Abstract
Threading a query protein sequence onto a library of weakly homologous structural templates remains challenging, even when sequence-based predicted contact or distance information is used. Contact- or distance-assisted threading methods utilize only the spatial proximity of the interacting residue pairs for template selection and alignment, ignoring their orientation. Moreover, existing threading methods fail to consider the neighborhood effect induced by the query-template alignment. We present a new distance- and orientation-based covariational threading method called DisCovER by effectively integrating information from inter-residue distance and orientation along with the topological network neighborhood of a query-template alignment. Our method first selects a subset of templates using standard profile-based threading coupled with topological network similarity terms to account for the neighborhood effect and subsequently performs distance- and orientation-based query-template alignment using an iterative double dynamic programming framework. Multiple large-scale benchmarking results on query proteins classified as weakly homologous from the Continuous Automated Model Evaluation (CAMEO) experiment and from the current literature show that our method outperforms several existing state-of-the-art threading approaches; and that the integration of the neighborhood effect with the inter-residue distance and orientation information synergistically contributes to the improved performance of DisCovER. DisCovER is freely available at https://github.com/Bhattacharya-Lab/DisCovER.
Keywords: protein threading, inter-residue orientation, inter-residue distance, template based modeling, protein structure prediction
INTRODUCTION
Accurate prediction of the three-dimensional (3D) structure of a protein from its sequence remains challenging1. Template-based modeling (TBM), one of the most reliable and accurate approaches for protein 3D structure prediction, uses homologous templates of analogous folds available in the Protein Data Bank2 to infer 3D structural models of unknown proteins. As such, the success of TBM intrinsically depends on the detection of homologous templates and the generation of accurate query-template alignments. However, in the absence of close sequence homology, template detection and alignment become challenging. Protein threading is a TBM approach that can address the challenge by leveraging multiple sources of information such as sequence profile, secondary structure, solvent accessibility, and torsion angles. Traditional threading methods such as HHpred3, MUSTER4, SparkX5, pGenThreader6, and CNFpred7,8 have shown noteworthy success by successfully modeling protein structures even in the absence of significant sequence homology. Nevertheless, the performance of traditional threading methods sharply declines when the evolutionary relationship between the query and templates becomes very low9, the so-called remote-homology threading scenario.
With significant recent progress in residue-residue contact or distance prediction mediated by sequence co-evolution and deep learning10–17, predicted contact or distance information becomes a valuable source of additional information for remote-homology threading. Consequently, predicted contact- or distance-based threading has attracted promising attention. For instance, EigenTHREADER18 utilizes contacts predicted by MetaPSICOV12 to perform contact map threading. map_align19 employs iterative double dynamic programming driven by coevolutionary contact predictor, GREMLIN11. Very recently, CEthreader9 and CATHER20 perform contact-assisted threading using contacts predicted by ResPre13 and MapPred21, respectively, together with sequence profile-based features. DeepThreader22 goes one step further by incorporating finer-grained distance information instead of contacts for boosting threading performance23(p13).
While these methods exploit predicted contact or distance information during threading often in conjunction with sequential information, they do not consider two key factors that can further improve threading accuracy. First, the recent extension of deep residual network architecture has resulted in accurate inter-residue orientations predicted from coevolution17 in addition to distances, but none of the threading methods incorporate orientation information. Second, most of the threading approaches do not include the effect of the residue pairs in the neighborhood of an aligned query-template residue pair. That is, they ignore the neighborhood effect induced by the query-template alignment.
In this article, we introduce a new distance- and orientation-based threading method DisCovER (Distance- and orientation-based Covariational threadER) that effectively integrates information from inter-residue distance and orientation along with the topological network neighborhood of a query-template alignment using an iterative double dynamic programming framework to greatly improve threading template selection and alignment. Experimental results show that our new method performs better than profile-based threading methods SparkX, HHpred, CNFpred, MUSTER, PPAS, and pGenThreader; as well as state-of-the-art contact-assisted approaches CEthreader, map_align, EigenTHREADER, and CATHER, especially on weakly homologous proteins. At one of the most challenging threading situations, DisCovER yields better performance than the RaptorX server22,24 participating in the Continuous Automated Model Evaluation (CAMEO) experiment25 and employing the distance-based threading method DeepThreader22. DisCovER is freely available at https://github.com/Bhattacharya-Lab/DisCovER.
MATERIALS AND METHODS
Feature sets and inter-residue geometries
Our feature set includes both sequential and pairwise features for the query protein and templates. For a query protein, we generate sequence profiles based on multiple sequence alignments (MSA)26, and predict profile-based features including secondary structure, solvent accessibility, and backbone dihedral angles using SPIDER327. We also predict inter-residue geometries including distances and orientations by feeding the MSAs into trRosetta17. The predicted distance map is then binned into 9 segments at 1Å interval: <6Å, <7Å, …, <14Å, by summing up probabilities for distance bins below specific distance thresholds. The predicted inter-residue orientations include two dihedral angles (ω, θ), both binned into 24 evenly spaced segments with a bin width of 15° each, and one planar angle (φ), binned into 12 evenly spaced segments with a bin width of 15° each. All distance orientation bins have associated likelihood values for the query protein predicted by trRosetta. For the templates, we use structure-derived profiles, extract secondary structures and solvent accessibility using DSSP28, and compute backbone dihedral angles, inter-residue distance maps, and orientation information including ω, θ dihedrals and φ angle from the structure.
Geometry-based scoring of a query-template alignment
DisCovER scores a query-template alignment as follows:
| (1) |
where Zfinal is the normalized alignment score for selecting the top-ranked template, Zwithout_geometry is the normalized alignment score based only on profile information with neighborhood effect, and Zwith_geometry is the normalized alignment score using profile information, neighborhood, and inter-residue geometries including distance and orientation. In the following, we describe each term in detail.
Stage 1 Scoring profile-based alignment with neighborhood effect:
A profile-based query-template alignment is scored for aligning the ith residue of the query and the jth residue of the template similar to our recent work29 as follows:
| (2) |
where the first term defines the sequence profile-profile alignment. fc(i,k) and fd(i,k) define the frequency of the kth residue at the ith query position of the MSA for “close” and “distant” homologous sequences, respectively. The frequency is determined using the Henikoff weighting scheme30. Lt is the log-odds profile of the template for the kth residue at the jth position, which is obtained by PSI-BLAST31 with an E-value of 0.001. The next term measures the consistency between the predicted and observed three-state secondary structures, such that the function δ returns 1 if two variables are matched and −1 otherwise. The next term is the agreement between the structure-derived profiles (fs) of the kth residue at the jth position of the template structure and the sequence profile (Lq) of the kth residue at the ith position of the query sequence. The function E in the next three terms is defined by: E(xi,xj) = (1 − 2|xi − xj|), where the variables are the predicted and the observed values of relative solvent accessibility (SA) and backbone dihedral angles (ϕ, ψ) of the ith position of the query and the jth position of the template, respectively. The seventh term corresponds to the match between the hydrophobic residues of the query and the template. c is the weighting parameter adopted from29.
To further improve the sensitivity of profile-based alignment, we borrow ideas from comparative network analysis. We adopt an approach similar to that was originally used in the IsoRank network alignment algorithm32 and very recently adopted in network-based structural alignment of RNA sequences33, in which two nodes in different networks are more likely to be aligned to each other if their neighbors are also aligned well to one another. This results in a similarity diffusion scheme to compute the agreement between the networks, leading to an improved alignment. Following similar principles, we integrate connected similarity, attempting to estimate the topological agreement between the query and the template by capturing the similarity between the neighborhoods of two residues.
Connected similarity (Sc) is based on the principle that one query-template residue pair is likely to be aligned if their neighboring residues are also aligned. It is calculated for the residue pair (i, j) as follows:
| (3) |
To further improve the sensitivity of the profile-based alignment score (Equation 2), the connected similarity (Sc) is then added to it as follows:
| (4) |
We use the Needleman-Wunsch34 global dynamic programming to score every query-template alignment. To select the top-fit templates, we compute the Zwithout_geometry based on the raw alignment score Swithout_geometry (Equation 4) to assess the quality of each query-template alignment as follows:
| (5) |
where is the larger one of the raw alignment score Swithout_geometry divided by the full alignment length (including query and template ending gaps) and the partial alignment length (excluding query ending gaps). is the average across all templates in the template library. A subset of fifty top-scoring templates is selected for the next stage.
Stage 2 Scoring distance- and orientation-based alignment:
A similarity score is calculated for each row of query (Q) and template (T) distance maps as follows:
| (6) |
where the first term calculates the similarity between the predicted distance map of the query and true distance map of the template at a given distance threshold of k, where k ∈{<6Å, <7Å, …, <14Å}. is the predicted likelihood value of the residue pair i and i′ of the query to be within a distance threshold of kÅ; is a Heaviside step function that has a value 1 if the residue pair j and j′ of the template is within the distance threshold of kÅ and 0 otherwise; wk is the corresponding weight parameter adapted from the literature20 with is the weight of the minimum of sequence separation of query residues and template residues defined as wsep(s) = 0.75 for s = 5 and log10(1+s) for s ≥ 6, similar to other studies19, and s = min (|i − i′|,|j − j′|); G(0,sd,sep) is a zero-mean Gaussian function, which is also adopted from the literature19 and defined as exp(−sep2/(2 sd2)), where sep is an absolute difference of sequence separation of query residues and sequence separation of template residues, and sd or standard deviation is a function of the smaller of the sequence separation of query residues and the sequence separation of template residues. The next three terms calculate dihedral (ω, θ) and planar (φ) angles similarities between the query and template residue pair. We treat the orientation information similar to distances and compute the similarity between the predicted ω, θ dihedrals or φ angle of the query and the corresponding true angles of the template at a specific angle bin of k, where k ∈{15°,30°,…,360°} for the ω, θ dihedrals and k ∈{15°,30°,…,180°} for the φ angle. Analogous to distances, , , and are the predicted likelihood values of the residue pair i and i′ of the query to be within an angle bin of k° for the angles ω, θ, and φ, respectively. Similarly, , , and are the box functions corresponding to the angles ω, θ, and φ, respectively, having values 1 if a residue pair j and j′ of the template is within an angle bin of k° and 0 otherwise. wsep(s) is the weight term described before.
Our double dynamic programming framework for computing the optimal alignment score between the query and each of the fifty top-scoring templates selected from the previous stage comprises of two dynamic programming steps. In the first step, we perform row-by-row comparisons between the query and template. Dynamic programming is used to find the alignment for the two rows being matched which maximizes the composite distance- and orientation-based alignment score described in equation 6. These scores are stored in a similarity matrix and are used to obtain the optimal alignment by using the Smith-Waterman35 algorithm.
At this step, however, the scores for individual row-row comparisons are overestimated since the alignments for each pair are independently computed in the first step. We subsequently update the similarity matrix using a second step based on the current alignment by employing a second dynamic programming. Such an iterative updating strategy is originally proposed in36 and later adopted in19, although our score is quite different. After obtaining the optimal alignment from the similarity matrix, the profile score and the gap-score are re-calculated to compute the raw alignment score (Swith_geometry). The similarity score for each query-template pair is normalized using Zwith_geometry as follows:
| (7) |
where is the larger one of the raw alignment score Swith_geometry divided by the full alignment length (including query and template ending gaps) and the partial alignment length (excluding query ending gaps). 〈…〉 denotes the average of all top-scoring templates.
Building full-length 3D models:
After selecting the first-ranked template using Equation (1), we use MODELLER (V9.22)37 to generate the full-length 3D model of a query protein using the associated query-template alignment. In addition to employing the standard automodel() class of MODELLER for model building purely through the satisfaction of spatial restraints from the query-template alignment, we also experiment with model building with additional restraints from predicted distances, orientations, and secondary structures by redefining the automodel.special_restraints() class. Specifically, we feed bounded harmonic restraints for the predicted distance thresholds corresponding to the 9 distance bins used in query-template alignment with a minimum likelihood cutoff of 0.85 using the physical.xy_distance() function, bounded harmonic restraints for the predicted orientation information derived from the highest likelihood bins having the minimum likelihood cutoff of 0.85 with the φ angle using physical.angle() function and ω, θ dihedrals using physical.dihedral() function, and secondary structure restraints for realizing the predicted secondary structure using the secondary_structure() module. Of note, all of these additional restraints are integrated to the list of spatial restraints derived from the query-template alignment to instruct MODELLER to satisfy them as best as it can.
Benchmark datasets, methods to compare, template libraries used, and threading performance evaluation
To evaluate remote-homology threading performance, we benchmark our new method DisCovER using targets from the Continuous Automated Model Evaluation (CAMEO) experiments consisting of 117 proteins classified as “hard”25, released between 8 December 2018 and 1 June 2019 having length between 50 and 500 residues (range is 51 to 487). On this dataset, DisCovER is compared against profile-based threading methods such as SparkX5, CNFpred7,8, MUSTER4, PPAS38, and pGenThreader6; as well as state-of-the-art contact-assisted methods including CEthreader9 utilizing ResPRE-predicted contact maps13 and EigenTHREADER18 utilizing DMPfold-predicted maps14. The template libraries for DisCovER, CEthreader, EigenTHREADER, SparkX, MUSTER, PPAS, pGenThreader are generated from the same set of 70,670 templates downloaded from https://zhanglab.ccmb.med.umich.edu/library/39, curated before the release of the CAMEO targets. The template library for CNFpred is downloaded from http://raptorx.uchicago.edu/download/. For all methods, we evaluate threading performance by comparing the top-ranked full-length predicted 3D models, built using the standard automodel() class of MODELLER from the query-template alignment, against the experimental structures of the target proteins using the TM-score metric40, which ranges from 0 to 1 with higher score indicating better performance and TM-score >0.5 indicating the attainment of correct overall fold41. Of note, DisCovER utilizes distances and orientations predicted from trRosetta17, which uses a training set collected from a snapshot as of 1 May 2018, older than the CAMEO test set used here. DisCovER also relies on secondary structure predictor SPIDER327, which uses a much older training dataset and thus independent of the CAMEO test set. We collect publicly available multiple sequence alignments (MSAs) independently generated using non-overlapping protein sequence databases from https://yanglab.nankai.edu.cn/trRosetta/benchmark/ to feed into trRosetta and SPIDER3. Furthermore, the template library used in DisCovER excludes any templates released after starting of CAMEO experiments (8 December 2018), free from any overlap. Finally, the “hard” target difficulty classification of the CAMEO test set defined by CAMEO25 warrants their non-overlapping and weakly homologous nature, thereby enabling us to focus on difficult targets in which existing methods have limitations22.
The definition of “hard” can be made even more stringent by requiring that TM-score of the HHpredB server3 participating in CAMEO to be less than 0.5. This reduces the number of targets to 60. This harder set simulates one of the most challenging threading situations while enabling a comparison between DisCovER and the distance-based threading method DeepThreader22. DeepThreader method is not publicly available, but the RaptorX server22,24 participates in CAMEO and employs the DeepThreader method according to the CAMEO assessment paper25. While RaptorX uses PDB90 as the template database and builds 3D models using Rosetta23(p13) as opposed to MODELLER, we compare the performance of DisCovER on this common set of 60 very hard targets to that of DeepThreader after downloading the predictions submitted by the RaptorX server from the official website of CAMEO (https://cameo3d.org/) and computing their TM-scores.
We are unable to directly compare DisCovER with two other state-of-the-art contact-assisted methods map_align19 and CATHER20 on the CAMEO test set, because map_align is too computationally expensive to run locally given our limited computational resources and CATHER is only available as a webserver and thus not suitable for large-scale benchmarking. However, the published work of CATHER reports the mean TM-scores of 3D models predicted using various threading methods including CATHER, map_align, EigenTHREADER, HHpred3, SparkX, and MUSTER over a dataset of 480 targets including 304 easy, 45 medium, and 131 hard targets with pairwise sequence identity <25% and length ranging from 50 to 500 residues20. We use this set to compare DisCovER against CATHER and map_align by running DisCovER locally after excluding templates with sequence identity >30% to the query proteins, and comparing its average performance against the reported results of CATHER and map_align, in addition to the other threading methods presented. Similar to DisCovER, CATHER employs MODELLER for 3D modeling, even though a detailed modeling protocol is not included in the published work of CATHER. In this dataset, DisCovER performs 3D modeling by employing MODELLER with additional restraints, as discussed before.
RESULTS AND DISCUSSION
Performance on 117 hard targets from CAMEO
Over the 117 hard targets from CAMEO, our distance- and orientation-based threading method DisCovER performs better than the two contact-assisted threading methods as well as all five profile-based approaches. As shown in Table 1, DisCovER attains a mean TM-score of 0.505, which is higher than the next best contact-assisted threading method CEthreader (0.483) and the best among profile-based threading methods CNFpred (0.464). The performance improvement for DisCovER is statistically significant at 95% confidence level compared to all other methods. DisCovER also predicts higher number of correct folds (TM-score >0.5) with a success rate of 57.3%, which is ~5% higher than the success rate of CEthreader and ~9% higher than that of CNFpred. Of note, the best contact-assisted threading method CEthreader falls short of achieving a mean TM-score of 0.5, whereas DisCovER exceeds this criterion.
Table 1.
Benchmark results of various threading methods on 117 hard targets from CAMEO. Values in bold represent the best performance.
| Methods | TM-score | p-value* | #Correct folds† |
|---|---|---|---|
|
| |||
| pGenThreader | 0.423 | 1.1E-08 | 46 |
| PPAS | 0.456 | 9.3E-05 | 54 |
| MUSTER | 0.459 | 0.0006 | 54 |
| SparkX | 0.461 | 0.0003 | 57 |
| CNFpred | 0.464 | 0.004 | 56 |
| EigenTHREADER | 0.461 | 1.9E-05 | 58 |
| CEthreader | 0.483 | 0.029 | 61 |
| DisCovER | 0.505 | - | 67 |
Column “p-value” represents one sample t-test’s p-value of the TM-score difference compared to DisCovER.
Column “#Correct folds” represents the number of models with TM-score >0.5
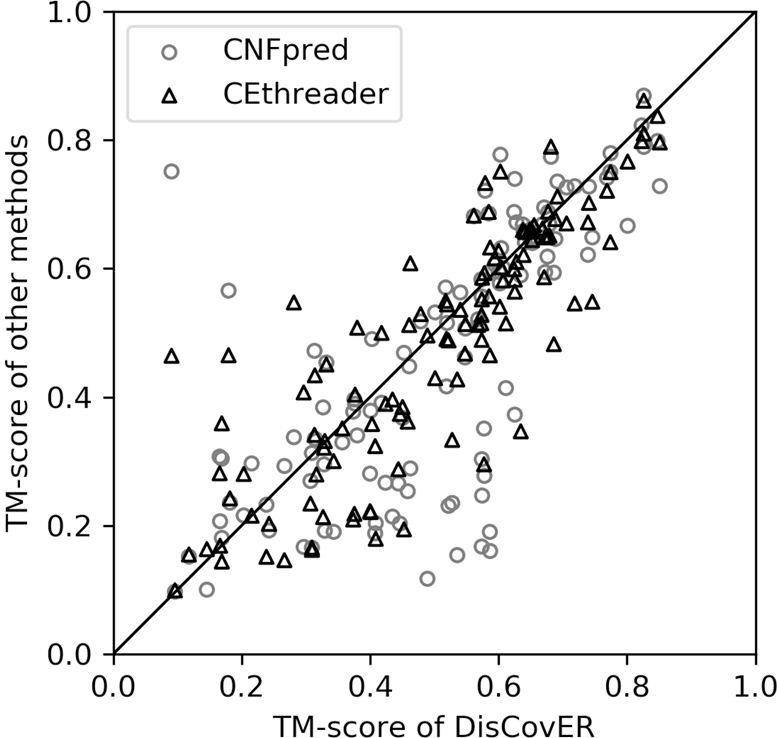
Figure 1 shows the head-to-head comparison between DisCovER and the best contact-assisted threading method CEthreader and the best profile-based threading method, CNFpred. DisCovER attains higher TM-score for 74 and 66 targets compared to CEthreader and CNFpred, respectively. In summary, the advantage of DisCovER in threading remote-homology proteins over the others is significant.
Figure 1.
Head-to-head performance comparison between DisCovER (x-axis) vs. CEthreader and CNFpred (y-axis) on 117 hard targets from CAMEO.
Contribution of individual components
In addition to profile-based alignment, DisCovER incorporates a) neighborhood effect, b) distance, and c) orientation. To investigate the contributions of each of these components to DisCovER performance, we perform ablation studies on the CAMEO test set as well as another independent dataset from the MUSTER paper4 by gradually removing each component from the full-fledged DisCovER method one at a time and evaluate the performance. The same template library, sequence databases, and same set of features are used in all cases to generate the query-template alignments that are then fed into the standard automodel() class of MODELLER to generate the 3D structures. While the study on the CAMEO test set exclusively focuses on hard and very hard targets, the study on the MUSTER dataset comprises of targets from easy to hard categories. Specifically, the original MUSTER test set contains 500 targets, including 203 easy and 255 hard targets as defined in the published work of MUSTER, with pairwise sequence identity <25% and length ranging from 50 to 633 residues. For the purpose of our study, targets with sequence identity >40% to the training set of trRosetta are excluded following the literature9 in order to make the test set free from any overlap, thus reducing the number of targets in the MUSTER test set to 86. Moreover, we exclude templates with sequence identity >20% to the query proteins or detectable by BLAST31 with an E-value <0.05 to remove the homologous templates, a practice adopted from the literature4.
As reported in Table 2, the full-fledged DisCovER attains the best performance (mean TM-score of 0.505) than any of its ablated variants over the 117 hard targets from CAMEO. Without orientation, the mean TM-score decreases to 0.503, which is further decreased to 0.488 without distance information. The performance of the ablated variant of DisCovER that only incorporates distance but no orientation outperforms the variant that only incorporates orientation but no distance. It is interesting to note that the average performance of either variant is still better than state-of-the-art contact-assisted methods CEthreader and EigenTHREADER, indicating that the incorporation of either distance or orientation information in DisCovER is sufficient to outperform top contact-assisted threading method such as CEthreader. When both distance and orientation terms are excluded, the mean TM-score drops to 0.472, but it is still better than the top profile-based threading method CNFpred. Upon further exclusion of neighborhood effect, the mean TM-score slightly reduces to 0.470. The results demonstrate that all components contribute to the improved performance of DisCovER, with distance and/or orientation information having significant contribution. We note that DisCovER incorporates distance and orientation information only in stage 2 for computing the optimal alignment score between the query and each of the fifty top-scoring templates selected from stage 1 based on profile-based threading in combination with neighborhood effect. That is, distance- and orientation-based alignment further improves the alignment accuracy over the top template recognition performance achieved by profile-based threading with topological network neighborhood. The trend remains very similar considering the subset of 60 very hard CAMEO targets, although the exclusion of neighborhood effect results in a noticeable performance decline in this case (a mean TM-score drop from 0.335 to 0.329) that shows the effectiveness of incorporating topological network neighborhood for challenging threading scenarios. Representative examples from CAMEO set 5ZER_B and 6D7Y_B further demonstrate the contribution of each component (see Supplementary Table S1). Target 5ZER_B is classified as hard and the target 6D7Y_B is classified as very hard. In both cases, the full-fledged DisCovER performs the best with a positive and significant contribution of each component with distance and orientation information having complementary effects. For instance, the exclusion of distance (or orientation) results in a TM-score drop from 0.446 to 0.330 (or 0.372) for the very hard target 6D7Y_B. A similar trend can also be seen for the target 5ZER_B. Moreover, a noticeable performance drop (0.015 TM-score) is also observed with the exclusion of the neighborhood effect for both targets. Overall, each component of DisCovER contributes synergistically, including the two novel contributions of this study, orientation and network neighborhood.
Table 2.
Contribution of individual features to DisCovER performance.
| Methods | Mean TM-score | ||
|---|---|---|---|
|
| |||
| CAMEO dataset | MUSTER dataset | ||
|
| |||
| 117 hard targets | 60 very hard targets | 86 easy and hard targets | |
|
| |||
| DisCovERNo neighborhood, No geometry† | 0.470 | 0.329 | 0.404 |
| DisCovERNo geometry† | 0.472 | 0.335 | 0.405 |
| DisCovERNo distance | 0.488 | 0.349 | 0.420 |
| DisCovERNo orientation | 0.503 | 0.372 | 0.416 |
| DisCovER | 0.505 | 0.376 | 0.432 |
| TM-align‡ | 0.636 | 0.542 | 0.562 |
geometry includes distance and orientation information.
TM-align results are included as a reference.
The ablation study on the MUSTER dataset amplifies the importance of incorporating orientation information and the complementarity of distance and orientation. As shown in Table 2 (last column), the ablated variant of DisCovER that only incorporates orientation but no distance (mean TM-score 0.420) outperforms the variant that only incorporates distance but no orientation (mean TM-score 0.416). That is, the incorporation of orientation information, which is a major contribution of this work, significantly contributes to threading performance. It is also interesting to note that the full-fledged DisCovER that integrates both distance and orientation attains a noticeably higher mean TM-score of 0.432 than its ablated variants using distance or orientation alone. Supplementary Figure S1 further shows the full-fledged DisCovER outperforms its ablated variants in more than half of the cases, demonstrating the complementarity of distance and orientation information and their significant contribution to the superior performance of the full-fledged DisCovER. We also notice that in the MUSTER dataset, which contains a mix of easy and hard targets, there is a positive but minor contribution of the neighborhood effect. This is consistent with our earlier observation in the CAMEO dataset that topological network neighborhood contributes the most for very hard targets. Overall, all components contribute to the improved threading performance of DisCovER with the orientation and distance information playing significant and complementary roles.
Table 2 also reports a reference oracle method that uses TM-align42 to structurally align the experimental structure of the query protein with each of the templates in the template library to select the structurally closest template and the resulting optimal query-template alignment is then fed into the standard automodel() class of MODELLER to generate the 3D structures. Not surprisingly, the TM-align-based oracle achieves much better performance with a mean TM-score > 0.5 even for the very hard CAMEO targets (Table 2, last row), indicating that there is still a large room for improvement.
3D model building using MODELLER from query-template alignment with additional restraints
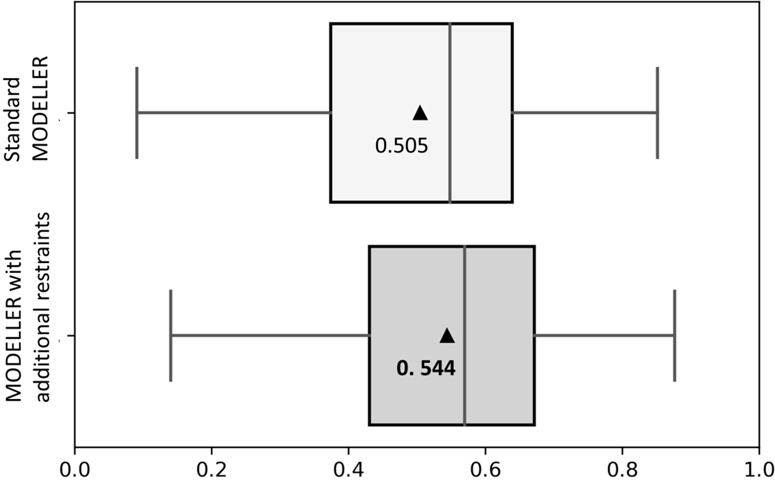
To examine whether the additional information used in DisCovER for threading template selection and alignment can further improve the full-length 3D model building, we compare the standard automodel() class of MODELLER that builds 3D models using spatial restraints collected from query-template alignment to another approach using MODELLER that integrates additional restraints from predicted distances, orientations, and secondary structures. As shown in Figure 2, the mean TM-score attained by MODELLER with additional restraints is 0.544 over the 117 hard targets from CAMEO, better than that of standard MODELLER. MODELLER with additional restraints also attains 75 correct folds while shifting the TM-score distribution towards higher accuracy. That is, 3D model building using MODELLER from query-template alignment with additional restraints can be an effective use of the additional information used in DisCovER. We follow this model building approach henceforth.
Figure 2.
3D model building performance using standard MODELLER and MODELLER with additional restraints for 117 hard targets from CAMEO. The x-axis represents TM-score. The mean TM-scores are represented by triangles.
Performance comparison with CAMEO server RaptorX employing DeepThreader
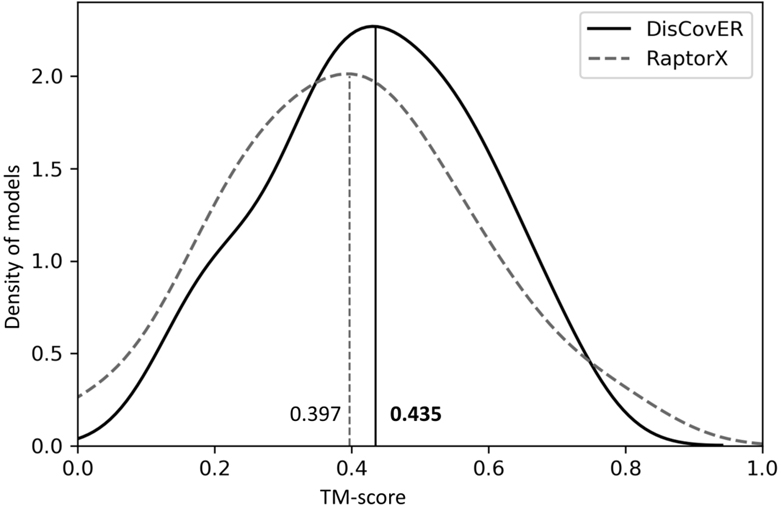
We compare the performance of DisCovER to that of DeepThreader22 on the 60 very hard targets from CAMEO after downloading the predictions submitted by the CAMEO server RaptorX22,24, which, according to the CAMEO assessment paper25, employs the DeepThreader method, otherwise not publicly available to run. As shown in Figure 3, DisCovER achieves a mean TM-score of 0.435, outperforming RaptorX (mean TM-score of 0.397), while skewing the overall TM-score distribution towards higher accuracy. DisCovER attains higher TM-scores for 36 targets (60%) compared to RaptorX. In summary, DisCovER attains better overall performance over the 60 very hard targets from CAMEO for which HHpredB has TM-score less than 0.5. That is, DisCovER outperforms DeepThreader at one of the most challenging threading situations.
Figure 3.
TM-score distribution of DisCovER and RaptorX on 60 very hard targets from CAMEO.
Performance on 480 targets from CATHER
The performance of DisCovER is further benchmarked against recent contact-assisted threading methods: CATHER20 and map_align19 by running DisCovER over 480 targets containing 304 easy, 45 medium, and 131 hard targets used in CATHER and directly comparing the mean TM-score with the results reported in the published work of CATHER20 over the same set. As shown in Table 3, DisCovER outperforms all the competing methods by attaining a mean TM-score of 0.683 over all targets in the dataset, which is about ~0.03 TM-score better than the next best method CATHER. The performance gap between DisCovER and CATHER and the other competing methods is more pronounced with increasing target difficulty. Specifically, while DisCovER outperforms the next-best method CATHER by 0.003 TM-score points for easy targets, the performance gap between DisCovER and CATHER steadily increases to ~0.07 and ~0.1 TM-score points for medium and hard targets, respectively, underscoring the advantage of DisCovER for weakly homologous protein targets. In particular, DisCovER significantly outperforms CATHER and map_align by attaining a mean TM-score of 0.551 as opposed to 0.456 of CATHER and 0.383 of map_align over 131 hard targets. DisCovER also greatly outperforms the reported mean TM-scores of other profile-based methods including HHpred (0.327), SparkX (0.349), and MUSTER (0.359) as well as the other contact-assisted approach EigenTHREADER (0.386) over the hard targets. DisCovER also delivers noticeably better performance than the other methods including contact-assisted approaches CATHER, map_align, and EigenTHREADER as well as the profile-based methods HHpred, SparkX, and MUSTER for the medium difficulty targets. We note that we are unable to perform a target-by-target analysis since the CATHER paper reports only the average performance. Nonetheless, the better average performance of DisCovER across all target categories, especially for the medium and the hard targets, continues to demonstrate its competitive advantage over current threading methods.
Table 3.
Benchmark results on 480 targets from CATHER. Mean TM-score are reported.
| Type | HHpred | SparkX | MUSTER | map_align | EigenTHR EADER | CATHER | DisCovER |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Easy | 0.691 | 0.692 | 0.728 | 0.678 | 0.682 | 0.747 | 0.750 |
| Medium | 0.376 | 0.456 | 0.500 | 0.418 | 0.442 | 0.543 | 0.615 |
| Hard | 0.327 | 0.349 | 0.359 | 0.383 | 0.386 | 0.456 | 0.551 |
| All | 0.562 | 0.576 | 0.606 | 0.573 | 0.579 | 0.649 | 0.683 |
Note: Except DisCovER, the results of other methods are taken from the reported results of CATHER. Values in bold represent the best performance.
Effect of homologous information
The performance of DisCovER is weakly correlated with the number of effective sequence homologs, as quantified by Nf26. As shown in Supplementary Figure S2, the Spearman correlation between TM-score attained by DisCovER and Nf are 0.23 over the 117 hard targets from CAMEO. We have similar observation (maximum Spearman correlation of 0.25) across various target categories from the CATHER dataset. Despite the weak correlation, DisCovER tends to perform suboptimally for targets with very low Nf values. Supplementary Table S1 shows the performance of DisCovER on two representative CAMEO targets: 5ZER_B with an Nf of 104.3 and 6D7Y_B with an Nf of 21.3. While 5ZER_B with a sufficiently high Nf is correctly folded by DisCovER with a TM-score of 0.826, 6D7Y_B with a low Nf results in an incorrect fold having a TM-score of 0.446. In fact, the mean TM-score of DisCovER on CAMEO targets with Nf <20 is 0.36. This is consistent with the current literature 9,22,43–45 and indicates a major limitation of the existing protein modeling methods.
Running time
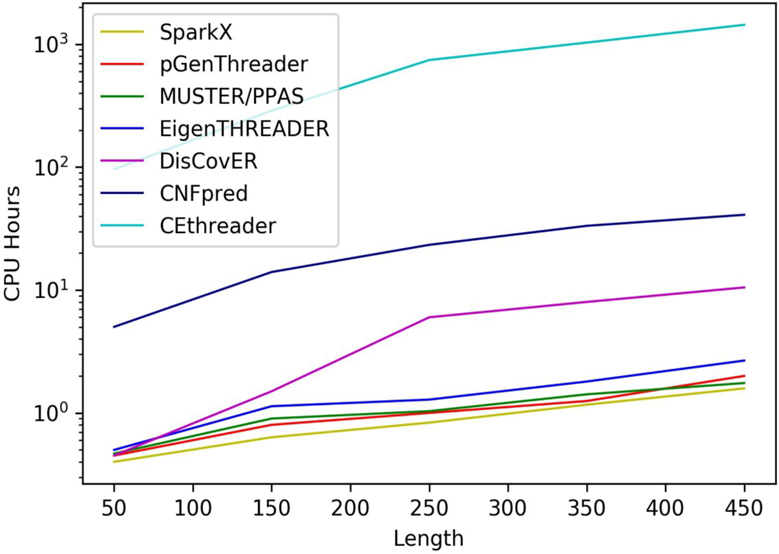
Figure 4 shows the running time of various threading methods with respect to the target length. All methods are run on the same Linux machine with 128 GB RAM and using a single CPU thread of Intel Xeon Processor (2.20 GHz). While it is expected that DisCovER is slower than most profile-based threading methods, but the running time of DisCovER is considerably faster than the top profile-based method CNFpred and orders of magnitude faster than the top contact-assisted approach CEthreader. Overall, DisCovER is reasonably efficient in terms of the running time.
Figure 4.
The running time of eight methods building alignments for 117 hard targets from CAMEO.
DISCUSSION
This article presents DisCovER, a new protein threading method that effectively integrates the covariational signal encoded in inter-residue distance and orientation information along with topological network neighborhood to significantly improve threading template selection and alignment for weakly homologous proteins. Experimental results show that our method yields better accuracy than existing threading methods, including profile-based methods and latest contact-assisted approaches such as CEthreader, EigenTHREADER, map_align, and CATHER. Controlled experiments reveal that distance and orientation information contributes significantly to the superior performance of DisCovER, complemented by the neighborhood effect particularly for weakly homologous proteins. At one of the most challenging threading situations, DisCovER outperforms the CAMEO server RaptorX employing the distance-based threading method DeepThreader. This suggests that our distance- and orientation-based coevolutionary threading method DisCovER is well-suited for remotely homologous targets. Being weakly correlated with the number of sequence homologs available for the query protein and reasonably efficient in terms of its running time, our study opens the possibility of successfully extending threading for many more protein sequences that were previously not amenable to template-based modeling.
The recent CASP14 experiment witnessed the remarkable performance of AlphaFold243 in predicting highly accurate protein 3D structures, outperforming the other participating groups by a large margin. A natural question to ask is whether methods such as DisCovER deliver some value over and beyond the highly accurate AlphaFold2 method. Since AlphaFold2 is an end-to-end deep learning-based ab initio protein structure prediction system that also incorporates templates information from the profile-based threading method HHsearch3,46, a direct comparison between AlphaFold2 and a pure template-based threading method such as DisCovER is not fair. Nonetheless, a case study on two recent CAMEO targets 7APJ_B and 7D8B_B, both having the submission date of August 21, 2021, provide some interesting perspective. 7APJ_B is officially classified as easy, and 7D8B_B is officially classified as medium by CAMEO. We run DisCovER on these two targets using a template library curated in 2018, which is significantly older than both targets’ submission dates. We also employ AlphaFold2 by submitting jobs to the Colab notebook released by DeepMind with default parameter settings. Since the full-fledged version of AlphaFold2 leverages template information by employing HHsearch, we also submit jobs to the HHpred webserver that is based on HHsearch. As shown in Supplementary Table S2, for the easy target 7APJ_B, DisCovER performs better (TM-score of 0.913) than AlphaFold2 (TM-score of 0.826) and much better than HHpred (TM-score of 0.769). For the medium target 7D8B_B, while HHpred fails to predict the correct fold (TM-score of 0.487), both DisCovER (TM-score of 0.847) and AlphaFold2 (TM-score of 0.873) deliver comparable performance. In both cases, DisCovER delivers much better threading performance than HHpred. In the future, it might be interesting to leverage improved threading methods such as DisCovER instead of HHsearch for incorporating template information into the AlphaFold2 system to potentially make it more sensitive and robust, particularly for homologous protein targets. Furthermore, inspired by the success of deep learning-based methods47–51, our future work shall focus on developing improved protein threading methods driven by deep learning, particularly for targets with very low Nf values. This should complement and supplement the state of the art of protein structure prediction.
Supplementary Material
ACKNOWLEDGEMENT
This work was partially supported by the National Institute of General Medical Sciences [R35GM138146 to DB] and the National Science Foundation [IIS-2030722, DBI-1942692 to DB].
Footnotes
Conflict of Interest: none declared.
REFERENCES
- 1.Dill KA, MacCallum JL. The Protein-Folding Problem, 50 Years On. Science. 2012;338(6110):1042–1046. doi: 10.1126/science.1219021 [DOI] [PubMed] [Google Scholar]
- 2.Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Söding J Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125 [DOI] [PubMed] [Google Scholar]
- 4.Wu S, Zhang Y. MUSTER: Improving protein sequence profile–profile alignments by using multiple sources of structure information. Proteins: Structure, Function, and Bioinformatics. 2008;72(2):547–556. doi: 10.1002/prot.21945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Faraggi E, Zhao H, Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics. 2011;27(15):2076–2082. doi: 10.1093/bioinformatics/btr350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobley A, Sadowski MI, Jones DT. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25(14):1761–1767. doi: 10.1093/bioinformatics/btp302 [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Peng J, Wang S, Xu J. A conditional neural fields model for protein threading. Bioinformatics. 2012;28(12):i59–i66. doi: 10.1093/bioinformatics/bts213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J, Wang S, Zhao F, Xu J. Protein threading using context-specific alignment potential. Bioinformatics. 2013;29(13):i257–i265. doi: 10.1093/bioinformatics/btt210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng W, Wuyun Q, Li Y, et al. Detecting distant-homology protein structures by aligning deep neural-network based contact maps. PLOS Computational Biology. 2019;15(10):e1007411. doi: 10.1371/journal.pcbi.1007411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones DT, Buchan DWA, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28(2):184–190. doi: 10.1093/bioinformatics/btr638 [DOI] [PubMed] [Google Scholar]
- 11.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue–residue contact predictions in a sequence- and structure-rich era. PNAS. 2013;110(39):15674–15679. doi: 10.1073/pnas.1314045110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones DT, Singh T, Kosciolek T, Tetchner S. MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics. 2015;31(7):999–1006. doi: 10.1093/bioinformatics/btu791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Hu J, Zhang C, Yu D-J, Zhang Y. ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics. 2019;35(22):4647–4655. doi: 10.1093/bioinformatics/btz291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greener JG, Kandathil SM, Jones DT. Deep learning extends de novo protein modelling coverage of genomes using iteratively predicted structural constraints. Nat Commun. 2019;10(1):1–13. doi: 10.1038/s41467-019-11994-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLOS Computational Biology. 2017;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J Distance-based protein folding powered by deep learning. PNAS. 2019;116(34):16856–16865. doi: 10.1073/pnas.1821309116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D. Improved protein structure prediction using predicted interresidue orientations. PNAS. 2020;117(3):1496–1503. doi: 10.1073/pnas.1914677117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan DWA, Jones DT. EigenTHREADER: analogous protein fold recognition by efficient contact map threading. Bioinformatics. 2017;33(17):2684–2690. doi: 10.1093/bioinformatics/btx217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ovchinnikov S, Park H, Varghese N, et al. Protein structure determination using metagenome sequence data. Science. 2017;355(6322):294–298. doi: 10.1126/science.aah4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du Z, Pan S, Wu Q, Peng Z, Yang J. CATHER: a novel threading algorithm with predicted contacts. Bioinformatics. 2020;36(7):2119–2125. doi: 10.1093/bioinformatics/btz876 [DOI] [PubMed] [Google Scholar]
- 21.Wu Q, Peng Z, Anishchenko I, Cong Q, Baker D, Yang J. Protein contact prediction using metagenome sequence data and residual neural networks. Bioinformatics. 2020;36(1):41–48. doi: 10.1093/bioinformatics/btz477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Wang S, Bu D, Xu J. Protein threading using residue co-variation and deep learning. Bioinformatics. 2018;34(13):i263–i273. doi: 10.1093/bioinformatics/bty278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Wang S. Analysis of distance-based protein structure prediction by deep learning in CASP13. Proteins: Structure, Function, and Bioinformatics. 2019;87(12):1069–1081. doi: 10.1002/prot.25810 [DOI] [PubMed] [Google Scholar]
- 24.Källberg M, Wang H, Wang S, et al. Template-based protein structure modeling using the RaptorX web server. Nature Protocols. 2012;7(8):1511–1522. doi: 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas J, Gumienny R, Barbato A, et al. Introducing “best single template” models as reference baseline for the Continuous Automated Model Evaluation (CAMEO). Proteins: Structure, Function, and Bioinformatics. 2019;87(12):1378–1387. doi: 10.1002/prot.25815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Zheng W, Mortuza SM, Li Y, Zhang Y. DeepMSA: constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins. Bioinformatics. 2020;36(7):2105–2112. doi: 10.1093/bioinformatics/btz863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan R, Yang Y, Paliwal K, Zhou Y. Capturing non-local interactions by long short-term memory bidirectional recurrent neural networks for improving prediction of protein secondary structure, backbone angles, contact numbers and solvent accessibility. Bioinformatics. 2017;33(18):2842–2849. doi: 10.1093/bioinformatics/btx218 [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya S, Bhattacharya D. Does inclusion of residue-residue contact information boost protein threading? Proteins: Structure, Function, and Bioinformatics. 2019;87(7):596–606. doi: 10.1002/prot.25684 [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S, Henikoff JG. Position-based sequence weights. Journal of Molecular Biology. 1994;243(4):574–578. doi: 10.1016/0022-2836(94)90032-9 [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh R, Xu J, Berger B. Global alignment of multiple protein interaction networks with application to functional orthology detection. PNAS. 2008;105(35):12763–12768. doi: 10.1073/pnas.0806627105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C-C, Jeong H, Qian X, Yoon B-J. TOPAS: network-based structural alignment of RNA sequences. Bioinformatics. 2019;35(17):2941–2948. doi: 10.1093/bioinformatics/btz001 [DOI] [PubMed] [Google Scholar]
- 34.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- 35.Smith TF, Waterman MS. Identification of common molecular subsequences. Journal of Molecular Biology. 1981;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5 [DOI] [PubMed] [Google Scholar]
- 36.Taylor WR. Protein structure comparison using iterated double dynamic programming. Protein Science. 1999;8(3):654–665. doi: 10.1110/ps.8.3.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb B, Sali A. Protein Structure Modeling with MODELLER. In: Kihara D, ed. Protein Structure Prediction. Methods in Molecular Biology. Springer; 2014:1–15. doi: 10.1007/978-1-4939-0366-5_1 [DOI] [PubMed] [Google Scholar]
- 38.Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins: Structure, Function, and Bioinformatics. 2004;57(4):702–710. doi: 10.1002/prot.20264 [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26(7):889–895. doi: 10.1093/bioinformatics/btq066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senior AW, Evans R, Jumper J, et al. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins: Structure, Function, and Bioinformatics. 2019;87(12):1141–1148. doi: 10.1002/prot.25834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng W, Li Y, Zhang C, Pearce R, Mortuza SM, Zhang Y. Deep-learning contact-map guided protein structure prediction in CASP13. Proteins: Structure, Function, and Bioinformatics. 2019;87(12):1149–1164. doi: 10.1002/prot.25792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger SJ, Söding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics. 2019;20(1):473. doi: 10.1186/s12859-019-3019-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu F, Xu J. Deep template-based protein structure prediction. PLOS Computational Biology. 2021;17(5):e1008954. doi: 10.1371/journal.pcbi.1008954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong L, Ju F, Zheng W-M, et al. ProALIGN: Directly learning alignments for protein structure prediction via exploiting context-specific alignment motifs. bioRxiv. Published online January 22, 2021:2020.12.28.424539. doi: 10.1101/2020.12.28.424539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H, Shen Y. Template-based prediction of protein structure with deep learning. BMC Genomics. 2020;21(11):878. doi: 10.1186/s12864-020-07249-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao M, Skolnick J. A novel sequence alignment algorithm based on deep learning of the protein folding code. Bioinformatics. 2021;37(4):490–496. doi: 10.1093/bioinformatics/btaa810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morton JT, Strauss CEM, Blackwell R, Berenberg D, Gligorijevic V, Bonneau R. Protein Structural Alignments From Sequence; 2020:20.20.11.03.365932. doi: 10.1101/2020.11.03.365932 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.