Abstract
Twenty-two vancomycin-resistant enterococcal (VRE) isolates of the VanA phenotype (21 Enterococcus faecium isolates and 1 E. faecalis isolate), representative of a large outbreak that occurred in a hospital in Gdańsk, Poland, were studied. All of the isolates demonstrated resistance to a wide variety of other antimicrobial agents in addition to glycopeptides. Several lines of evidence suggested that the outbreak most probably consisted of two epidemics that followed the independent introduction of VanA determinants into two separate hematological wards of the hospital. This hypothesis is supported by the fact that isolates recovered in these wards possessed two different polymorphs of the highly conserved DNA region encompassing the vanRSHAX genes and two distinct polymorph types of Tn1546-like transposons, which contain these genes. According to pulsed-field gel electrophoresis data, the outbreak in the adult hematology ward (HW) was highly polyclonal, which suggested a major role for the horizontal transmission of Tn1546-like elements among nonrelated strains of E. faecium and E. faecalis in this environment. On the other hand, the outbreak in the pediatric hematology ward (PHW) was most probably due to the clonal spread of two epidemic E. faecium strains, which had exchanged a plasmid carrying the Tn1546-like transposon. Restriction fragment length polymorphism studies of transposons and their insertion loci in plasmid DNA have suggested that numerous isolates from both HW and PHW contained two or more copies of Tn1546-like elements that underwent diversification due to various genetic modifications. The reported data demonstrated a very complex epidemiology of the first, and up to now the only, VanA VRE outbreak characterized in Poland.
Over the last decade, enterococci have emerged as very important nosocomial pathogens (19, 21), and this has been attributed, among other factors, to their broad natural and acquired resistance to antimicrobial agents, including glycopeptides (vancomycin and teicoplanin). The first vancomycin-resistant enterococci (VRE), Enterococcus faecium and E. faecalis, were identified in 1986 in France (17) and in the United Kingdom (30), and since then VRE strains have been reported in many countries worldwide (3, 5, 27). Five different phenotypes of glycopeptide resistance (VanA to VanE) have been described to date, and the highest clinical relevance has been assigned to phenotypes VanA and VanB (9, 11, 17, 18, 24). Enterococcal strains of the VanA phenotype are highly resistant to vancomycin and moderately or highly resistant to teicoplanin. This phenotype is inducible and determined by a cluster of genes, three of which, vanH, vanA, and vanX, are critical for resistance determination. Together with two regulatory genes, vanR and vanS, they form a highly conserved vanRSHAX region of active transposons of the Tn1546 type (1, 2). Tn1546-like elements are often carried by plasmids (1, 2), and various modifications in different positions of the transposons cause their remarkable polymorphism (12, 13, 23, 32). The location of genes responsible for the VanA phenotype within transposons and plasmids strongly facilitates their horizontal spread among enterococcal strains (1, 2, 12, 13, 23, 32). Numerous nosocomial VRE outbreaks, attributed either to horizontal transfer of resistance determinants or to clonal dissemination of epidemic strains, have been reported to date (6, 20, 27, 29, 34).
Here we present the results of the detailed epidemiological analysis of VRE isolates representative of the outbreak that occurred in the University Hospital in Gdańsk, Poland, from 1997 to 1999. Isolates recovered at the beginning of the epidemic were the first VRE strains described in Poland (14, 25), and to date this has been the only outbreak caused by VanA enterococci identified and characterized in Poland.
MATERIALS AND METHODS
Clinical isolates.
The first VRE isolate was cultured in the University Hospital in Gdańsk from the urinary tract infection of a patient located in the hematology ward (HW) in December 1996. From the beginning of 1997 to the end of 1999, VRE isolates were recovered from 128 infected or colonized patients who were hospitalized in the HW (112 patients), the pediatric hematology ward (PHW; 12 patients), and the intensive care unit (ICU; 4 patients) (26; A. Samet, personal communication).
Of these, 22 isolates (21 E. faecium and 1 E. faecalis isolates) were sent to the National Reference Center for Antibiotic Resistance (NRCAR) in Warsaw, Poland, and these were included in the study reported here. Selected clinical data concerning these isolates are shown in Table 1. A total of 9 isolates were recovered from patients in the HW (3 isolates in 1997 and 6 isolates in 1998 and 1999), 12 isolates were collected from patients (7 isolates) or environmental samples (5 isolates) in the PHW (all in 1998), and the remaining isolate was obtained from a patient in the ICU (in 1999). Identification of the isolates was confirmed by the NRCAR; genus identification was performed according to the method of Facklam and Collins (10), and species were identified using the API Rapid ID32 STREP test (bioMérieux, Charbonnieres-les-Bains, France) supplemented by potassium tellurite reduction, motility, and pigment production tests (10).
TABLE 1.
Clinical Enterococcus isolates: selected clinical data, typing by PFGE, MICs, and mating results
| Isolatea | Species | Ward | Date of isolation (yr.mo.day) | Sourceb | PFGE group | MIC [μg/ml]c of
|
Matingd | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pen | Amp | Gen | Str | Van | Tei | Chl | Tet | Cip | |||||||
| 1639 | faecium | HW | 97.03.21 | Blood | A | >128 | >128 | 16 | 2,048 | 512 | 32 | 4 | 0.25 | 128 | + |
| 1640 | faecium | HW | 97.05.02 | Blood | B | 128 | 32 | >1,024 | >2,048 | >512 | 64 | 16 | 64 | 128 | + |
| 1641 | faecium | HW | 97.07.08 | Stool | C | >128 | >128 | >1,024 | >2,048 | 256 | 32 | 4 | 64 | 128 | − |
| 3131 | faecium | PHW | 98.03.25 | Urethra | D1 | >128 | 64 | 32 | 2,048 | 512 | 32 | 4 | 0.5 | 32 | + |
| 3132 | faecium | PHW | 98.03.26 | Stool | D2 | 128 | 64 | 32 | 2,048 | 512 | 32 | 4 | 0.5 | 32 | + |
| 3133 | faecium | PHW | 98.03.28 | Stool | D1 | 128 | 64 | 32 | 2,048 | 512 | 32 | 4 | 1 | 32 | + |
| 3134 | faecium | PHW | 98.03.28 | Stool | D3 | 128 | 64 | 64 | >2,048 | 512 | 32 | 4 | 1 | 64 | + |
| 3135 | faecium | PHW | 98.03.28 | Stool | E1 | >128 | 128 | 512 | 2,048 | >512 | 128 | 4 | 64 | 64 | + |
| 3136 | faecium | PHW | 98.03.29 | Stool | E1 | >128 | 128 | 1,024 | 64 | >512 | 128 | 4 | 64 | 64 | + |
| 3137 | faecium | PHW | 98.03.29 | Stool | D4 | >128 | 128 | 64 | >2,048 | >512 | 128 | 4 | 1 | 64 | + |
| 3138 | faecium | PHW | 98.03.28 | Ward environ. | E2 | >128 | 128 | >1,024 | 128 | >512 | 128 | 16 | 128 | 128 | + |
| 3139 | faecium | PHW | 98.03.28 | Ward environ. | E1 | >128 | 128 | 512 | 64 | >512 | 128 | 4 | 64 | 64 | + |
| 3140 | faecium | PHW | 98.03.28 | Ward environ. | E3 | >128 | 128 | 512 | 64 | >512 | 128 | 4 | 64 | 64 | + |
| 3141 | faecium | PHW | 98.03.28 | Ward environ. | E4 | >128 | >128 | 512 | 64 | >512 | 128 | 4 | 64 | 64 | + |
| 3142 | faecium | PHW | 98.03.28 | Ward environ. | D5 | >128 | 64 | 64 | 2,048 | >512 | 128 | 4 | 0.25 | 64 | + |
| 7946 | faecalis | HW | 98.09.22 | Surgical wound | I | 4 | 2 | >1,024 | >2,048 | >512 | 128 | 8 | 32 | 128 | − |
| 7947 | faecium | HW | 99.02.26 | Surgical wound | F1 | >128 | >128 | >1,024 | >2,048 | 512 | 128 | 8 | 32 | 128 | + |
| 7948 | faecium | HW | 99.03.01 | Catheter | G | >128 | 128 | 32 | >2,048 | 512 | 256 | 16 | 64 | 32 | + |
| 7949 | faecium | ICU | 99.03.08 | Blood | F2 | >128 | >128 | >1,024 | >2,048 | 512 | 128 | 8 | 32 | 128 | + |
| 7950 | faecium | HW | 99.03.09 | Surgical wound | G | 128 | 64 | 32 | >2,048 | 512 | 256 | 16 | 64 | 64 | + |
| 7952 | faecium | HW | 99.05.18 | Urine | H | >128 | 128 | >1,024 | 32 | 512 | 256 | 8 | 128 | 64 | + |
| 7953 | faecium | HW | 99.05.19 | Stool | F1 | >128 | >128 | >1,024 | >2,048 | 512 | 256 | 8 | 32 | 256 | − |
Isolates 3131 and 3133 and isolates 7948 and 7950 were recovered from two single patients.
Ward environ., environmental samples collected in the ward.
Pen, penicillin; Amp, ampicillin; Gen, gentamicin; Str, streptomycin; Van, vancomycin; Tei, teicoplanin; Chl, chloramphenicol; Tet, tetracycline; Cip, ciprofloxacin.
Isolates that produced (+) and did not produce (−) transconjugants are indicated.
Results of the preliminary analysis of three E. faecium isolates, 1639, 1640, and 1641, identified in the HW in 1997 (susceptibility testing, pulsed-field gel electrophoresis [PFGE] typing, VanA phenotype identification) were published separately (14).
Antimicrobial susceptibility testing.
MICs of different antimicrobial agents were evaluated by the agar dilution method according to NCCLS guidelines (22). The following agents were tested: penicillin, ampicillin, gentamicin, streptomycin, tetracycline, and chloramphenicol (Polfa Tarchomin, Warsaw, Poland); vancomycin (Eli Lilly, Indianapolis, Ind.); teicoplanin (Marion Merrell, Denham, United Kingdom); and ciprofloxacin (Bayer, Wuppertal, Germany). E. faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and E. faecium BM4147 VanA standard strain (1) were used as reference strains.
Resistance transfer (mating).
The vancomycin resistance transfer experiment was carried out according to the filter-mating procedure described by Klare et al. (16). E. faecium 64/3 resistant to rifampin and fusidic acid (31) was used as a recipient strain. Transconjugants were selected on the brain heart infusion agar (Oxoid, Basingstoke, United Kingdom) containing rifampin (64 μg/ml; Polfa Tarchomin) or fusidic acid (64 μg/ml; Leo Pharmaceutical Products, Ballerup, Denmark) and vancomycin (32 μg/ml).
PFGE typing.
Total DNA preparations embedded in 0.75% agarose plugs (InCert Agarose; FMC Bioproducts, Rockland, Maine) were digested with SmaI restriction enzyme (Takara, Otsu, Japan) and separated in a 1% agarose gel (Pulsed Field-Certified; Bio-Rad, Hercules, Calif.) using a CHEF DRII system (Bio-Rad). DNA purification and digestion were performed according to the method of Clark et al. (5), and the electrophoresis was run under conditions described by de Lencastre et al. (7). PFGE results were interpreted in accordance with the criteria proposed by Tenover et al. (28).
Detection of the vanA gene.
Total DNA was purified from the isolates using the Genomic DNA Prep Plus kit (A&A Biotechnology, Gdańsk, Poland). The vanA gene was detected by specific PCR with two different pairs of primers (5, 8) in a GeneAmp 2400 thermocycler (Perkin-Elmer, Norwalk, Conn.). DNA isolated from the E. faecium BM4147 VanA standard strain (1) was used as a control.
Amplification of Tn1546-like transposons and vanRSHAX regions by L-PCR.
Tn1546-like elements were amplified using the Expand Long Template PCR System (Boehringer-Mannheim, Mannheim, Germany), and total DNA preparations of the isolates as templates. A single oligonucleotide, primer 1 (23), complementary to the transposon-flanking inverted repeats was used as a primer. The buffering conditions were as recommended by the manufacturer, and cycling was performed as described by Palepou et al. (23) with the annealing temperature decreased to 60°C. Subsequently, the amplified Tn1546-like transposons served as templates for long PCR (L-PCR) of vanRSHAX gene-containing regions. Primers 2 and 3 (23) were used in the reactions, and these were run under conditions according to the method of Palepou et al. (23), with the annealing temperature decreased to 56°C and the initial elongation time reduced to 4 min. The amplifications were carried out in a GeneAmp 2400 thermocycler (Perkin-Elmer), and L-PCR products were electrophoresed in 0.8% agarose gels (SeaKem; FMC Bioproducts). DNA purified from the E. faecium BM4147 VanA standard strain (1) was used as a control.
Restriction fragment length polymorphism (RFLP) analysis of Tn1546-like transposons and their vanRSHAX regions.
The L-PCR products containing Tn1546-like elements were digested independently with ClaI (Takara) (23) and EcoRI (MBI Fermentas, Vilnius, Lithuania) restriction enzymes, and the resulting DNA fragments were separated in 1% agarose gels (SeaKem). The vanRSHAX amplicons were cut with the DdeI restrictase (Promega, Madison, Wis.) (23) and electrophoresed in 2% agarose gels (SeaKem).
RFLP analysis of Tn1546 loci.
For plasmid DNA purification, bacterial cells were predigested with lysozyme (0.5 mg/ml; Sigma Chemical Co., St. Louis, Mo.), and plasmids were isolated from the spheroplasts by the alkaline lysis method (4) using a Plasmid Midi Kit (Qiagen, Hilden, Germany). DNA preparations were digested with EcoRI (MBI Fermentas), electrophoresed in 1% agarose gels (SeaKem), and blotted onto a Hybond-N membrane (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) for hybridization with the Tn1546 probe. The L-PCR amplicon of the Tn1546 transposon present in the E. faecium BM4147 VanA standard strain (1) was used as the probe. Prior to labeling, the PCR product was purified using the QIAquick PCR Purification Kit (Qiagen); probe labeling, hybridization, and signal detection were performed with the ECL Direct Nucleic Acid Labeling and Detection systems (Amersham Pharmacia Biotech). Plasmid DNA isolated from the E. faecium BM4147 strain (1) was used as a control.
RESULTS
Antimicrobial susceptibility testing.
MICs are presented in Table 1. All of the isolates were characterized by high MICs of vancomycin (MICs, 256 to >512 μg/ml) and teicoplanin (MICs, 32 to 256 μg/ml), and all were found to be resistant to ciprofloxacin (MICs, 32 to 256 μg/ml). The E. faecium isolates were uniformly resistant to penicillin (MICs, ≥128 μg/ml) and ampicillin (MICs, 32 to >128 μg/ml); none of the isolates was resistant to chloramphenicol (MICs, 4 to 16 μg/ml). Various susceptibility phenotypes were observed regarding aminoglycosides and tetracycline. Nine isolates demonstrated the HLSR (high-level streptomycin resistance) phenotype (streptomycin MICs, ≥2,048 μg/ml; gentamicin MICs, 16 to 64 μg/ml), six isolates were of the HLGR (high-level gentamicin resistance) phenotype (gentamicin MICs, ≥512 μg/ml; streptomycin MICs, 32 to 128 μg/ml), and seven isolates expressed the HLAR (high-level aminoglycoside resistance) phenotype (streptomycin MICs, ≥2,048 μg/ml; gentamicin MICs, ≥512 μg/ml). Seven isolates were susceptible to tetracycline (MICs, 0.25 to 1 μg/ml), whereas the remaining ones were resistant to this antibiotic (MICs, 32 to 128 μg/ml).
Mating.
Results of mating are presented in Table 1. All but three isolates (E. faecium 1641 and 7953 and E. faecalis 7946) produced vancomycin-resistant transconjugants. The efficiency of conjugation ranged from 10−3 to 10−9 and was significantly higher in the group of isolates from the PHW (data not shown). Evaluation of MICs has revealed that in several cases resistance to streptomycin (isolates 1639, 7947, and 7948) or to tetracycline (isolates 7950 and 7952) was cotransferred with the glycopeptide resistance (data not shown).
Typing.
Results of PFGE typing are presented in Table 1. The analysis distinguished eight different PFGE types among all 21 E. faecium isolates (types A to H). Eight isolates obtained from patients in the HW were classified into six distinct PFGE types. Only two pairs of isolates from the HW produced identical PFGE patterns, including one from the isolates obtained from the same patient (7948 and 7950, PFGE type G). Isolates of the second pair (7947 and 7953, PFGE type F1), were found to be closely related to the only studied isolate from the ICU (7949, PFGE type F2).
On the other hand, 12 E. faecium isolates collected in the PHW were split into two equal groups of closely related isolates (PFGE types D and E) and could be further classified to several subtypes (D1 to D5 and E1 to E4). Two isolates from different patients (3135 and 3136) and one from the environment (3139) were found to be indistinguishable by PFGE (subtype E1).
Detection of the vanA gene.
The vanA gene was detected in all the isolates with the use of two pairs of specific PCR primers. The amplification carried out with primers A1 and A2 (8) produced DNA molecules of the expected size of about 730 bp, whereas amplicons of the expected size of about 1 kb were obtained in the reaction performed according to the method of Clark et al. (5) (results not shown).
RFLP analysis of Tn1546-like transposons.
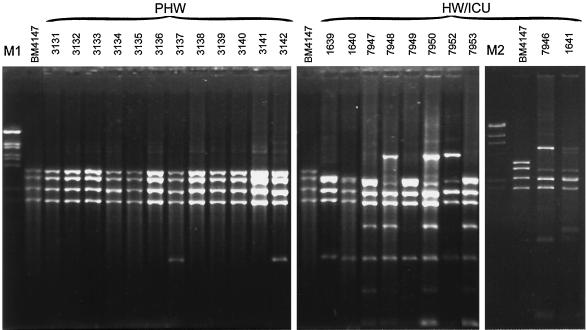
Tn1546-like elements were amplified by L-PCR, and products of the expected size of about 10 kb were obtained for all of the analyzed isolates. Results of their restriction analysis (ClaI digests) are shown in Fig. 1. Transposons amplified from 10 isolates recovered in the HW and the ICU (E. faecium and E. faecalis) produced RFLP patterns that were different from that obtained for the original Tn1546 (23). In several cases (isolates 1641, 7946, 7948, 7950, and 7952) these patterns were complex and contained DNA bands of a nonproportional intensity, which could suggest the presence of two or more transposon copies of different RFLP types in a single isolate. Nevertheless, obvious similarities between the patterns could be observed. Transposons of E. faecium isolates 1639, 1640, and 7949 were characterized by identical, relatively simple RFLP patterns, and all of their bands were also included in the patterns specific for isolates 7947 and 7953. Transposons amplified from the E. faecium isolate 7952 and the E. faecalis isolate 7946 produced the same RFLP patterns, which were very similar to those characteristic for the E. faecium isolates 7948 and 7950. Several common DNA bands could be found in all RFLP patterns of Tn1546-like elements of the isolates from the HW and the ICU.
FIG. 1.
RFLP analysis of Tn1546-like elements amplified by L-PCR and digested with ClaI. For E. faecium 1641 and E. faecalis 7946 the L-PCR products were repeatedly obtained with a low efficiency; therefore, these samples were processed separately. M1 (λ/BstEII) and M2 (λ/HindIII) are DNA molecular weight standards (Kucharczyk TE, Warsaw, Poland).
Transposons specific for 12 isolates collected in the PHW revealed ClaI RFLP patterns that were identical to each other and to the original Tn1546 element. Single additional bands were observed in three cases (isolates 3134, 3137, and 3142), and this variation was confirmed by the EcoRI digestion (results not shown).
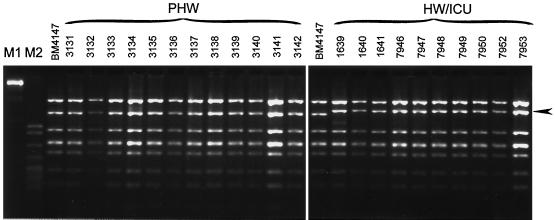
RFLP analysis of vanRSHAX regions.
Figure 2 presents results of DdeI digestion of DNA amplicons of about 4.4 kb encompassing vanRSHAX genes, which were obtained in nested L-PCR reactions with the use of transposon templates. These regions of all the isolates from the HW and the ICU produced an identical RFLP pattern that was different than that obtained for the homologous fragment of the original Tn1546 element (23). The vanRSHAX DNA amplified from all the PHW isolates were found to be of the other DdeI RFLP type, and this one was indistinguishable from Tn1546.
FIG. 2.
RFLP of the vanRSHAX region digested with DdeI. The arrow indicates the DNA band of reduced mobility common for all the HW and ICU isolates. M1 (λ/BstEII) and M2 (pBR322/MspI) are DNA molecular weight standards (Kucharczyk TE).
RFLP analysis of the Tn1546 locus.
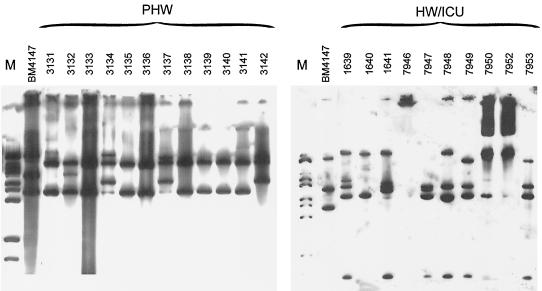
EcoRI-digested plasmids purified from clinical isolates or their transconjugants were used to study the polymorphism of DNA regions (or entire plasmids), in which Tn1546-like elements were inserted. The isolates (as well as transconjugants) were found to contain numerous plasmid molecules of different size, sequence and abundance, and their isolate-specific combinations have determined a high complexity and diversity of restriction patterns of plasmid DNA even among isolates belonging to the same PFGE type (results not shown). Results of hybridization of plasmid DNA with the Tn1546 probe are presented in Fig. 3. The Tn1546 hybridization patterns of the isolates recovered in the HW and the ICU revealed a high degree of diversity. A single band common for the majority of patterns was most probably the internal EcoRI restriction fragment of one of the Tn1546-like element variants that was spread within this population. Similarities of the entire RFLP patterns could also be observed between particular isolates (e.g., isolates 7949 and 7953 and isolates 1639 and 7948).
FIG. 3.
Hybridization of plasmid DNA purified from the isolates with the Tn1546 probe. M (λ/BstEII) is a DNA molecular weight standard (Kucharczyk TE). The plasmid purification procedure repeatedly did not work for the E. faecalis 7946 isolate; plasmids purified from E. faecium 7950 and 7952 were not completely digested.
Isolates collected from the PHW demonstrated remarkable homogeneity of Tn1546 hybridization patterns, which were much different from those of the HW and ICU isolates. The majority of the PHW isolates produced the relatively simple RFLP pattern with one of the bands that was shared also by the pattern specific for the plasmid purified from the BM4147 VanA standard strain (1) (the internal EcoRI fragment of transposons). The three isolates (3134, 3137, and 3142), characterized by single additional bands in their transposon ClaI RFLPs, produced distinct though related Tn1546 hybridization patterns, too. These patterns suggested that two different copies of Tn1546-like elements could exist in the isolates.
DISCUSSION
In a 1996 survey of antimicrobial susceptibility of enterococci isolated in Polish hospitals, no vancomycin-resistant strains of E. faecium or E. faecalis were detected (15). Numerous isolations of VRE in the University Hospital in Gdańsk in 1997 to 1999 indicated that the first nosocomial outbreak of VRE had occurred in the country (14, 25). The outbreak was caused by E. faecium as only a single vancomycin-resistant isolate of E. faecalis was identified in the hospital during this time. All of the collected isolates demonstrated the VanA phenotype (9) of glycopeptide resistance, and this was confirmed by PCR detection of the vanA gene in groups of representative isolates (references 14 and 25 and this study). The isolates were found to be multidrug resistant, uniformly demonstrating additional resistance to penicillins, ciprofloxacin, and high concentrations of aminoglycosides.
Several lines of evidence obtained in this study of representative isolates suggested that the VanA phenotype was most likely selected by at least two independent events within the hospital enterococcal population. This hypothesis was mostly supported by the observation of two distinct RFLP types of the highly conserved DNA region encompassing the vanRSHAX genes that are critical for the resistance phenotype. The revealed dichotomy of vanRSHAX regions strictly correlated with two general similarity groups in the RFLP of the entire Tn1546-like transposons containing van genes; however, these elements demonstrated a remarkable degree of diversity. The distribution of isolates containing the two main types of VanA determinants suggested that the selection events had occurred in two different hematological wards (HW and PHW), which are located in separate buildings of the hospital. Therefore, it is very likely that the collected isolates represented two rather distinct VRE outbreaks that occurred at the same time in the center.
The outbreak in the HW was highly polyclonal, as demonstrated by the diversity of the distinguished E. faecium PFGE types and by the isolation of the single vancomycin-resistant E. faecalis strain. The observed relatedness of some of the E. faecium isolates (PFGE type F) suggested, however, that clonal spread has also occurred in the HW and that one of the strains was probably exported from the HW to the ICU. (The small number of isolates studied has precluded the possibility of evaluating the extent of the clonal dissemination in the ward.) It may be postulated that originally a single variant of the Tn1546-like transposon was spread among the nonrelated enterococcal strains circulating in the HW. This hypothesis is supported by the fact that the HW isolates, collected over a period of approximately 3 years, produced identical RFLP patterns for the vanRSHAX region and related RFLP patterns of Tn1546-like transposons. The certain degree of diversity observed within the transposon RFLP most probably reflected multiplication of the elements, in particular strains and differentiation of the resulting copies by various genetic events such as point mutations, insertions, or deletions within their less-conserved parts (13, 23, 31, 32). The remarkable heterogeneity of transposon loci RFLP patterns characteristic for the HW isolates has been most likely due to transposition of the Tn1546-like elements to a high variety of plasmid molecules, some of which could be then further transmitted by conjugation to other enterococcal strains.
The outbreak in the PHW was most probably due to the clonal spread of two different E. faecium strains (represented by isolates of PFGE types D and E) that were not related to those in the HW. Minor differences in PFGE patterns observed in both types revealed the ongoing evolutionary diversification process within their populations. Isolates of the two types produced identical RFLP patterns of vanRSHAX regions, entire Tn1546-like transposons, and transposon loci in plasmid DNA. This indicated that most probably a single variant of the transposon was transmitted along with a plasmid from one of the epidemic strains to the other. This element is likely identical or closely related to the original Tn1546 transposon (23); however, it is located in a different plasmid than the one specific for the BM4147 VanA standard strain (1). Modifications of the transposon RFLP observed in some isolates of the group were probably due to the element's duplication and recombination, and this was reflected also by the heterogeneity of Tn1546 insertion loci RFLP patterns in these isolates.
Numerous previous reports have documented the majority of the epidemiological phenomena demonstrated by the present study. Multiple selection events (33), transposition to different replicons (1, 2, 6), plasmid-mediated horizontal transfer (34), and clonal dissemination of epidemic strains (20, 29) were revealed as major factors of vancomycin resistance spread in enterococcal populations. However, in most cases the parallel occurrence of all of these mechanisms was reported either in multicenter studies (12, 23, 29) or studies in which isolates of both human and nonhuman sources were compared (31, 32). Data presented here demonstrate the complexity of the epidemiological situation concerning VRE that may occur in a single medical center. Analysis of a small group of representative isolates has shown that concurrent outbreaks in two different wards of the hospital commenced following more than one independent selection event for acquisition of the VanA determinants. Thus, the developing outbreaks consisted of transmission of plasmids carrying the transposon-located resistance genes, followed by clonal dissemination of the strains. Our understanding of these outbreaks has been subsequently getting more complicated due to transposon multiplication and modification, insertion of copies to other plasmids, and their consequent further spread.
ACKNOWLEDGMENTS
We are grateful to L/ukasz Naumiuk, Marek Bronk, and Alfred Samet from the University Hospital in Gdańsk for the VRE isolates and clinical data. We also thank Patrice Courvalin who kindly provided E. faecium BM4147, Wolfgang Witte for the E. faecium 64/3 strain, Teresa Kamińska for her excellent technical assistance, and Stephen Murchan for critical reading of the manuscript.
This study was partially supported by the U.S.–Poland Maria Skl/odowska-Curie Joint Fund II, MZ/NIH–98-324.
REFERENCES
- 1.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell J M, Paton J C, Turnidge J. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J Clin Microbiol. 1998;36:2187–2190. doi: 10.1128/jcm.36.8.2187-2190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark N, Cooksey R, Hill B, Swenson J, Tenover F. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lencastre H, Brown A E, Chung M, Armstrong D, Tomasz A. Role of transposon Tn5482 in the epidemiology of vancomycin-resistant Enterococcus faecium in the pediatric oncology unit of a New York City hospital. Microb Drug Resist. 1999;5:113–129. doi: 10.1089/mdr.1999.5.113. [DOI] [PubMed] [Google Scholar]
- 7.de Lencastre H, Severina E P, Roberts R B, Kreiswirth B N, Tomasz A. Testing the efficacy of a molecular surveillance network: methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) genotypes in six hospitals in the metropolitan New York City area. The BARG Initiative Pilot Study Group. Bacterial Antibiotic Resistance Group. Microb Drug Resist. 1996;2:343–351. doi: 10.1089/mdr.1996.2.343. [DOI] [PubMed] [Google Scholar]
- 8.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evers S, Reynolds P E, Courvalin P. Sequence of the vanB and ddl genes encoding d-alanine:d-lactate and d-alanine:d-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene. 1994;140:97–102. doi: 10.1016/0378-1119(94)90737-4. [DOI] [PubMed] [Google Scholar]
- 10.Facklam R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fines M, Perichon B, Reynolds P, Sahm D F, Courvalin P. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother. 1999;43:2161–2164. doi: 10.1128/aac.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39:2446–2453. doi: 10.1128/aac.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hryniewicz W, Szczypa K, Bronk M, Samet A, Hellmann A, Trzciński K. First report of vancomycin-resistant Enterococcus faecium isolated in Poland. Clin Microbiol Infect. 1999;5:503–505. doi: 10.1111/j.1469-0691.1999.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 15.Hryniewicz W, Zarȩba T, Kawalec M. Susceptibility patterns of Enterococcus spp. isolated in Poland during 1996. Int J Antimicrob Agents. 1998;10:303–307. doi: 10.1016/s0924-8579(98)00058-2. [DOI] [PubMed] [Google Scholar]
- 16.Klare I, Collatz E, Al-Obeid S, Wagner J, Rodloff A C, Witte W. Glykopeptidresistenz bei Enterococcus faecium aus Besiedlungen und Infectionen von Patienten aus Intensivstationen Berliner Kliniken und einem Transplantationszentrum. ZAC Zschr Antimikrob Antineoplast Chemother. 1992;10:45–53. [Google Scholar]
- 17.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 18.Leclercq R, Dutka-Malen S, Duval J, Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother. 1992;36:2005–2008. doi: 10.1128/aac.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moellering R C., Jr Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173–1178. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 20.Morrison D, Woodford N, Cookson B. Epidemic vancomycin-resistant Enterococcus faecium in the UK. Clin Microbiol Infect. 1995;1:146–147. doi: 10.1111/j.1469-0691.1995.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 21.Murray B E. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4:37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M100-S9, 5th ed. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement, vol. 19, no. 1. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 23.Palepou M I, Adebiyi A A, Tremlett C H, Jensen L B, Woodford N. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J Antimicrob Chemother. 1998;42:605–612. doi: 10.1093/jac/42.5.605. [DOI] [PubMed] [Google Scholar]
- 24.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samet A, Bronk M, Hellmann A, Kur J. Isolation and epidemiological study of vancomycin-resistant Enterococcus faecium from patients of a haematological unit in Poland. J Hosp Infect. 1999;41:137–143. doi: 10.1016/s0195-6701(99)90051-8. [DOI] [PubMed] [Google Scholar]
- 26.Samet A, Bronk M, Hellmann A, Juszczyk J, Kur J. Epidemiology of glycopeptide resistant Enterococcus faecium in a hematology unit. Presented at the 10th European Congress of Clinical Microbiology and Infectious Diseases [ESCMID], Stockholm, Sweden 28–31 May 2000. Clin Microbiol Infect. 2000;6(Suppl. 1):128. (MoP206). [Google Scholar]
- 27.Suppola J P, Kolho E, Salmenlinna S, Tarkka E, Vuopio-Varkila J, Vaara M. vanA and vanB incorporate into an endemic ampicillin-resistant vancomycin-sensitive Enterococcus faecium strain: effect on interpretation of clonality. J Clin Microbiol. 1999;37:3934–3939. doi: 10.1128/jcm.37.12.3934-3939.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover F C, Arbeit R, Goering V, Mickelsen P, Murray B M, Pershing D, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thal L, Donabedian S, Robinson-Dunn B, Chow J W, Dembry L, Clewell D B, Alshab D, Zervos M J. Molecular analysis of glycopeptide-resistant Enterococcus faecium isolates collected from Michigan hospitals over a 6-year period. J Clin Microbiol. 1998;36:3303–3308. doi: 10.1128/jcm.36.11.3303-3308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uttley A H C, George R C, Naidoo J, Woodford N, Johnson A P, Collins C H, Morrison D, Gilfillan A J, Fitch L E, Heptonstall J. High-level vancomycin-resistant enterococci causing hospital infections. Epidemiol Infect. 1989;103:173–181. doi: 10.1017/s0950268800030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner G, Klare I, Witte W. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol Lett. 1997;155:55–61. doi: 10.1111/j.1574-6968.1997.tb12685.x. [DOI] [PubMed] [Google Scholar]
- 32.Woodford N, Adebiyi A M, Palepou M I, Cookson B D. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodford N, Chadwick P R, Morrison D, Cookson B D. Strains of glycopeptide-resistant Enterococcus faecium can alter their van genotypes during an outbreak. J Clin Microbiol. 1997;35:2966–2968. doi: 10.1128/jcm.35.11.2966-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodford N, Morrison D, Johnson A P, Briant V, George R C, Cookson B. Application of DNA probes for rRNA and vanA genes to investigation of a nosocomial cluster of vancomycin-resistant enterococci. J Clin Microbiol. 1993;31:653–658. doi: 10.1128/jcm.31.3.653-658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]