Abstract
A quantitative competitive reverse transcription-PCR assay was developed to quantify dengue virus RNA in this study. The main features include a primer pair targeting a highly conserved region in the capsid and the addition of competing RNA that contains an internal deletion to provide a stringent internal control for quantification. It can be utilized to quantify RNA isolated from the four dengue virus serotypes but not RNA isolated from other flaviviruses, including Japanese encephalitis virus and hepatitis C virus, both prevalent in Asia. It can also be used to quantify dengue virus RNA isolated from the plasma of infected individuals. The sensitivity of the assay was estimated to be 10 to 50 copies of RNA per reaction, and twofold differences in virus titer are distinguishable. This assay is a convenient, sensitive, and accurate method for quantification and can be used to further understanding of the pathogenesis of dengue virus infection.
Among the 70 or so arthropod-born flaviviruses, epidemics of the four dengue viruses (DEN-1, DEN-2, DEN-3, and DEN-4) continue to be a major public health problem in tropical and subtropical regions (1, 8, 11, 21). It has been estimated that about 100 million cases of dengue fever (DF) and 250,000 cases of dengue hemorrhagic fever occur annually worldwide (8, 21).
The clinical presentations of the four dengue virus serotypes are similar. They range from mild, self-limited DF to severe and potentially life-threatening dengue hemorrhagic fever-dengue shock syndrome (8, 11, 30). Following an incubation period, there are fever and a variety of symptoms, which concur with the appearance of dengue virus in blood (8). Detection and quantification of dengue virus in plasma are not only crucial for rapid diagnosis but can also add to our understanding of the pathogenesis of dengue (8, 11). Several techniques have been developed to detect dengue viremia (2, 4, 8, 9, 14, 26, 29). The conventional method is virus isolation, although it is time-consuming and has variable isolation rates (4, 8, 29). The reverse transcription (RT)-PCR assays are more rapid and sensitive, but many of them require separate procedures for RT and PCR (4, 8, 26, 29). The recently reported TaqMan amplification system employs the one-step RT-PCR protocol and is a sensitive method for quantification (16).
Quantification based on the standard RT-PCR method, however, has certain limitations (5, 22, 25). First, the amount of product does not consistently reflect the amount of the initial target. This is probably due to differential efficiencies and kinetics of PCR and/or RT depending on the abundance of the target and the presence of various inhibitors, particularly in clinical samples (5, 22). Second, comparison of the amount amplified from the specimen to that from a titrated standard in separate reactions is not ideal for quantification. Third, normalization by coamplifying a heterologous target (such as actin or β-globulin) cannot provide a good internal control because of differences in abundance and the priming efficiency of the heterologous target (5, 22).
In the quantitative competitive RT-PCR (QC-RT-PCR) assay, increasing amounts of competing RNA that is largely identical, but somewhat distinguishable (containing an internal deletion) from the target sequence, are added to replicate tubes that contain the same amounts of the target sequence (5, 22). The competing RNA and target RNA are in the same tube and provide a stringent internal control. After electrophoresis, the different-size RT-PCR products are quantified. The amounts of the target sequence are determined based on the relative, not the absolute, amounts of the products by either direct or interpolated assessment of the equivalent point (5, 22). In this report, we describe a sensitive and convenient QC-RT-PCR assay that can quantify dengue virus RNA of all four serotypes.
MATERIALS AND METHODS
Generation of the construct and competitor RNA.
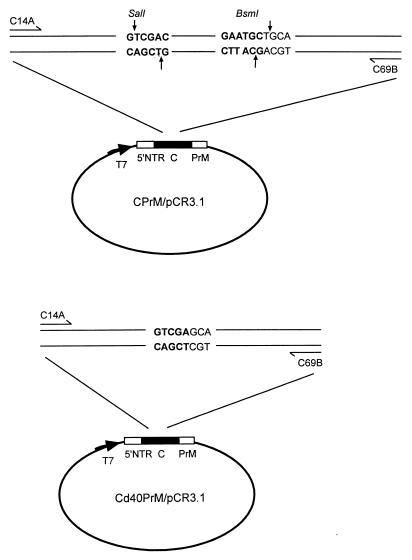
CPrM/pCR3.1 is a plasmid containing the entire capsid (C) region and the N-terminal 54 amino acids of the precursor membrane (PrM) region of DEN-2 strain PL046 in the vector pCR3.1 (Invitrogen, San Diego, Calif.) (17) (Fig. 1). The competitor construct Cd40PrM/pCR3.1 was modified from CPrM/pCR3.1 by digestion with two restriction enzymes, SalI and BsmI, in the C region, followed by filling of the 3′ recessed end (from the SalI cut) and removal of the 5′ protruding end (from the BsmI cut) with T4 DNA polymerase (Fig. 1). Compared to CPrM/pCR3.1, Cd40PrM/pCR3.1 had a 40-bp internal deletion in the C region, as was verified by DNA sequencing. Competitor RNA, Cd40PrM, which was generated from in vitro transcription (Promega, Madison, Wis.) of linearized Cd40PrM/pCR3.1, was purified by phenol-chloroform extraction and quantified by spectrophotometer. The copy number of Cd40PrM was calculated based on the concentration measured and its molecular weight. Wild-type RNA, CPrM, was similarly generated from linearized CPrM/pCR3.1 and quantified by spectrophotometer. The copy number of CPrM was thus calculated, and known amounts of CPrM were used as target RNA in the QC-RT-PCR assay.
FIG. 1.
Schematic diagram of the construct containing wild-type dengue virus CPrM sequences, CPrM/pCR3.1 (top) and the competitor construct containing an internal deletion of CPrM, Cd40PrM/pCR3.1 (bottom). The relative positions of the primers used for the QC-RT-PCR assay, C14A and C69B, are shown. 5′ NTR, 5′ nontranslated region.
Isolation of viral RNA.
Dengue virus RNA was isolated from aliquots of culture supernatants or plasma using the QIAamp viral RNA mini kit (Qiagen). Plasma samples were obtained within 24 h of the onset of fever from two confirmed DF patients during a DEN-3 outbreak in southern Taiwan in 1998. Plasma samples were also obtained from two hepatitis C virus (HCV) carriers and two healthy individuals. Stock viruses of the four dengue virus serotypes, the Hawaii (DEN-1), New Guinea (DEN-2), H-87 (DEN-3), and H-241 (DEN-4) strains, had titers of 2.1 × 106, 1.3 × 106, 2 × 106, and 1 × 106 PFU/ml, respectively. They were obtained from culture supernatants of infection of mosquito C6/36 cells and then titrated on BHK-21 cells by standard plaque-forming assay. Three Japanese encephalitis virus (JEV) strains (the Nakayama vaccine strain, the Beijing vaccine strain, and the CH1949 Taiwan local strain isolated from Changhwa County) with titers of 105 to 106 PFU/ml were also used for comparison, since JEV is another prevalent flavivirus circulating in Taiwan. Stock virus dilutions of 1- to 100-fold were used to isolate viral RNA for RT-PCR detection. For QC-RT-PCR quantification, 1- to 10,000-fold dilutions of the Hawaii strain virus and serial 2-fold dilutions thereafter were also prepared and subjected to virus RNA isolation. Briefly, a 140-μl volume of plasma or diluted stock virus was mixed with the buffer AVL-carrier RNA and loaded onto the spin column. This was followed by a wash with the buffer AW and elution with the buffer AVE to a final volume of 50 μl, as recommended by the manufacturer (Qiagen).
RT-PCR primers.
Despite the sequence variation observed among different dengue viruses, certain regions in the dengue virus genome are more conserved than other regions (12, 14, 20). Through an analysis of all of the dengue virus sequences available in the GenBank database, a region in the C that is highly conserved in the four dengue viruses, but not in other flaviviruses, was identified. A primer pair covering this region, C14A and C69B, was thus designed for the QC-RT-PCR assay. The sequences of C14A and C69B are 5′-AATATGCTGAAACGCGAGAGAAACCGCG-3′ (corresponding to genome positions 136 to 163 of the DEN-2 Jamaica strain) and 5′-CCCATCTCITCAIIATCCCTGCTGTTGG-3′ (corresponding to genome positions 278 to 305 of the DEN-2 Jamaica strain), respectively (3, 14). They were designed to amplify a 170-bp product in the C region for dengue virus RNA or a 130-bp product in the same region for competitor RNA Cd40PrM.
QC-RT-PCR.
For adequate quantification, eluates containing RNA templates isolated from plasma were further diluted 10-fold. Equal amounts (2 μl) of the RNA eluates or diluted eluates were mixed with increasing copy numbers of competitor RNA Cd40PrM (0, 10, 50, 100, 500, 1,000, 5,000, and 10,000 copies) and subjected to RT-PCR using the Superscript one-step RT-PCR system (Gibco/BRL, Life Technologies). The RT-PCR conditions were 55°C for 40 min and 94°C for 2 min, followed by 40 cycles of 94°C for 45 s, 65°C for 45 s, and 68°C for 45 s. For the control reaction of PCR only, RNA eluate (2 μl) was subjected to PCR using super Taq DNA polymerase (HT Biotechnology, Cambridge, England) under PCR conditions identical to those used in the RT-PCR, except that the step using 55°C for 40 min was omitted.
Sample analysis and quantification.
The QC-RT-PCR products were electrophoresed through 2% agarose gel and stained with ethidium bromide. The amounts of the 170- and 130-bp products were measured under UV light using a digital gel documentation and analysis system consisting of a UV-transilluminator light box, a darkroom cabinet, and a DigiPix (Ultralum, Paramount, Calif.) digital camera. The image acquired with the digital camera was sent to the computer directly and analyzed by the 1D scan (Scanalytics, Fairfax, Va.) software. The intensity of the ethidium fluorescence associated with the DNA band is proportional to the amount of DNA. The RT-PCR product of the wild-type target RNA is 170 bp, and that of the competitor RNA, Cd40PrM, is 130 bp. Since the comparisons in the QC-RT-PCR were based on molar amounts, the fluorescence intensity of the 130-bp product was corrected by a factor of 170/130 to enable direct comparison of the corrected intensity of the 130-bp competitor (Cor. Int. 130) with the fluorescence intensity of the 170-bp target (Int. 170). The ratio of the Cor. Int. 130 to the Int. 170 (log scale) was plotted against the copy number of the competitor RNA (log scale). A regression line with the coefficient of determination, R2, was generated. The copy numbers of dengue virus RNA per reaction were determined by interpolated assessment of the equivalence point of the above curve. The 95% confidence intervals (CI) of the copy numbers were calculated using the software SPSS base 8.0 (SPSS Inc., Chicago, Ill.). Since 2 μl out of 50 μl of RNA eluates, which were derived from 140 μl of culture supernatant or plasma, was used in each reaction, the number of dengue virus RNA copies per reaction was divided by 5.6 μl (140 μl × 2 μl/50 μl) and multiplied by 1,000 to obtain the number of RNA copies per milliliter of supernatant or plasma.
Sequencing of RT-PCR products.
The 170-bp products derived from QC-RT-PCR were cloned into the TA cloning vector pCRII-TOPO using the procedures recommended by the manufacturer (Invitrogen, Carlsbad, Calif.). Plasmids containing the inserts were sequenced using the BigDye terminator cycle sequencing kit under the conditions recommended by the manufacturer (PE Applied Biosystems, Foster City, Calif.). Samples were loaded onto 4.75% polyacrylamide gel of ABI automated sequencers (Applied Biosystems ABI-373A; Perkin-Elmer).
RESULTS
Detection of the four dengue virus serotypes.
A close examination of the dengue virus sequences available in the GenBank database revealed a small region in the C that was highly conserved in isolates of all four dengue viruses but not in other flaviviruses. This region was chosen as our target sequence, and primers covering this region, C14A and C69B, were designed for the QC-RT-PCR assay. Deoxyinosines, which are known to pair with all four bases except G, were incorporated into primer C69B at three positions where variations of A, T, and C were noted to maintain comparable priming efficiency.
To examine whether the designed primer pair can detect dengue virus RNA, RNA templates derived from culture supernatants of viruses representing the four dengue virus serotypes, including Hawaii (DEN-1), New Guinea (DEN-2), H-87 (DEN-3), and H-241 (DEN-4), were subjected to RT-PCR. As shown in Fig. 2, amplified products of the expected size of 170 bp were seen in the reactions using the RNA template derived from the four dengue viruses. There was no product seen in the reactions containing no RNA or when the RT step was omitted (Fig. 2 and data not shown). RNA templates derived from other flaviviruses prevalent in Taiwan, including three JEV and two HCV, as well as from the plasma of two healthy individuals, were also subjected to the RT-PCR assay. None of these resulted in any amplified product of the expected size (data not shown). It was of note that the RNA templates of JEV and HCV can be amplified by their homologous primers (data not shown). These results were consistent with our sequence analysis of the C region and indicated that the designed primers, C14A and C69B, can detect the four dengue virus serotypes but not the other flaviviruses tested.
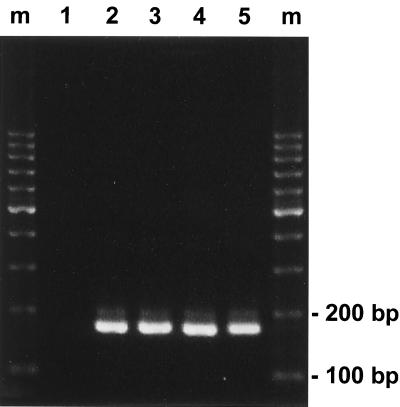
FIG. 2.
Detection of the four dengue virus serotypes using the designed primer pair. Dengue virus RNAs isolated from stock viruses of the four serotypes were subjected to RT-PCR analysis as described in Materials and Methods. Products with an expected size of 170 bp were seen in the reactions using RNA templates derived from the DEN-1 Hawaii strain (lane 2), the DEN-2 New Guinea strain (lane 3), the DEN-3 H-87 strain (lane 4), and the DEN-4 H-241 strain (lane 5) but not seen in the reaction containing no RNA (lane 1). Lanes m, molecular size marker.
Quantification of dengue virus RNA by QC-RT-PCR.
To assess the feasibility of the QC-RT-PCR assay for quantification, known amounts of in vitro-transcribed target RNA, CPrM, which contained the wild-type dengue virus sequence, and Cd40PrM competitor RNA were used in the QC-RT-PCR assay. As shown in Fig. 3, addition of increasing amounts of Cd40PrM (from 0 to 10,000 copies) to replicate reactions containing identical amounts of CPrM (300 copies) resulted in a gradual increase in the intensity of RT-PCR products of the competitor RNA (130 bp) and a gradual decrease in the Int. 170 of the target RNA. The point where the intensity of the 130-bp product derived from a known amount of competitor RNA corresponds to the Int. 170 in molar equivalence indicates the amount of target RNA present in the sample. After correction of the fluorescence intensity of the 130-bp product by a factor of 170/130, the ratios of the Cor. Int. 130 to the Int. 170 on a log scale [log (Cor. Int. 130/Int. 170)] were plotted against the copy number of competitor RNA on a log scale. A regression line was obtained. The point at which the Cor. Int. 130/Int. 170 ratio equals 1 represents the amount of target RNA present in the sample. The amount of target RNA, CPrM, was thus determined to be 302 copies (95% CI, 224 to 388 copies), which is very close to the actual amount of 300 copies added in each reaction.
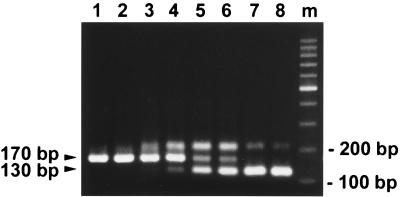
FIG. 3.
Quantification of dengue virus RNA by the QC-RT-PCR assay. An ethidium bromide-stained gel of the QC-RT-PCR assay is shown. Equal amounts (300 copies) of in vitro-transcribed target RNA, CPrM, mixed with increasing copy numbers of competitor RNA, Cd40PrM, were subjected to RT-PCR analysis as described in Materials and Methods. Lanes: 1 to 8, 0, 10, 50, 100, 500, 1,000, 5,000, and 10,000 copies of Cd40PrM, respectively; m, molecular size markers. The positions of the expected products of CPrM (170 bp) and Cd40PrM (130 bp) are indicated.
It should be noted that for reactions near the equivalent points (lanes 4 to 6, Fig. 3), there was a third band migrating more slowly than the product of the target RNA CPrM (170 bp) and the product of the competitor RNA Cd40PrM (130 bp). The slower migration pattern and the prominence of the bands seen only in reactions near the equivalent points between the target and competitor RNAs suggested that these bands were heteroduplex complexes of the two different-size products. The formation of such heteroduplexes should not affect the accuracy of our quantification, as the quantification in the QC-RT-PCR is based on the amount of the target RNA product relative to that of the competitor RNA and not the absolute amount. In addition, both the target and competitor products will, in principle, be similarly affected by formation of the heteroduplex complex (5, 22).
To demonstrate the accuracy of quantification of the QC-RT-PCR assay, different amounts of target RNA CPrM, ranging from 10 to 2,000 copies, were tested in the QC-RT-PCR assay and the number of copies determined was very close to the actual number of copies added to each reaction mixture (data not shown). From continuing optimization of the assay, its sensitivity was estimated to be 10 to 50 copies of RNA per reaction.
Quantification of dengue virus RNA in culture supernatants.
To evaluate the QC-RT-PCR assay with samples more biologically relevant than in vitro-transcribed RNA, RNA isolated from the Hawaii strain of the DEN-1 virus was subjected to the QC-RT-PCR assay. As shown in Fig. 4, a gradual decrease in the intensity of the products of the expected size (170 bp) and a gradual increase in intensity of the 130-bp products of competitor RNA were seen as the amounts of the competitor increased from 0 to 5,000 copies. The ratios of the Cor. Int. 130 to the Int. 170 on a log scale [log (Cor. Int. 130/Int. 170)] were plotted against the copy number of the competitor RNA on a log scale. Based on the regression line obtained, the amount of RNA was determined to be 205 copies (95% CI, 109 to 388 copies) per reaction, which corresponds to 36,607 copies of RNA per ml of supernatant.
FIG. 4.
Quantification of dengue virus RNA derived from culture supernatants. An ethidium bromide-stained gel of the QC-RT-PCR assay is shown. Equal amounts of RNA eluates derived from culture supernatants of a DEN-1 virus (Hawaii strain) were mixed with increasing copy numbers of competitor RNA, Cd40PrM, and subjected to RT-PCR analysis as described in Materials and Methods. Lanes: 1 to 7, 0, 10, 50, 100, 500, 1,000, and 5,000 copies of Cd40PrM, respectively; m, molecular size markers. The positions of the expected products of dengue virus RNA (170 bp) and Cd40PrM (130 bp) are indicated.
To further evaluate this QC-RT-PCR assay for the quantification of dengue virus, serial twofold dilutions of the Hawaii strain of the DEN-1 virus were subjected to viral RNA isolation and the RNA was employed in the QC-RT-PCR assay. The RNA copy number was determined for each dilution and plotted against the virus titer of each dilution. A linear relationship was found between the determined RNA copy numbers per milliliter of supernatant and the virus titer, expressed in PFU per milliliter, and a twofold difference in virus titer can be differentiated by this QC-RT-PCR assay. Taken together, these results demonstrate the feasibility and accuracy of this assay for the quantification of dengue viruses.
Quantification of plasma dengue virus RNA.
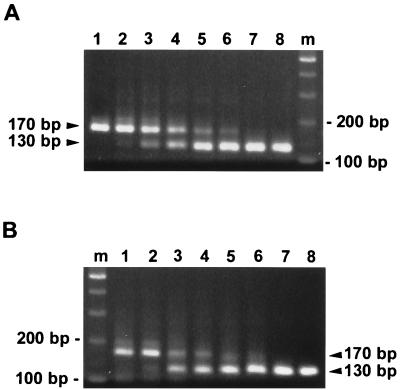
To examine whether the QC-RT-PCR assay can be utilized to quantify dengue virus RNA present in clinical specimens, plasma samples obtained from two DEN-3 virus-infected individuals were subjected to viral RNA isolation and the RNA was subjected to the QC-RT-PCR assay. As shown in Fig. 5A, a gradual decrease in the intensity of the 170-bp product of viral RNA obtained from patient 1 and a gradual increase in the intensity of the 130-bp products of competitor RNA were seen as the amount of the competitor increased from 0 to 10,000 copies. The ratios of the Cor. Int. 130 to the Int. 170 on a log scale [log (Cor. Int. 130/Int. 170)] were plotted against the copy number of competitor RNA on a log scale. A regression line was obtained with the equation y = 0.8787x − 1.9601, R2 = 0.9384. The amount of RNA was determined to be 170 copies (95% CI, 89 to 359 copies) per reaction, which corresponded to 303,570 copies of RNA per ml of plasma for patient 1. The results of the QC-RT-PCR assay for plasma virus RNA obtained from another patient, patient 2, are shown in Fig. 5B. Based on the plot generated and the regression line (y = 0.5124x − 0.8156, R2 = 0.9359), the amount of RNA was calculated to be 39 copies (95% CI, 18 to 115 copies) per reaction, corresponding to 69,640 copies of RNA per ml of plasma. The identity of the 170-bp RT-PCR products was confirmed to be dengue virus C region after cloning and sequencing of the 170-bp bands for both patients (data not shown).
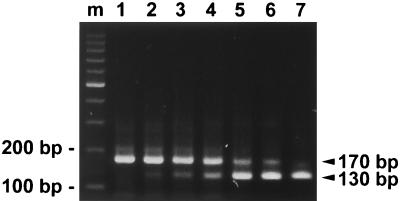
FIG. 5.
Quantification of dengue virus RNA in plasma. (A and B) Ethidium bromide-stained gels of the QC-RT-PCR assay for two DEN-3 virus-infected individuals, patients 1 (A) and 2 (B). Equal amounts of RNA eluates derived from plasma were mixed with increasing copy numbers of competitor RNA, Cd40PrM, and subjected to RT-PCR analysis as described in Materials and Methods. Lanes: 1 to 8, 0, 10, 50, 100, 500, 1,000, 5,000, and 10,000 copies of Cd40PrM, respectively; m, molecular size markers. The positions of the expected products of dengue virus RNA (170 bp) and Cd40PrM (130 bp) are indicated.
DISCUSSION
In this study, we developed a sensitive QC-RT-PCR assay that can quantify dengue virus RNA derived from culture supernatants and plasma samples of dengue virus-infected individuals. Since outbreaks of dengue frequently involve more than one serotype (8, 11, 18), a quantitative method that can detect all four dengue virus serotypes would be ideal and practical. Based on our sequence analysis, a primer pair (C14A and C69B) targeting a region in the capsid highly conserved by dengue viruses but not by other flaviviruses was designed in this study. Among the 38 dengue virus C sequences available in the GenBank database only 2 had two mismatches in the C14A primer region and another 2 had three mismatches in the C69B primer region. The mismatches were located primarily at the 5′ half of the primers and are therefore less likely to affect the RT-PCR significantly. Whether strain variability of all field isolates would affect the performance of the assay remains to be examined.
C14A overlaps a previously reported consensus primer, D1, which, in combination with another primer in the CPrM region, has been shown to be dengue virus specific (14). While determination of the specificity of our primer pair for dengue virus requires further testing with many different flaviviruses, our findings that they can detect the four dengue virus serotypes but not the other flaviviruses tested were consistent with our original primer design. The impact and magnitude of viral loads in concurrent infections with the multiple dengue virus serotypes that have been documented in both mosquitoes and humans could therefore be addressed using the QC-RT-PCR assay reported here (7, 13, 18).
Using in vitro-transcribed RNA as a template, this assay can reliably detect 10 to 2,000 copies of dengue virus RNA per reaction. The sensitivity of the assay was estimated to be 10 to 50 copies per reaction. This assay can be utilized for accurate quantification of dengue virus RNA derived from stock viruses in the laboratory. The Hawaii strain of DEN-1 virus was examined, and the results are shown here. The RNA copy numbers thus determined were higher than the titers of the virus, with a ratio of around 170. The QC-RT-PCR assay has also been employed in the quantification of DEN-2, DEN-3, and DEN-4 stock viruses, and the ratios were found to be in the same range (data not shown). These findings were in agreement with a recent publication by Houng et al. in which each infectious PFU of dengue virus was found to represent at least 100 genomic equivalents (10). Several factors may account for the discrepancy, one possibility being the presence of genetically defective viruses due to the error-prone nature of viral RNA polymerase (20). The sensitivity of flavivirus envelope to changes in the pH of the medium and the instability or deterioration of other viral components may also contribute to it (20). Higher ratios of genome copy numbers to infectious units have been observed in other viruses, such as human immunodeficiency virus type 1, where the ratios were found to range from 104 to 107 (23). This assay can also be utilized to quantify dengue virus RNA isolated from the plasma of infected individuals. In this case, plasma samples from two DEN-3-infected individuals collected during an outbreak in southern Taiwan in 1998 were tested. The applicability of this assay to the quantification of a larger number of samples awaits further evaluation.
Compared with RT-PCR-based quantitative methods such as the TaqMan amplification system, the most important feature of this assay is the competitive nature of the procedures, which provide stringent internal control. This is particularly critical for clinical samples, in which the presence of inhibitors or other variables might affect the kinetics and efficiency of amplification. Although the TaqMan system was reported to have a wide range of detection and good sensitivity, it required four sets of primers, fluorescent dye-labeled probes, and protocols for different serotypes (16). Our QC-RT-PCR assay utilized a single primer pair and was able to distinguish between twofold differences in viral titers. These features make this assay attractive and suitable for quantitative analysis of clinical samples. Moreover, combining the RT and PCR into one step is convenient and can reduce the chance of contamination between samples.
Accurate quantification of plasma viral loads by QC-RT-PCR has been successfully utilized for other viruses, such as human immunodeficiency virus type 1 and HCV, and this method has been shown to be useful for assessing both clinical status and response to therapy (15, 19, 23, 24). Using the virus isolation method, higher dengue viremia titers were recently reported to correlate with increased disease severity (28). This is consistent with both the role of viral virulence and the presence of enhancing antibody in determining disease severity (8, 11). The relationship between dengue virus loads in plasma during the course of infection and disease severity, along with various clinical manifestations, remains to be elucidated by the more convenient QC-RT-PCR assay developed in this study. This assay can also be utilized to monitor sequential changes in viral loads in plasma and to investigate their relationship to the levels of several immune activation markers that have been reported recently (6, 8, 11, 27). Findings from such studies may provide new insights into the pathogenesis of dengue virus infection.
ACKNOWLEDGMENTS
We thank Shih-Chung Lin at the Kao General Hospital, Ben Jam-Ming Lee at the Chi-Mei Foundation Hospital, and Shin C. Chang at the College of Medicine, National Taiwan University, for kindly providing clinical samples and Dai-Yu Chao for administrative assistance. We also thank D. J. Gubler for the DEN-1 Hawaii strain and the DEN-2 New Guinea strain and the National Institute of Preventive Medicine, Department of Health, Taiwan, for the DEN-4 H-241 strain.
This work was supported in part by the National Science Council (NSC89-2320-B-002-035) and by the National Health Research Institute (NHRI-CN-CL8903P), Taiwan, Republic of China.
REFERENCES
- 1.Centers for Disease Control. Dengue surveillance—United States 1986–1992. Morbid Mortal Weekly Rep. 1994;43(SS-2):7–19. [PubMed] [Google Scholar]
- 2.Chang G-J J, Trent D W, Vorndam A V, Vergne E, Kinney R M, Mitchell C J. An integrated target sequence and signal amplification assay, reverse transcriptase–PCR–enzyme-linked-immunosorbent assay, to detect and characterize flaviviruses. J Clin Microbiol. 1994;32:477–483. doi: 10.1128/jcm.32.2.477-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deubel V, Kinney R M, Trent D W. Nucleotide sequence and deduced amino sequence of the structural proteins of dengue type 2 virus, jamaica genotype. Virology. 1986;155:365–377. doi: 10.1016/0042-6822(86)90200-x. [DOI] [PubMed] [Google Scholar]
- 4.Deubel V. The contribution of molecular techniques to the diagnosis of dengue infection. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. London, United Kingdom: CAB International; 1997. pp. 335–366. [Google Scholar]
- 5.Freeman W M, Walker S J, Vrana K E. Quantitative RT-PCR: pitfalls and potential. BioTechniques. 1999;26:112–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 6.Green S, Vaughn D W, Kalayanarooj S, Nimmannitya S, Suntayakorn S, Nisalak A, Lew R, Innis B L, Kurane I, Rothman A L, Ennis F A. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J Infect Dis. 1999;179:755–762. doi: 10.1086/314680. [DOI] [PubMed] [Google Scholar]
- 7.Gubler D J, Kuno G, Sather G E, Waterman S H. A case of natural concurrent human infection with two dengue viruses. Am J Trop Med Hyg. 1985;34:170–173. doi: 10.4269/ajtmh.1985.34.170. [DOI] [PubMed] [Google Scholar]
- 8.Gubler D J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris E, Roberts G, Smith L, Selle J, Kramer L D, Valle S, Sandoval E, Balmaseda A. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998;36:2634–2639. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houng H S H, Hritz D, Kanesa-thasan N. Quantitative detection of dengue 2 virus using fluorogenic RT-PCR based on 3′-noncoding sequence. J Virol Methods. 2000;86:1–11. doi: 10.1016/s0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- 11.Innis B L. Dengue and dengue hemorrhagic fever. In: Porterfield J S, editor. Exotic viral infections—1995. London, United Kingdom: Chapman & Hall; 1995. pp. 103–146. [Google Scholar]
- 12.Kuno G, Chang G-J J, Tsuchiya R, Karabatsos N, Cropp C B. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laille M, Deubel V, Sainte-Marie F F. Demonstration of concurrent dengue 1 and dengue 3 infection in six patients by the polymerase chain reaction. J Med Virol. 1991;34:51–54. doi: 10.1002/jmv.1890340109. [DOI] [PubMed] [Google Scholar]
- 14.Lanciotti R S, Calisher C H, Gubler D J, Chang G-J, Vorndam A V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau J Y N, Davis G L, Kniffen J, Qian K P, Urdea M S, Chan C S, Mizokami M, Neuwald P D, Wilber J C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 16.Laue T, Emmerich P, Schmitz H. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J Clin Microbiol. 1999;37:2543–2547. doi: 10.1128/jcm.37.8.2543-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y-L, Liao C-L, Chen L-K, Yeh C-T, Liu C-I, Ma S-H, Huang Y-Y, Huang Y-L, Kao C-L, King C-C. Study of dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorono-Pino M A, Cropp C B, Farfan J A, Vorndam A V, Rodriguez-Angulo E M, Rosado-Paredes E P, Flores-Flores L F, Beaty B J, Gubler D J. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am J Trop Med Hyg. 1999;61:725–730. doi: 10.4269/ajtmh.1999.61.725. [DOI] [PubMed] [Google Scholar]
- 19.Mellors J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kinsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 20.Monath T P, Heinz F X. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 961–1034. [Google Scholar]
- 21.Monath T P. Dengue, the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piatak M, Luk K C, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–79. [PubMed] [Google Scholar]
- 23.Piatak M, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 24.Shiratori Y, Kato N, Yokosuka O, Hashimoto E, Hayashi N, Nakamura A, Asada M, Kuroda H, Ohkubo H, Arakawa Y, Iwama A, Omata M. Quantitative assays for hepatitis C virus in serum as predictors of the long-term response to interferon. J Hepatol. 1997;27:437–444. doi: 10.1016/s0168-8278(97)80346-7. [DOI] [PubMed] [Google Scholar]
- 25.Souaze F, Ntodou-Thome A, Tran C Y, Rostene W, Forgez P. Quantitative RT-PCR: limits and accuracy. BioTechniques. 1996;21:280–285. doi: 10.2144/96212rr01. [DOI] [PubMed] [Google Scholar]
- 26.Sudiro T M, Ishiko H, Green S, Vaughn D W, Nisalak A, Kalayanarooj S, Rothman A L, Raengsakulrach B, Janus J, Kurane I, Ennis F A. Rapid diagnosis of dengue viremia by reverse transcriptase-polymerase chain reaction using 3′-noncoding region universal primers. Am J Trop Med Hyg. 1997;56:424–429. doi: 10.4269/ajtmh.1997.56.424. [DOI] [PubMed] [Google Scholar]
- 27.Vaughn D W, Green S, Kalayanarooj S, Innis B L, Nimmannitya S, Suntayakorn S, Rothman A L, Ennis F A, Nisalak A. Dengue in the early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- 28.Vaughn D W, Green S, Kalayanarooj S, Innis B L, Nimmannitya S, Suntayakorn S, Endy T P, Raengsakulrach B, Rothman A L, Ennis F A, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 29.Vorndam V, Kuno G. Laboratory diagnosis of dengue virus infections. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. London, United Kingdom: CAB International; 1997. pp. 313–334. [Google Scholar]
- 30.World Health Organization. Dengue hemorrhagic fever, diagnosis, treatment and control. Geneva, Switzerland: World Health Organization; 1986. [Google Scholar]