)b | E. coli TG1 (TEM-1) | E. coli TG1 with pBMB-1 (OXA-24) | |
| Ampicillin | >1,024 | 4 | 128 | >1,024 | 128 |
| Ampicillin + clavulanic acidc | 512 | 4 | 64 | 16 | 64 |
| Ticarcillin | >1,024 | 4 | 32 | >1,024 | 256 |
| Cefazolin | >256 | 8 | >256 | 16 | 32 |
| Cefuroxime | >256 | 4 | >256 | 4 | 4 |
| Cefoxitin | >256 | 4 | 4 | 4 | 4 |
| Cefotaxime | >256 | 0.06 | 4 | 0.06 | 0.06 |
| Ceftazidime | >256 | 0.125 | 16 | 0.125 | 0.25 |
| Cefepime | 256 | 0.06 | 0.25 | 0.06 | 0.125 |
| Aztreonam | >256 | 0.06 | 1 | 0.06 | 0.125 |
| Imipenem | 128 | 0.125 | 0.125 | 0.125 | 1 |
| Meropenem | 256 | 0.03 | 0.03 | 0.03 | 0.125 |
All the β-lactamase genes were cloned into the same pBGS18− plasmid.
See reference 7.
Concentration of the inhibitor was 4 μg/ml.
PFGE and REP-PCR analysis.
PFGE patterns of the representative A. baumannii IMRAB and IMSAB isolates are shown in Fig. 1. All IMRAB isolates analyzed had an identical band pattern and were classified in genotype A. In contrast, preoutbreak (strains 31 to 33), at-outbreak (strains 34 to 37), and postoutbreak (strains 38 to 40) IMSAB isolates belonging to a different genotype on the basis of the band profile and were assigned to genotypes B-C, D-E, and F-H, respectively (Table 1). A. baumannii ATCC strains showed a different band pattern than those of the IMRAB and IMSAB isolates (data not shown).
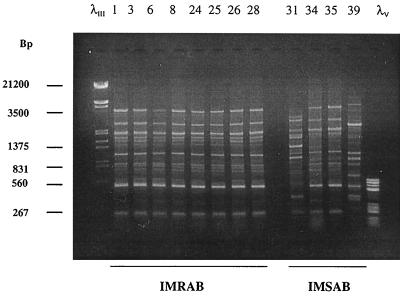
FIG. 1.
Patterns obtained by PFGE with SmaI. Lanes 1, 10, 14, and 19, low-range PFG DNA marker (New England Biolabs, United Kingdom). Lanes 2 through 9, IMRAB strains 1, 3, 6, 8, 24, 25, 26, and 28, respectively, in Table 1. Lanes 11 through 13, IMSAB PRE strains 31 to 33, respectively; lanes 15 through 18, IMSAB AT strains 34 through 37, respectively; lanes 20 through 22, IMSAB POST strains 38 through 40, respectively.
An interesting subject is at-outbreak carbapenem-susceptible A. baumannii strains 34 to 36. The genotypic data obtained with these strains led us to classify them into a different genotype (PFGE group D); however, comparing their profile with that of the IMRAB strains indicated that less than six different bands were obtained by PFGE. These results suggest the possibility of a genetic relationship between IMSAB strains 34 to 36 and the IMRAB isolates.
The agarose gel electrophoresis of the amplified fragments by REP-PCR of representative A. baumannii IMRAB and IMSAB isolates is shown in Fig. 2. The patterns of eight IMRAB isolates (strains 1, 3, 6, 8, 24, 25, 26, and 28 in Table 1) obtained from different patients and wards of the hospital were identical, and strains were assigned to PCR group 1, thus confirming the PFGE results. In the same gel, for comparison, the pattern of four A. baumannii isolates susceptible to the carbapenems obtained pre- (strain 31), at (strains 34 to 35), and post- (strain 39) outbreak is shown. As observed in Fig. 2, strains 31 and 39 showed a very different band pattern than that of the IMRAB profile. However, in the case of strains 34 to 35, a similar band pattern was observed compared to those of the IMRAB strains (up to two different bands). Therefore, this result suggests the possibility of a genetic relationship between IMSAB strains 34 and 35 and the IMRAB isolates.
FIG. 2.
Patterns obtained by REP-PCR. The numbers above the figure indicate the corresponding strains (Table 1). Lanes λIII and λV, DNA molecular weight markers III and V, respectively (Boehringer GmbH, Mannheim, Germany).
Isoelectric focusing analysis.
The sonicated extract of the strain A. baumannii RYC 52763/97 contained three β-lactamase bands: one with a pI of 5.4 (TEM-1 like) was plasmid mediated because it was cloned as described in Materials and Methods; another focused at pI 9.0 and failed to be transferred by conjugation experiments; and the third, focused at pI 9.4, corresponded to the A. baumannii chromosomal cephalosporinase previously described (7).
Cloning and sequencing the β-lactamase genes.
In order to determine the presence of a carbapenemase responsible for the high levels of carbapenem resistance, we attempted to clone all of the β-lactamase genes of the A. baumannii RYC 52763/97 strain. Neither antibiotic resistance gene transfer was obtained by conjugation experiments with A. baumannii RYC 52763/97, although a blaTEM-1 gene was cloned from the plasmid of the A. baumannii RYC 52763/97 strain as described in Materials and Methods. Moreover, carbapenem MICs for the E. coli transformants harboring this blaTEM-1 gene were not affected (Table 2).
Chromosomal DNA of A. baumannii strain RYC 52763/97 was independently digested with HindIII and BglII as previously described (7, 8). The resulting fragments were ligated into the pBGS18− plasmid digested with HindIII and BglII, respectively, and transformants were selected on kanamycin (10 μg/ml) and ampicillin (25 μg/ml) plates. When chromosomal DNA was digested with HindIII and transformants were selected, an insert of 2.2 kb was obtained. Afterwards, the nucleotide sequencing revealed the presence of a bla gene, which showed a close homology with different chromosomal and plasmid-mediated AmpC β-lactamases and likely corresponds to the A. baumannii AmpC β-lactamase that has been previously reported (7). Carbapenem MICs for E. coli TG1 transformants harboring A. baumannii AmpC β-lactamase were identical to that for the E. coli host strain (Table 2).
When chromosomal DNA was digested with BglII, an insert of 4.2 kb harboring another bla gene was cloned. The bla gene of this plasmid was subcloned by enzymatic digestion with XbaI, yielding the plasmid pBMB-1 with an insert of 1.5 kb. After nucleotide sequencing, the amino acid sequence of this β-lactamase (OXA-24) showed a close homology with OXA-10 and OXA-7 β-lactamases (40% identity). The molecular characterization of this OXA-24 β-lactamase has recently been reported (8). Interestingly, the MICs of imipenem (1 μg/ml) and meropenem (0.125 μg/ml) were moderately increased for the E. coli transformants harboring the pBMB-1 plasmid with the OXA-24 enzyme (Table 2) with respect to that for the E. coli TG1 host strain.
Kinetic experiments and carbapenem inactivation.
Imipenem and meropenem hydrolysis was detected spectrophotometrically with a sonicated extract of the strain A. baumannii RYC 52763/97. In addition, a positive Masuda test with imipenem was obtained (data not shown). This result strongly suggested the presence of a carbapenem-hydrolyzing enzyme in the epidemic IMRAB strain. Moreover, the addition of EDTA at different concentrations did not affect the imipenem hydrolysis, thus suggesting the absence of a class B β-lactamase.
Biochemical experiments performed with the semipurified OXA-24 enzyme showed imipenem and meropenem hydrolysis (Table 3). In addition, a positive Masuda test was also performed with imipenem and the semipurified OXA-24 β-lactamase (data not shown). These results correlated well with the increase in the carbapenem MICs observed with the E. coli TG1 strain harboring the OXA-24 enzyme (plasmid pBMB-1 in Table 2).
TABLE 3.
Biochemical parameters of the OXA-24 enzyme
| Antibiotic | Km (μM) | Vmax (μmol/min/μl) | Relative Vmaxa |
|---|---|---|---|
| Benzylpenicillin | 65 | 370 | 100 |
| Cephaloridine | 395 | 200 | 54 |
| Imipenem | 20 | 15 | 4 |
| Meropenem | 775 | 280 | 75 |
Obtained by normalizing the value for each antibiotic to that for benzylpenicillin (set to 100).
OMP analysis.
OMP analysis of the A. baumannii RYC 52763/97 strain and one IMSAB isolate (number 34 in Table 1) showed a reduction in the expression of two porins at 22 and 33 kDa in the IMRAB strain (Fig. 3A). In the same figure, the densitometry pattern of the gel is shown (Fig. 3B).
FIG. 3.
(A) OMP profile of the IMSAB strain 34 (Table 1) (lane 1) and IMRAB strain RYC 52763/97 (lane 2). SDS—12.5% polyacrylamide gel electrophoresis and silver staining. Arrows, the different expression of porins at 22 and 33 kDa. (B) Densitometric analysis of the gel shown in panel A. 1, IMSAB protein pattern; 2, IMRAB protein pattern. Arrows indicate the different expression of porins at 22 and 33 kDa.
Efflux mechanism.
No differences in the MICs for the A. baumannii RYC 52763/97 strains was observed when reserpine was added, thus suggesting that a putative efflux mechanism was not present in this strain.
Detection of the OXA-24 gene in the epidemic IMRAB strains but not in carbapenem-susceptible A. baumannii strains.
Six epidemic IMRAB strains (strains 1, 3, 6, 24, 25, and 28 in Table 1) and 10 genetically unrelated A. baumannii strains (strains 31 to 40 in Table 1) with different levels of susceptibility to carbapenems (imipenem MIC range, 0.1 to 4 mg/liter) were used to investigate the presence of the OXA-24 gene by using specific OXA-24 primers. A positive amplification band was observed in all carbapenem-resistant epidemic IMRAB strains, while no amplification was observed in all preoutbreak, at-outbreak, and postoutbreak IMSAB strains (Fig. 4). These results suggest that the OXA-24 β-lactamase may play a role in the resistance of carbapenems in the IMRAB strains in combination with other resistance mechanisms. Moreover, the OXA-24 gene can be used as a marker of the epidemic outbreak strain.
FIG. 4.
Absence of the OXA-24 gene in different carbapenem-susceptible IMSAB strains by PCR amplification with P1 and P2 OXA primers. Lanes 1 and 11, molecular weight marker λIII (Boehringer); lane 2, A. baumannii RYC 52763/97 as the positive control; lanes 3 and 4, two epidemic genetically related IMRAB strains; lanes 5 through 10, different genetically unrelated carbapenem-susceptible IMSAB strains.
DISCUSSION
In the last 2 decades, a significant number of Acinetobacter nosocomial infection outbreaks, caused mainly by A. baumannii strains, have been reported, causing increasing concern in hospitals (5, 6). In order to investigate the origin of infection, the route of spread, and the prevalence of isolates in a bacterial population, several phenotypic and molecular typing methods have been described. Although antibiotyping may alert us to the emergence of a multiresistant A. baumannii outbreak, distinguishing between strains with slight differences in their resistance profiles may be difficult. Therefore, genotypic methods including plasmid typing, ribotyping, PFGE of chromosomal DNA restriction fragments, and PCR fingerprinting have been used to investigate nosocomial A. baumannii outbreaks (13, 17, 28, 33, 34, 38, 39; Bou et al., 38th ICAAC). We report here the molecular typing of a nosocomial outbreak caused by a multiresistant A. baumannii strain. By using PFGE and REP-PCR, we demonstrated the spread of one epidemic strain between 29 patients during a period of 10-months. On the other hand, seven different genotypes (B through H) were observed in the preoutbreak, at-outbreak, and postoutbreak A. baumannii strains included in this study.
Regarding the resistance to carbapenems in A. baumannii, different mechanisms have been involved. PBP with reduced affinity and loss of porins, besides several class D and class B β-lactamases, have been associated with resistance to carbapenems in A. baumannii clinical strains (2, 4, 7, 8, 9, 11, 12, 15, 18, 27, 29, 32). In addition, we describe here the molecular mechanism associated with resistance to the carbapenems in an epidemic A. baumannii strain. Three different β-lactamases have been characterized in the epidemic IMRAB strain, a TEM-1-type plasmid-mediated enzyme, the A. baumannii cephalosporinase AmpC-type enzyme, and a novel OXA-derived chromosomally mediated enzyme (7, 8).
The three β-lactamase genes were cloned into pBGS18− plasmid, and the protein products were expressed in E. coli TG1 cells. As shown in Table 2, the MICs of the carbapenems conferred by each β-lactamase did not reach the carbapenem MICs for the original A. baumannii RYC 52763/97 strain, suggesting that other mechanisms are involved in the carbapenem resistance of A. baumannii strains, in addition to the β-lactamases. An increase in the carbapenem MICs was observed only for OXA-24 β-lactamase; in addition, E. coli extracts expressing the OXA-24 enzyme yielded a carbapenem hydrolysis that was very similar to that of the IMRAB strain (data not shown). Also, it is important to mention the imipenem and meropenem hydrolysis detected in the spectrophotometer with the semipurified OXA-24 enzyme (Vmax, 4% and 75% that of benzylpenicillin, respectively). By cloning and kinetic experiments, no other β-lactamase with carbapenem hydrolysis was detected in the A. baumannii RYC 52763/97 strain (index case), suggesting that carbapenem hydrolysis is associated only with the presence of this OXA enzyme. Moreover, protein extracts of the RYC 52763/97 strain showed imipenem hydrolysis that was not inhibited by the addition of EDTA, thus suggesting that a class B β-lactamase was not present in the epidemic IMRAB strain.
Therefore, the high levels of carbapenem resistance were not due solely to the presence of β-lactamases. This result prevents the use of β-lactamase inhibitors (MIC of sulbactam, 16 to 32 μg/ml) in the treatment of the infections caused by IMRAB strains and leaves as a unique alternative the use of colistin and polymyxin B, nephrotoxic and neurotoxic drugs used at the beginning of the 1950s (35).
In general, the emergence of carbapenem-hydrolyzing enzymes in A. baumannii has been limited compared to the prevalence of other β-lactamases. Recently the production of carbapenem-hydrolyzing enzymes in different A. baumannii strains has been reported (12, 15, 18, 29). Several of these β-lactamases showed characteristics of metalloenzymes (class B β-lactamases by the classification method of Bush et al. [10]) because their enzymatic activity was inhibited in the presence of 1 mM EDTA and activated in the presence of 1 mM ZnCl2 solution. In contrast, we report here the presence of an OXA β-lactamase (class D β-lactamases by the same classification system) with carbapenem-hydrolyzing activity in an epidemic outbreak strain. The fact that this OXA enzyme is chromosomally mediated makes the spread of the OXA gene to other microorganisms or A. baumannii strains difficult. Thus, the PCR assay using the OXA primers with different IMSAB strains isolated before, during, and after outbreak did not show a positive band in either of the A. baumannii strains. In addition, a transfer of the imipenem resistance was not detected in filter mating experiments by using A. junii and E. coli BM21 as the recipient cells.
On the other hand, three at-outbreak IMSAB isolates (34 to 36 [Table 1]) obtained from different patients at different wards displayed a different genotype (PFGE group D) than that of IMRAB isolates. Following the criteria of Tenover et al. (40), these isolates are possibly part of the outbreak, as revealed by the band pattern (less than six different bands by PFGE when compared with the IMRAB pattern). In addition, a REP-PCR assay performed with the same strains (Fig. 2) yielded a very similar band pattern to that of the IMRAB strains, thus suggesting a genetic relationship. The first at-outbreak IMSAB strain (isolate 34 [Table 1]) was isolated in a surgical ICU when the outbreak started. Supporting this view, the results obtained with the OMP analysis between the IMRAB strain and this strain 34 showed that a single loss of porins might be involved in addition to other mechanisms in the resistance to carbapenems (Fig. 3); moreover, by PCR assay, the OXA-24 gene was not detected in the IMSAB 34 to 36 strains. These results strongly suggest that both carbapenem hydrolysis by the OXA-enzyme and loss of OMPs are involved in the carbapenem resistance of the epidemic IMRAB strain. It is also important to note that reserpine experiments failed to detect any efflux mechanism in the IMRAB strains. The data obtained between the 34 to 36 isolates and the IMRAB strains may suggest an evolutionary relationship between these strains. Also, it is important to emphasize that the amount of carbapenem consumed remained practically unchanged during the months previous to the outbreak in our hospital.
In summary, we report here the characterization of a nosocomial outbreak caused by an A. baumannii multiresistant strain harboring a novel class D enzyme (OXA-24) with carbapenem-hydrolyzing activity besides a reduction in the expression of two outer membrane proteins at 22 and 33 kDa. A correct antibiotic policy should be addressed at the hospitals to avoid the dissemination of this class of strains.
ACKNOWLEDGMENTS
We thank Antonio Oliver for his help in kinetic experiments and Dolores Malpica for technical assistance.
REFERENCES
- 1.Afzal M S, Livermore D. Worldwide emergence of carbapemem-resistant Acinetobacter spp. J Antimicrob Chemother. 1998;41:576–577. doi: 10.1093/jac/41.5.576. [DOI] [PubMed] [Google Scholar]
- 2.Amyes S G B, Young H-K. Mechanism of antibiotic resistance in Acinetobacter spp.—genetics of resistance. In: Bergogne-Berezin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 185–223. [Google Scholar]
- 3.Bakken J S, Sanders C C, Clark R B, Hori M. β-Lactam resistance in Aeromonas spp. caused by inducible beta-lactamases active against penicillins, cephalosporins, and carbapenems. Antimicrob Agents Chemother. 1988;32:1314–1319. doi: 10.1128/aac.32.9.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin E. Resistance of Acinetobacter spp. to antimicrobials—overview of clinical resistance patterns and therapeutic problems. In: Bergogne-Berezin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 133–183. [Google Scholar]
- 5.Bergogne-Bérézin E, Joly-Guillou M L, Vieu J F. Epidemiology of nosocomial infections due to Acinetobacter calcoaceticus. J Hosp Infect. 1987;10:105–113. doi: 10.1016/0195-6701(87)90135-6. [DOI] [PubMed] [Google Scholar]
- 6.Bergogne-Bérézin E, Towner K J. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bou G, Martinez-Beltran J. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2000;44:428–432. doi: 10.1128/aac.44.2.428-432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bou G, Oliver A, Martinez-Beltran J. OXA-24, a novel class D β-lactamase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000;44:1556–1561. doi: 10.1128/aac.44.6.1556-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown S, Bantar C, Young H K, Amyes S G B. Limitation of Acinetobacter baumannii treatment by plasmid-mediated carbapenemase ARI-2. Lancet. 1998;351:186–187. doi: 10.1016/S0140-6736(05)78210-6. [DOI] [PubMed] [Google Scholar]
- 10.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark R B. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33–36 kDa outer membrane protein. J Antimicrob Chemother. 1996;38:245–251. doi: 10.1093/jac/38.2.245. [DOI] [PubMed] [Google Scholar]
- 12.Da Silva G J, Leitao G J, Peixe L. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J Clin Microbiol. 1999;37:2109–2110. doi: 10.1128/jcm.37.6.2109-2110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkshoom L, Aucken H M, Gerner-Smidt P, et al. Correlation of typing method for Acinetobacter isolates for hospital outbreaks. J Clin Microbiol. 1993;31:702–705. doi: 10.1128/jcm.31.3.702-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkshoorn L. Acinetobacter—microbiology. In: Bergogne-Bérézin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 37–69. [Google Scholar]
- 15.Donald H M, Scaife W, Amyes S G B, Young H K. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–199. doi: 10.1128/aac.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouby A, Neuwirth C, Bourg G, Bouziges N, Carles-Nurit M J, Despaux E, Ramuz M. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended spectrum beta-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994;32:301–305. doi: 10.1128/jcm.32.2.301-305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graser Y, Klare I, Halle E, et al. Epidemiological study of an Acinetobacter baumannii outbreak by using polymerase chain reaction fingerprinting. J Clin Microbiol. 1993;31:2417–2420. doi: 10.1128/jcm.31.9.2417-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornstein M, Sautjeau-Rostoker C, Peduzzi J, Vessieres A, Hong L T, Barthelemy M, Scavizzi M, Labia R. Oxacillin-hydrolyzing β-lactamase involved in resistance to imipenem in Acinetobacter baumannii. FEMS Microbiol Lett. 1997;153:333–339. doi: 10.1111/j.1574-6968.1997.tb12593.x. [DOI] [PubMed] [Google Scholar]
- 19.Joly-Guillou M L, Bergogne-Bérézin E, Philippon A. Distribution of β-lactamases and phenotypic analysis in clinical strains of Acinetobacter calcoaceticus. J Antimicrob Chemother. 1988;22:597–604. doi: 10.1093/jac/22.5.597. [DOI] [PubMed] [Google Scholar]
- 20.Joly-Guillou M L, Brun-Buisson C. Epidemiology of Acinetobacter spp.: surveillance and management of outbreaks. In: Bergogne-Bérézin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 71–100. [Google Scholar]
- 21.Kämpfer P, Tjernberg I, Ursing J. Numerical classification and identification of Acinetobacter genomic species. J Appl Bacteriol. 1993;75:259–268. doi: 10.1111/j.1365-2672.1993.tb02775.x. [DOI] [PubMed] [Google Scholar]
- 22.Levi I, Rubinstein E. Acinetobacter infections—overview of clinical features. In: Bergogne-Bérézin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 101–115. [Google Scholar]
- 23.Martínez-Beltrán J, Alvarez M, Sierra M P. Acinetobacter spp.: patrón de sensibilidad y resistencia a los antimicrobianos In Acinetobacter baumannii en pacientes críticos. Merck Sharp & Dohme, Madrid, Spain. 1998. pp. 21–43. [Google Scholar]
- 24.Masuda G, Tomioka S, Hasegawa M. Detection of β-lactamase production by gram-negative bacteria. J Antibiot (Tokyo) 1976;29:662–664. doi: 10.7164/antibiotics.29.662. [DOI] [PubMed] [Google Scholar]
- 25.Matthew M, Harris A M, Marshall M, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 27.Paton R, Miles R S, Hood J, Amyes S G B. ARI-1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2:81–88. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 28.Reboli A C, Houston E D, Monteforte J S, Wood C A, Hamill R J. Discrimination of epidemic and sporadic isolates of Acinetobacter baumannii by repetitive element PCR-mediated DNA fingerprinting. J Clin Microbiol. 1994;32:2635–2640. doi: 10.1128/jcm.32.11.2635-2640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riccio M L, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, Amicosante G, Rossolini G M. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existance of blaIMP allelic variants carried by gene casettes of different phylogeny. Antimicrob Agents Chemother. 2000;44:1229–1235. doi: 10.1128/aac.44.5.1229-1235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato K, Nakae T. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J Antimicrob Chemother. 1991;28:33–45. doi: 10.1093/jac/28.1.35. [DOI] [PubMed] [Google Scholar]
- 33.Seifert H, Gerner-Smidt P. Comparison of ribotyping and pulsed-field gel electrophoresis for molecular typing of Acinetobacter isolates. J Clin Microbiol. 1995;33:1402–1407. doi: 10.1128/jcm.33.5.1402-1407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seifert H, Schulze A, Baginski R, Pulverer G. Comparison of four different methods for epidemiologic typing of Acinetobacter baumannii. J Clin Microbiol. 1994;32:1816–1819. doi: 10.1128/jcm.32.7.1816-1819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snavely S R, Hodges G R. The neurotoxicity of antibacterial agents. Ann Intern Med. 1984;101:92–104. doi: 10.7326/0003-4819-101-1-92. [DOI] [PubMed] [Google Scholar]
- 36.Spratt B G, Hedge P J, Heesen T S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 37.Stone J W, Das B C. Investigation of an outbreak of infection with Acinetobacter calcoaceticus in a special baby care unit. J Hosp Infect. 1985;6:42–48. doi: 10.1016/0195-6701(86)90025-3. [DOI] [PubMed] [Google Scholar]
- 38.Struelens M J, Carlier E, Maes N, Serruys E, Quint W G. Nosocomial colonization and infection with multiresistant Acinetobacter baumannii: outbreak delineation using DNA macrorestriction analysis and PCR fingerprinting. J Hosp Infect. 1993;25:15–32. doi: 10.1016/0195-6701(93)90005-k. [DOI] [PubMed] [Google Scholar]
- 39.Tankovic J, Legrand P, De Gatines G, Chemineau V, Brun-Buisson C, Duval J. Characterization of a hospital outbreak of imipenem-resistant Acinetobacter baumannii by phenotypic and genotypic typing methods. J Clin Microbiol. 1994;32:2677–2681. doi: 10.1128/jcm.32.11.2677-2681.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover F C, Arbeit R, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vila J, Marcos M A, Jiménez de Anta M T. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J Med Microbiol. 1996;44:482–489. doi: 10.1099/00222615-44-6-482. [DOI] [PubMed] [Google Scholar]
- 42.West S E, Schweizer H P, Dall C, Sample A K, Runyen Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]