Abstract
Ryanodine receptor 1 (RyR1) is an intracellular calcium ion (Ca2+) release channel required for skeletal muscle contraction. Although cryo-electron microscopy identified binding sites of three coactivators Ca2+, ATP and caffeine (CFF), the mechanism of co-regulation and synergy of these activators is unknown. Here, we report allosteric connections among the three ligand binding sites and pore region in (i) Ca2+ bound-closed, (ii) ATP/CFF bound- closed, (iii) Ca2+/ATP/CFF bound-closed, and (iv) Ca2+/ATP/CFF bound-open RyR1 states. We identified two dominant networks of interactions that mediate communication between the Ca2+ binding site and pore region in Ca2+ bound-closed state, which partially overlapped with the pore communications in ATP/CFF bound-closed RyR1 state. In Ca2+/ATP/CFF bound-closed and -open RyR1 states, co-regulatory interactions were analogous to communications in the Ca2+ bound-closed and ATP/CFF bound-closed states. Both ATP- and CFF- binding sites mediate communication between the Ca2+ binding site and the pore region in Ca2+/ATP/CFF bound - open RyR1 structure. We conclude that Ca2+, ATP, and CFF propagate their effects to the pore region through a network of overlapping interactions that mediate allosteric control and molecular synergy in channel regulation.
Keywords: Ryanodine receptor, allosteric coupling, percolation theory, computational analysis, single channel measurements, allosteric interactions
Introduction
RyRs are homo-tetrameric intracellular Ca2+ channels that mediate the rapid release of Ca2+ ions from the sarcoplasmic reticulum (SR) into the cytoplasm for muscle contraction (1-5). RyRs are the largest known ion channels with molecular mass of ~2.2 megadaltons comprised of four 560-kDa RyR subunits and four ~12-kDa FK506 binding protein subunits. There are three RyR isoforms in mammalian cells: RyR1 in skeletal muscle, RyR2 in heart muscle, and RyR3 in many tissues but in lower concentrations. RyR1 is controlled by direct interaction with Cav1.1 voltage-gated Ca2+ channels, and by Ca2+ through an unknown mechanism (3, 6, 7). In contrast, influx of extracellular Ca2+ through voltage-gated Ca2+ channels activate RyR2 and RyR3 and triggers the release of Ca2+ by Ca2+-induced Ca2+ release mechanism. Endogenous modulators, Ca2+, ATP, Mg2+, and calmodulin along with the exogenous effectors, CFF and Ryanodine regulate the function of RyR1 (3). Despite the presence of activator binding sites in close proximity to the pore region of RyRs and the possibility of allosteric communications within the channels, the mechanism of allosteric regulation is still unclear (8-12).
Recent advances in single-particle cryo-electron microscopy (cryo-EM) have captured RyR1 in multiple states that implicate Ca2+, ATP and CFF as key activators in RyR1 regulation (8-10, 13-17). High resolution cryo-EM structures emphasized binding of Ca2+, ATP, and CFF as trigger for local and global structural changes in RyR1 to induce channel opening (10, 17). Mutagenesis and single-channel recordings have revealed the role of Ca2+ binding site residues Glu-3893, Glu-3967, and Thr-5001 in Ca2+-dependent activation of RyR1 (18). Further, Murayama et al demonstrated that Trp-4716 in the CFF-binding pocket regulate Ca2+ sensitivity by controlling the structure of the Ca2+-binding site (19). Additionally, recent experimental and computational studies suggested that allosteric gating of RyR1 is influenced by extensive structural changes in the RyR1 peripheral and central domains (11, 12, 17) due to the binding of activating ligands. The association of these modulators specifically alter the conformation of cytoplasmic portion of the core region, which is also known as the “activation module”. The activation module comprises EF-hand pair (EF1&2) domain, Thumb and Forefingers (TaF) domain, Helical-bundle domain between S2 and S3 (S2S3), S6c pore helix, and the Carboxy-terminal domain (CTD). While the cryo-EM structures and experimental studies provide insights into ligand binding and structural states, the allosteric mechanisms that couple and synergize communication among the ligand binding sites and pore region in RyR1 are not well understood.
In this study we present the structural basis for allosteric control of Ca2+-binding site and pore region in open and closed RyR1 channels. We also show how binding of ATP and CFF alters the allosteric network and affects Ca2+-free and Ca2+-occupied channel activities. Conformational changes related to channel opening or closing appear to alter the allosteric network. Synergistic interactions among the three activators Ca2+, ATP and CFF propagate their effects to the pore region through a network of connecting amino acid residues.
Materials and Methods
Mapping allosteric communication in different functional states of RyR1
We analyzed the cryo-EM structures of different RyR1 states: Ca2+ bound-closed RyR1 (PDB ID: 5T15), ATP/CFF bound-closed RyR1 (PDB ID: 5TAP), Ca2+/ATP/CFF bound-closed RyR1 (PDB ID: 5TAQ), and Ca2+/ATP/CFF bound-open RyR1 (PDB ID: 5TAL) to map allosteric interactions among the activator binding sites using our recently developed methodology (20-23). We chose Ca2+, ATP, CFF Gly-4934 the channel gating residue (24) to annotate the pore region. The detailed methodology of allosteric interaction prediction among the chosen sites is described below.
Construction of protein graphs.
Graph representations G(N, E) were constructed for each channel state, where ni are nodes corresponding to each protein residue, eij are edges connecting each pair of nodes i and j and i,j=1…N corresponds to residue number, where N is the protein size (25). To further incorporate structural information of the protein, we assigned weight (eij) for each graph edge as shown below.
| [1] |
and
| [2] |
where H(x) is a Heaviside step function and r0 =3.4 Å is a cutoff distance. The summation is over all pairs of heavy atoms ai and aj (amino acid atoms excluding hydrogens, i.e. C, O, N, S) in two residues of the protein i and j. The parameter denotes pair distance between two heavy atoms in residues i and j. Equation [2] represents a simplified method to quantify the strength of interaction between amino acid residues in the protein. In this case, wij favors edges of the graph, which connect nodes ni and nj corresponding to stronger interacting residues, i and j, of the protein. If two residues have a distance between any of their atoms greater than r0, corresponding edge weight for those residues was set as zero. The detailed methodology on construction and evaluation of protein structure graphs based on edge weights can be found in the ref: (23).
Shortest pathway and paths clustering.
To map allosteric interactions between two residues, the shortest paths between two nodes (either ligand–ligand or ligand–Gly-4934) on the graph were initially identified. Yen’s algorithm was used to compute rank-ordered loop-less optimal interactions (26). For each iteration, Yen’s algorithm uses Dijkstra algorithm (26, 27), which calculates the optimal connection responsible for propagation of communication through the network. The product of edge weights along the path was used as a metrics to quantify communication strength along the pathway:
| [3] |
Representative allosteric interactions (optimal paths) were predicted by hierarchical clustering of the top 10,000 optimal communication pathways using the measure for the dissimilarity between them,
| [4] |
where, nl and nm denote the number of nodes in path l and m, and (0 ≤ nlm ≤ min{nl,nm}) is the number of common nodes in both paths. Clustering according to this distance allows to combine pathways with minimal deviations from each other (large nlm). After each step of clustering, we consider that the representative path carries the allosteric communication equal to the sum of those carried by all merged pathways. There are several ways to choose the cutoff for hierarchical clustering (28). The detailed methodology on construction and evaluation of protein structure graphs based on edge weights can be found in the ref: (23).
In this work, we observed that the allosteric networks always had several strongest distinctive pathways that were carried through the clustering procedure and were always becoming representative interactions in the end of clustering. Based on this observation, for simplicity of representation we show and analyzed only top five pathways (Figs. 2-5). We performed allosteric interaction analyses on tetrameric rather than monomeric conformations of RyR1 in different functional states as stated above. All structural figures were rendered using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
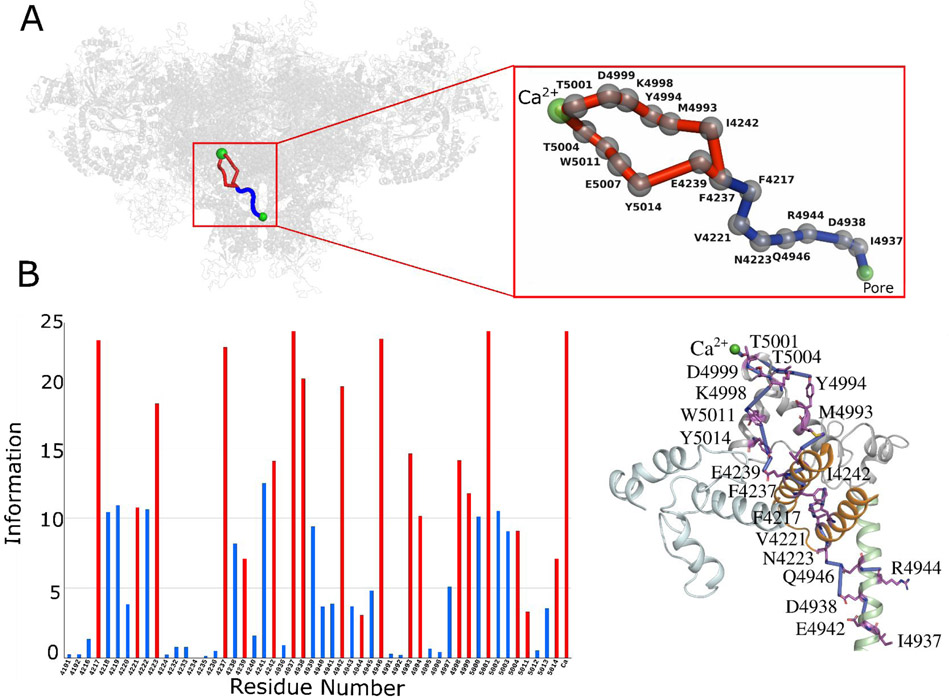
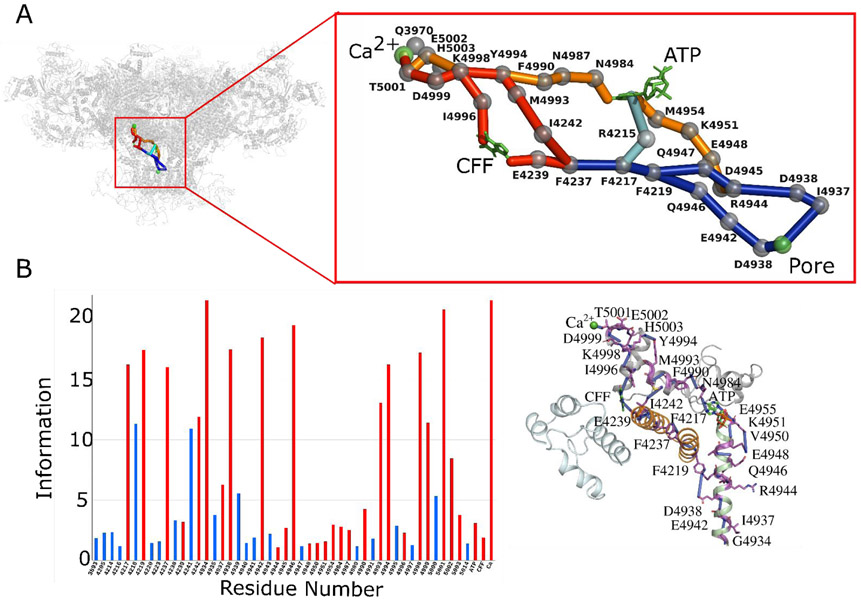
Fig. 2. Allosteric pathway analysis on Ca2+ bound-closed RyR1 structure (PDB ID: 5T15).
(A) The network of allosteric connections between Ca2+ binding site and pore region in Ca2+ bound-closed RyR1 structure. Protein is shown in transparent gray cartoon representation and Ca2+ and pore lining residue Gly-4934 as green sphere. The allosteric pathways between Ca2+ and pore regions are shown in red and blue with Cα atoms of allosteric information passing residues (hot spot residues) as gray spheres. (B) Allosteric communication between Ca2+ binding site and pore region. Bars corresponding to residues participating in most optimal allosteric pathway between Ca2+ binding site and pore region are highlighted in red. Bars for residues participating in other possible but least significant pathways are colored in blue. The height of bars in each chart represents the extent of allosteric information transmitted through the corresponding residue. Inter-residue interactions are rendered as blue cylinders. Color code for RyR1 domains participating in allosteric communication among the ligand sites is as follows: TaF domain (4175–4253) in orange, helical-bundle domain between S2 and S3 (4666–4786) in cyan, channel pore domain (4820–4956) in green, and CTD (4957–5037) in gray. ATP and CFF are shown in green licorice representation, and Ca2+ is shown in green van der Waals representation.
Fig. 5. Mapping allosteric pathways in Ca2+/ATP/CFF bound-open RyR1 structure (PDB ID: 5TAL).
(A) The detailed network of allosteric connections among the three ligand sites (CFF, ATP, and Ca2+) and the pore region in Ca2+/ATP/CFF bound-open RyR1 structure. Protein is shown in transparent gray cartoon representation, ATP and CFF are shown in green licorice, and Ca2+ and pore lining residue Gly-4934 are as green spheres. Color code for allosteric pathways shown in this figure is as follows: Ca2+-pore and Ca2+-CFF-pore in red and blue, Ca2+-ATP-pore in orange and blue, and CFF-ATP in cyan. Cα atoms of allosteric information passing residues (hot spot residues) are as gray spheres. (B) Allosteric communication between Ca2+, ATP and CFF binding sites and pore region. Bars corresponding to residues participating in most optimal allosteric pathway between Ca2+ binding site and pore region are highlighted in red. Bars for residues participating in other possible but least significant pathways are colored in blue. The height of bars in each chart represents the extent of allosteric information transmitted through the corresponding residue. The interactions between residues participate in allosteric communications between ligand sites are rendered as blue cylinders. Color code for RyR1 domains participating in allosteric communication among the ligand sites is as follows: TaF domain (4175–4253) in orange, helical-bundle domain between S2 and S3 (4666–4786) in cyan, channel pore domain (4820–4956) in green, and CTD (4957–5037) in gray. ATP and CFF are shown in green licorice representation, and Ca2+ is shown in green van der Waals representation.
Results
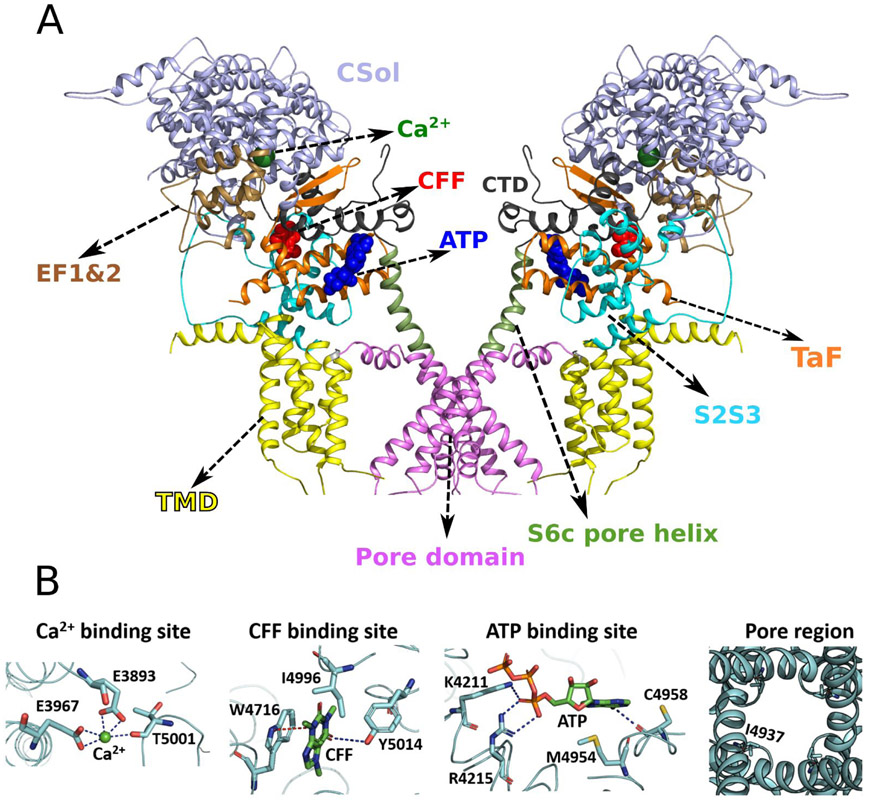
Cryo-EM structures of RyR1 solved in different functional states have identified the binding sites for channel activators Ca2+, ATP and CFF at inter-domain interfaces of the C-terminal domain (CTD) (Fig. 1) (10). Comparison of cryo-EM density maps in the presence of Ca2+ but without ATP/CFF, and in the presence of ATP/CFF without Ca2+ indicate variations in densities of core solenoid (residues 3667-4174) and CTD (residues 4957-5037) of RyR1, that suggest the synergy in the activation of RyR1 by Ca2+ and ATP/CFF (10). Co-localization of ATP, CFF, and Ca2+ binding sites at interfaces of the CTD suggests allosteric communication between these binding sites during channel opening and/or closing.
Fig. 1. Domain organization and location and structure of ligand binding sites in RyR1 (PDB ID: 5TAL).
(A) Co-localization of ATP, CFF, and Ca2+ binding sites at interfaces of CTD suggests allosteric communication between these binding sites during channel opening and/or closing. Only two monomers out of four are shown for better visualization of ligand binding sites and RyR1 domains. Protein is shown in cartoon representation and ATP, CFF, and Ca2+ are shown in blue, red, and green van der Waals representation, respectively. The important domains of RyR1 that participate in allosteric communication are colored as follows: Core Solenoid (CSol) domain in light blue, EF-hand pair (EF1&2) domain in brown, Thumb and Forefingers (TaF) domain in orange, Helical-bundle domain between S2 and S3 (S2S3) in cyan, Transmembrane domain (TMD) in yellow, Pore domain in magenta, S6c pore helix in green, and Carboxy-terminal domain (CTD) in black. (B) Structural conformations of ligand binding sites (Ca2+, CFF, and ATP) and pore region of Ca2+/ATP/CFF bound-open RyR1 (PDB ID: 5TAL). The side chain of Ile-4937 (constriction site) is shown in licorice to demonstrate the open pore conformation.
To establish an allosteric network among Ca2+, ATP, and CFF binding sites during channel activation, allosteric interaction analysis was performed on tetrameric conformations of four structures: Ca2+ bound-closed RyR1 (PDB ID: 5T15), ATP/CFF bound-closed RyR1 (PDB ID: 5TAP), Ca2+/ATP/CFF bound-closed RyR1 (PDB ID: 5TAQ), and Ca2+/ATP/CFF bound-open RyR1 (PDB ID: 5TAL). A graph-theoretical approach was used to depict allosteric interactions among the chosen ligand binding sites in RyR1 structures by comparing relative allosteric communication transmitted through each residue in a specific sequence of interactions. The bar charts specific to each pathway illustrate the amount of relative allosteric communication transmitted by each residue specific to that pathway and subsequently highlight allosteric hot spot residues crucial for the communication (Figs. 2-5). Thus, communication between ligand binding sites and pore region was predicted with maximal contribution to the allosteric couplings.
Ca2+ binding site to pore communication propagate through unoccupied CFF-binding site in Ca2+ bound- closed RyR1.
The binding site for Ca2+ is constituted by side chains Glu-3893 and Glu-3967 (core solenoid) and the backbone of the CTD residue Thr-5001 (Fig. 1B and Fig. S1). Cryo-EM data indicate the Ca2+ binding site is situated in close proximity to the ATP and CFF binding sites and the three sites are likely coupled to regulate binding or release of Ca2+ either directly or indirectly (Fig. 1A) (10). The influence of unoccupied CFF and ATP binding sites on Ca2+ to pore communication was determined by analyzing Ca2+ only bound-closed RyR1 structure (PDB ID: 5T15) using percolation theory and computational modeling techniques. The allosteric network of the Ca2+ bound-closed RyR1 structure contained two main Ca2+ binding site pore interactions, a primary connection propagating through CFF binding site residues Tyr-5014 and Glu-4239, and a secondary one independent of CFF and ATP binding sites (Fig. 2). The primary interactions initiated from Ca2+-binding pocket residue Thr-5001 progressed through CTD residues Thr-5004, Trp-5011, Glu-5007 to CFF binding site residues Tyr-5014 (CTD) and Glu-4239 (TaF domain). Allosteric communication was further propagated via hydrophobic interactions between Glu-4239 and Phe-4237 (TaF domain), aromatic interactions between Phe-4237 and Phe-4217 (TaF domain), and hydrophobic interactions among TaF domain residues Phe-4217, Val-4221, Asn-4223, and S6c helix residues Gln-4946, Arg-4944, Asp-4938, and Ile-4937 (constriction site of the Ca2+ bound-closed RyR1 channel) (10, 14, 30). The secondary interactions also initiated from Ca2+ binding pocket residue Thr-5001 but progressed along CTD residues Asp-4999, Lys-4998, Tyr-4994, and Met-4993 through covalent or hydrophobic interactions (Fig. 2). Allosteric signaling advanced from the CTD to the TaF domain through tightly packed Ile-4242 and Met-4993 sidechains. As observed in the primary interaction, the TaF domain residues: Phe-4237, Phe-4217, Val-4221, Asn-4223 along with the S6c helix residues: Gln-4946, Arg-4944, Asp-4938 and Ile-4937 mediated the signal to the pore region. Thus, in Ca2+ bound-closed RyR1, Ca2+ binding site communicate to the pore region via the CTD, the TaF, and the S6c domains. Interestingly, the Ca2+ - pore interactions propagated through the unoccupied CFF binding site but evade the unoccupied ATP binding site.
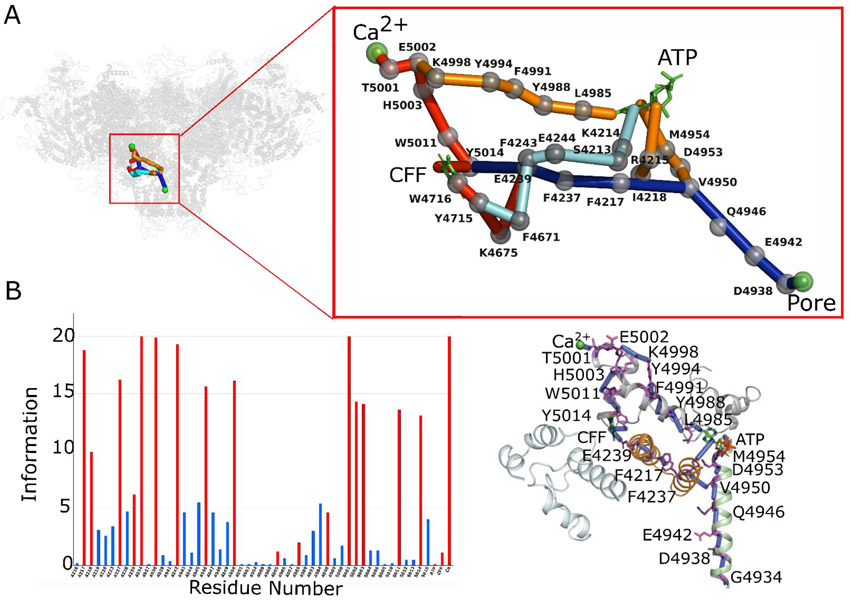
ATP- and CFF- binding sites communicate with the pore region in ATP/CFF bound-closed RyR1.
Previous studies suggest that ATP and CFF activate RyR1 in the absence or presence of Ca2+ (18). In this work, a potential network of allosteric connections among ATP-,CFF- binding sites, and pore region in the absence of Ca2+ was investigated using the ATP/CFF bound-closed RyR1 (PDB ID: 5TAP) (Fig. 3). Three major interactions were predicted involving polar, aromatic, and non-covalent residues in the CTD, TaF, S2S3, and S6c helix domains (Fig. 3). The major pathway initiated from the CFF binding site residue Glu-4239 resembled allosteric interactions in Ca2+ bound-closed RyR1 structure that connected the TaF domain residues Phe-4237, Phe-4217, Phe-4219, and S6c helix residues Gln-4947, Arg-4944, Asp-4938 and Ile-4937. A second interaction originated from the CFF binding site residue Trp-4716 proceeded through the S2S3 domain residues Lys-4675, Phe-4671 and Lys-4672 and merged with the major pathway. A third pathway initiated from the CFF site residue Ile-4996 passed through ATP binding site residue Met-4954 and merged with previously solved pathways at S6c residue: Gln-4947 (Fig. 3). Finally, the merged pathways passed through the CTD residues: Leu-4992, Met-4989, Ala-4986, Leu-4985 and S6c residues: Met-4954, Glu-4955, Lys-4951, Glu-4948, Gln-4947, Arg-4944, Asp-4938 and communicated the pore region (Figs. 3, S1, S4A). Altogether, three different networks of allosteric interactions originate from the CFF binding site progressed through the activation module to communicate with the pore region. During the progression of the allosteric signal from the CFF-binding site, while one pathway interacted the ATP-binding site, remaining two pathways avoided it.
Fig. 3. Mapping allosteric pathways in ATP/CFF bound-closed RyR1 structure (PDB ID: 5TAP).
(A) Network of allosteric connections among ATP and CFF ligand binding sites and pore region in ATP/CFF bound-closed RyR1 structure. Protein is shown in transparent gray cartoon representation, ATP and CFF are shown in green licorice, and pore lining residue Gly-4934 is shown as green sphere. Color code for allosteric pathways shown in this figure is as follows: CFF-pore in red and blue, ATP-pore in orange and blue, and CFF-ATP in cyan. Cα atoms of allosteric information passing residues (hot spot residues) are as gray spheres. (B) Allosteric communications between CFF - pore and ATP - pore regions. Bars corresponding to residues participating in most optimal allosteric pathway between Ca2+ binding site and pore region are highlighted in red. Bars for residues participating in other possible but least significant pathways are colored in blue. The height of bars in each chart represents the extent of allosteric information transmitted through the corresponding residue. Inter-residue interactions are rendered as blue cylinders. Color code for RyR1 domains participating in allosteric communication among the ligand sites is as follows: TaF domain (4175–4253) in orange, helical-bundle domain between S2 and S3 (4666–4786) in cyan, channel pore domain (4820–4956) in green, and CTD (4957–5037) in gray. ATP and CFF are shown in green licorice van der Waals representations.
Three diverse networks of allosteric interactions communicate the effects of Ca2+ binding to the pore region in the Ca2+/ATP/CFF bound-closed RyR1 structure.
Three sets of allosteric interactions predicted in the Ca2+/ATP/CFF bound-closed RyR1 structure (PDB ID: 5TAQ) involve mostly polar and aromatic residues to mediate the communication between the Ca2+ binding site and pore region. While two networks of interactions passed through the CFF- (Fig. 4, Fig. S2A, and Fig. S5A) and ATP- binding sites (Fig. 4, Fig. S3A, and Fig. S6A), a third network bypassed these two binding sites (Fig. 4). The three predicted allosteric networks of interactions between the Ca2+ binding site and pore region are as follows: (a) CTD residues Thr-5001, Asp-4999, Lys-4998, Ile-4996 (CFF binding site), TaF residues Glu-4239 (CFF binding site), Phe-4237, Phe-4217, Phe-4219, and S6c residues Gln-4946, Glu-4942, Asp-4938, Ile 4937, Gly-4934, (b) Core Solenoid (CSol) residue Glu-3970, CTD residues Glu-5002, Asn-5003, Lys-4998, Tyr-4994, Met-4993, TaF residues Ile-4242, Phe-4237, Phe-4217, Phe-4219, and S6c residues Gln-4946, Glu-4942, Asp-4938*, Ile-4937*, Gly-4934, and (c) Core Solenoid residue Glu-3970, CTD residues Glu-5002, Asn-5003, Lys-4998, Tyr-4994, Phe-4990, Asn-4987, Asn-4984, and S6c residues Met-4954 (ATP binding site), Lys-4951, Glu-4948, Asp-4945, Arg-4944, Asp-4938*, Ile-4937* and Gly-4934. It is worth mentioning here that the allosteric signal in network-b and network-c passed from S6c helix in one monomer to S6c helix in another monomer before terminating at the pore region (denoted by * above) (Fig. 4). Interestingly, the TaF domain residues Glu-4239, Phe-4237, Phe-4217 and Arg-4215 provided direct communication between the ATP- and CFF- binding sites in Ca2+/ATP/CFF bound-closed RyR1 (Fig. 4 and Fig. S4B).
Fig. 4. Co-regulation interactions in Ca2+/ATP/CFF bound-closed RyR1 structure (PDB ID: 5TAQ).
(A) The detailed network of allosteric connections among the three ligand sites (CFF, ATP, and Ca2+) and the pore region in Ca2+/ATP/CFF bound-closed RyR1 structure. Protein is shown in transparent gray cartoon representation, ATP and CFF are shown in green licorice, and Ca2+ and pore lining residue Gly-4934 are as green spheres. Color code for allosteric pathways shown in this figure is as follows: Ca2+-pore and Ca2+-CFF-pore in red and blue, Ca2+-ATP-pore in orange and blue, and CFF-ATP in cyan. Cα atoms of allosteric information passing residues (hot spot residues) are as gray spheres. (B) Allosteric communication between Ca2+, ATP and CFF binding sites and pore region. Bars corresponding to residues participating in most optimal allosteric pathway between binding sites and pore region are highlighted in red. Bars for residues participating in other possible but least significant pathways are colored in blue. The height of bars in each chart represents the extent of allosteric information transmitted through the corresponding residue. The interactions between residues participate in allosteric communication between ligand sites are rendered as blue cylinders. Color code for RyR1 domains participating in allosteric communication among the ligand sites is as follows: TaF domain (4175–4253) in orange, Helical-bundle domain between S2 and S3 (4666–4786) in cyan, channel pore domain (4820–4956) in green, and CTD (4957–5037) in gray. ATP and CFF are shown in green licorice representation, and Ca2+ is shown in green van der Waals representation.
The comparison of allosteric interactions in different RyR1 functional states denote that several partial overlapped networks were evident, specifically among the closed Ca2+ bound-closed, ATP/CFF bound-closed and Ca2+/ATP/CFF bound-closed states (Fig. S2-S6). Furthermore, the interactions that bypassed the ATP- and CFF- binding sites partially overlapped with the network of interactions in Ca2+ bound-closed state. Partial alignment of interactions between ATP/CFF bound-closed and Ca2+/ATP/CFF bound-closed states emphasize the regulatory role of CFF and ATP in Ca2+-dependent RyR1 activation. Further, the binding of ATP and CFF molecules altered the orientation of the CTD and S6c domains and amplified the communication between Ca2+ and pore region to modulate pore opening and closing. Hence, ATP and CFF act as modulators for Ca2+ to pore communication and subsequent RyR1 activation.
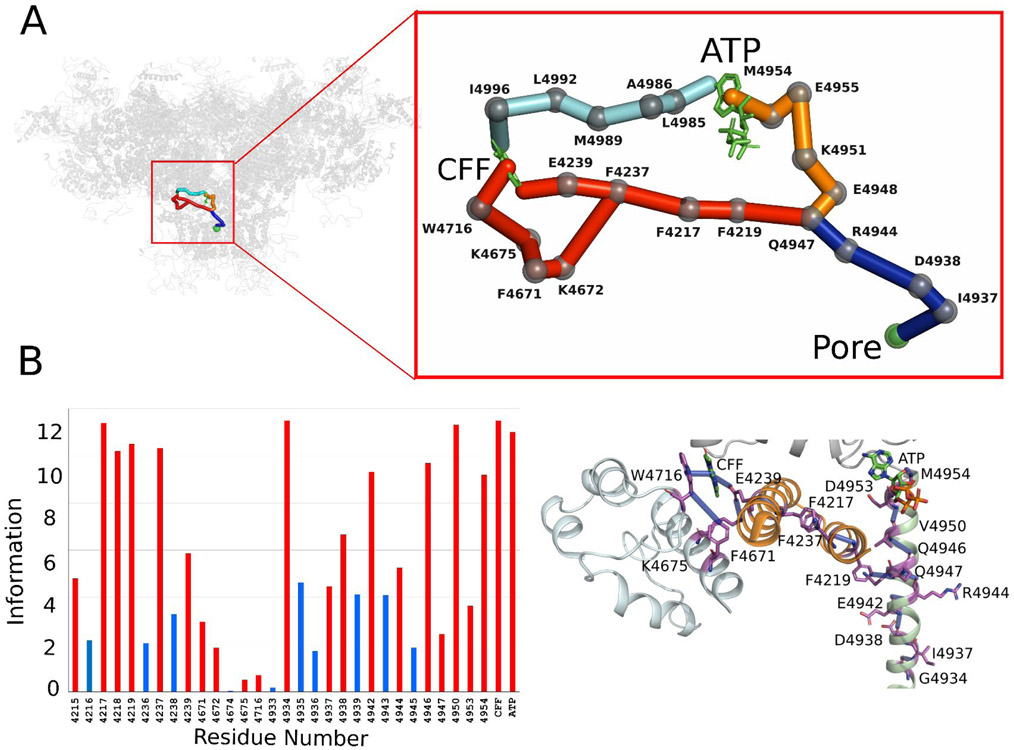
ATP- and CFF- binding sites mediate the communication between Ca2+- and the pore in Ca2+/ATP/CFF bound - open RyR1 structure.
Three allosteric interactions were predicted to transduce the signal from the Ca2+ binding site to the pore region through either CFF or ATP binding sites in Ca2+/ATP/CFF bound-open RyR1 structure (PBD ID: 5TAL) (Fig. 5). Two networks of interactions passed through the CFF site initiated from Ca2+ binding pocket residue Thr-5001 and proceeded through CTD residues Glu-5002, His-5003, Trp-5011 to Tyr-5014 (Fig. S1). Allosteric communication between the CTD and the TaF domain was propagated through a series of overlapping interactions among the CTD (Tyr-5014) and the S2S3 (Trp-4716, Tyr-4715 and Lys-4675 to Glu-4239) domains. Subsequently, the allosteric signal from the TaF reside Glu-4239 progressed through Phe-4237, Phe-4217, Ile-4218, and S6c residues Val-4950, Gln-4946, Glu-4942 and Asp-4938 to reach the pore region. Hence, in the Ca2+/ATP/CFF bound-open RyR1 structure CFF binding site residues Tyr-5014 and Glu-4239 were predicted to propagate the signal between the Ca2+ binding site and pore region (Figs. S2B and S5B) by transferring the signal to TaF residues Phe-4237 and Phe-4217. The other connection that passed the signal from the Ca2+ binding site to pore through the ATP binding site, instead of the CFF binding site, was predicted to proceed through the CTD residues Thr-5001, Glu-5002, Lys-4998, Tyr-4994, Phe-4991, Tyr-4998, Leu-4985 and S6c residues Met-4954, Asp-4953, Val-4950, Gln-4946, Glu-4942, Asp-4938, and Ile-4937. S2S3 residues Tyr-4715 and Phe-4671 and TaF residues Phe-4243, Glu-4244, Ser-4213, and Lys-4214 provided a direct link between the CFF and ATP binding sites. Notably, the communication that progressed through the S2S3 domain is most dominant due to its strength of interactions and synergistic communication among TaF, S2S3, CTD and pore domains.
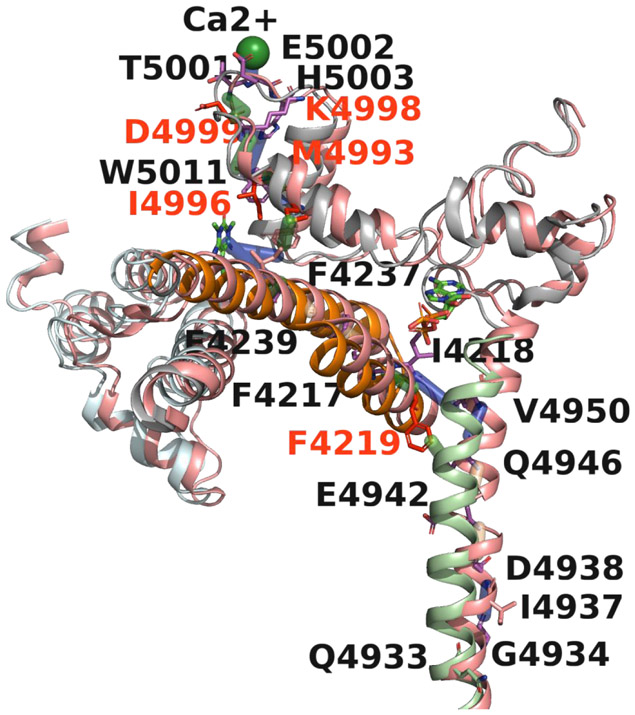
High resolution cryo-EM structures (10) revealed that binding of Ca2+, ATP, and CFF initiate structural changes in peripheral and central domains associated with transition between open and closed states of RyR1. Fig. 6 as an example provides a visual illustration for the Ca2+ binding site to pore communication in closely related primary Ca2+/ATP/CFF bound-open and Ca2+/ATP/CFF bound-closed structures passing through the CFF binding site, CTD, TaF, and S6c domains encompassing the two allosteric interactions. Specifically, the twisting and outward tilting of the S6c helix from the vertical axis opens up the channel to Ca2+ by rotating the Cα atoms of constriction site residues Ile-4937 (closed) and Gln-4933 (open) away from the pore (10, 30).
Fig 6. Structural basis of RyR1 channel gating.
Shown is a visual illustration of Ca2+ to pore communication in Ca2+/ATP/CFF bound - open and Ca2+/ATP/CFF bound- closed structures. The structure of open RyR1 is shown in salmon and closed RyR1 in color based on domains for better visualization. The pathway passing through the CFF binding site, CTD (gray), TaF (orange) and S6c (light green) domains encompassing the two allosteric interactions. Ile-4937 and Gln-4933 are the constriction sites of the closed and open channel, respectively. The differential interactions in closed-RyR1 are colored green and corresponding residues are shown in red. Twisting and outward tilting of the S6c helix from the vertical axis opened the channel to Ca2+ by rotating the Cα atoms of Ile-4937 and Gln-4933 away from the pore.
Discussion
The assessment of allosteric interactions between Ca2+ binding site and pore region in various RyR1 functional states highlighted structural differences in the organization of activation module. Specifically, the CTD exhibited distinct differences in transducing the effects of Ca2+ binding on the ATP or CFF binding sites. The TaF domain residues Phe-4217 and Phe-4237 were identified as part of an allosteric network in all four structures that mediated the signal from the CFF binding site to the pore region. Involvement of the S2S3 domain in transducing the effects of Ca2+ to the TaF domain was predicted to be limited to the ATP/CFF bound-closed and Ca2+/ATP/CFF bound-open structures. Structural differences in the organization of the CTD and S6c domains in ATP/CFF bound-closed, Ca2+/ATP/CFF bound-closed, and Ca2+/ATP/CFF bound-open states demonstrate that significant structural changes occur in the activation module during the Ca2+ dependent RyR1 activation. We also predicted major differences in the communication between the ATP and CFF sites in the ATP/CFF bound-closed and Ca2+/ATP/CFF bound-closed and -open states. Previous studies showed that the binding of CFF in the presence of an adenine nucleotide altered the orientation of Trp-4716 in the TaF domain and induced conformational changes in CTD to modulate Ca2+ binding pocket structure for more favorable binding of Ca2+ (19). The subsequent binding of Ca2+ initiated reorientation of the CTD and thereby induced flexible alterations in the activation module, which comprises EF1&2, TaF, S2S3, S6c helix, and CTD. The activation module thus acquired a conformation and orientation responsible for pore enlargement as evident from the cryo-EM data of Ca2+/ATP/CFF bound-open RyR1 structure (10).
Several amino acid variants in the predicted allosteric interactions were shown to affect RyR1 function. RyR1-T5001A and RyR1-Q3970K in the Ca2+ binding site exhibited an altered Ca2+-dependent single channel activity, and RyR1-Q3970K displayed reduced activation by CFF or ATP (18, 31). RyR1-W4716A and RyR1-I4996A variants in the CFF-binding pocket of RyR1 and RyR2 increased Ca2+ sensitivity in [3H]ryanodine binding ligand assay by controlling the structure of the Ca2+ binding site (19). Mutations in the S6c helix such as RyR1 residues Gln-4933, Ile-4937, Asp-4838, Gln-4946, Gln-4947 and Gly-4934 impaired Ca2+-gated activity and/or reduced Ca2+ conductance (30, 32, 33). Thus, combined impaired Ca2+ activation and ion conductance upon mutating pore-lining helix residues suggested allosteric coupling between the Ca2+ binding site and pore region.
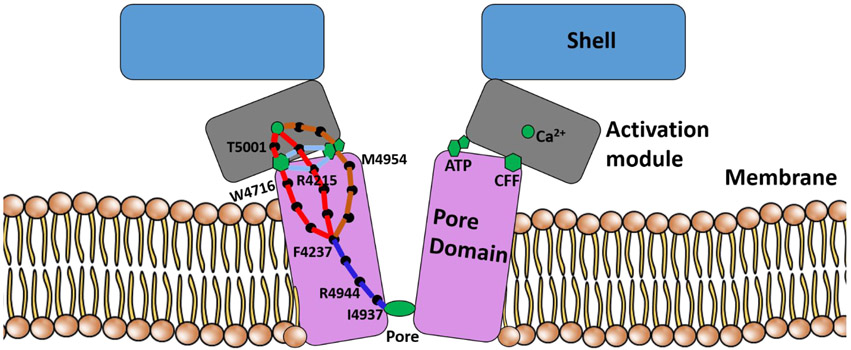
To summarize, the allosteric interactions among three ligand binding sites and the pore region of RyR1 were investigated in four functional states. The studies predicted that the opening and closing of the pore is allosterically coupled to Ca2+, ATP and CFF binding sites and that the communication between Ca2+ binding to pore region is mediated by CFF binding and ATP binding site residues in both open and closed RyR1 structures. Previous studies showed that the addition of ATP and CFF increased channel opening probability in RyR1-WT single channel measurements in the presence and absence of Ca2+, and the combined addition of ATP and CFF further increased channel activity. Thus, the enhanced channel opening probability in presence of Ca2+, ATP, and CFF suggests allosteric coupling among Ca2+, ATP, CFF and the pore region (Fig. 7). Studies in the absence of CFF will provide more direct insights into the allostery mediated by the two physiological activators Ca2+ and ATP. An extension to other endogenous ligands such as Mg2+ and calmodulin or exogenous effectors may corroborate a complex network of interacting domains and assist in designing therapeutics for skeletal muscle myopathies such as core diseases and malignant hyperthermia or catecholaminergic polymorphic ventricular tachycardia in the heart.
Fig 7. Molecular basis of synergy of channel regulation by multiple activators in closed RyR1.
Schematic representation of residues responsible for initiating allosteric signal from each ligand-binding site and residues that modulate and propagate the allosteric communication to the pore region. Ile-4937 obstructs ion conductance in closed RyR1.
Supplementary Material
Statement of Significance.
Ryanodine receptors (RyRs) are a group of Ca2+ channels that bind to endogenous and exogenous modulators to regulate Ca2+ release through closed-to-open gating transitions. Recent advances in single-particle cryo-electron microscopy (cryo-EM) have captured RyR1 in multiple states and provide insights into RyR1 regulation by endogenous activators Ca2+ and ATP and exogenous activator caffeine (CFF). Despite the high-resolution structural data available for RyR1, the allosteric mechanism of RyR1 gating is not well understood. In this study, we used a graph-theoretical approach to predict the allosteric networks of interactions among various co-activators in RyR1 that control opening/closing of the pore region. Such synergistic coordination appears to be necessary for channel regulation by multiple activators, which includes co-regulation among various activators in RyR1 that regulate the closed-to-open gating transitions. The findings may assist in the design of strategies to control allosteric regulation of RyR1. Understanding of allosteric coupling among the ligand sites and pore region in RyR1 may enable the design of advanced therapeutics for several debilitating diseases.
Acknowledgements
We thank Dr. Jian Wang for developing an interactive web server to depict graph-inspired pathways in RyR1. N.V.D. acknowledges NIH support grants R35GM134864 and RO1GM123247. N.V.D. and G.M. are supported by NIH grant AR018687.
Footnotes
Conflict of interest
The authors declare no conflicts of interest with the contents of this article.
References:
- 1.Fill M, and Copello JA. 2002. Ryanodine Receptor Calcium Release Channels. Physiol. Rev 82:893–922. [DOI] [PubMed] [Google Scholar]
- 2.Bers DM 2004. Macromolecular complexes regulating cardiac ryanodine receptor function. J. Mol. Cell. Cardiol 37:417–429. [DOI] [PubMed] [Google Scholar]
- 3.Meissner G 2017. The structural basis of ryanodine receptor ion channel function. J. Gen. Physiol 149:1065 LP – 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzini-Armstrong C, and Protasi F. 1997. Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions. Physiol. Rev 77:699–729. [DOI] [PubMed] [Google Scholar]
- 5.Van Petegem F 2015. Ryanodine Receptors: Allosteric Ion Channel Giants. J. Mol. Biol 427:31–53. [DOI] [PubMed] [Google Scholar]
- 6.Endo M 2009. Calcium-Induced Calcium Release in Skeletal Muscle. Physiol. Rev 89:1153–1176. [DOI] [PubMed] [Google Scholar]
- 7.Ríos E 2018. Calcium-induced release of calcium in muscle: 50 years of work and the emerging consensus. J. Gen. Physiol 150:521 LP – 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai X-C, Yan Z, Wu J, Li Z, and Yan N. 2016. The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening. Cell Res. 26:995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei R, Wang X, Zhang Y, Mukherjee S, Zhang L, Chen Q, Huang X, Jing S, Liu C, Li S, Wang G, Xu Y, Zhu S, Williams AJ, Sun F, and Yin C-C. 2016. Structural insights into Ca(2+)-activated long-range allosteric channel gating of RyR1. Cell Res. 26:977–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, Hendrickson WA, Marks AR, and Frank J. 2016. Structural Basis for Gating and Activation of RyR1. Cell. 167:145–157.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng W 2015. Toward decrypting the allosteric mechanism of the ryanodine receptor based on coarse-grained structural and dynamic modeling. Proteins. 83:2307–2318. [DOI] [PubMed] [Google Scholar]
- 12.Steele TWE, and Samsó M. 2019. The FKBP12 subunit modifies the long-range allosterism of the ryanodine receptor. J. Struct. Biol 205:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efremov RG, Leitner A, Aebersold R, and Raunser S. 2014. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 517:39. [DOI] [PubMed] [Google Scholar]
- 14.Yan Z, Bai X, Yan C, Wu J, Li Z, Xie T, Peng W, Yin C, Li X, Scheres SHW, Shi Y, and Yan N. 2014. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 517:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zalk R, Clarke OB, des Georges A, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, and Marks AR. 2015. Structure of a mammalian ryanodine receptor. Nature. 517:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samsó M 2017. A guide to the 3D structure of the ryanodine receptor type 1 by cryoEM. Protein Sci. 26:52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dashti A, Mashayekhi G, Shekhar M, Ben Hail D, Salah S, Schwander P, des Georges A, Singharoy A, Frank J, and Ourmazd A. 2020. Retrieving functional pathways of biomolecules from single-particle snapshots. Nat. Commun 11:4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu L, Chirasani VR, Carter JS, Pasek DA, V Dokholyan N, Yamaguchi N, and Meissner G. 2018. Ca2+-mediated activation of the skeletal-muscle ryanodine receptor ion channel. J. Biol. Chem . 293:19501–19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murayama T, Ogawa H, Kurebayashi N, Ohno S, Horie M, and Sakurai T. 2018. A tryptophan residue in the caffeine-binding site of the ryanodine receptor regulates Ca2+ sensitivity. Commun. Biol 1:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dokholyan NV 2016. Controlling Allosteric Networks in Proteins. Chem. Rev 116:6463–6487. [DOI] [PubMed] [Google Scholar]
- 21.Proctor EA, Kota P, Aleksandrov AA, He L, Riordan JR, and V Dokholyan N. 2015. Rational Coupled Dynamics Network Manipulation Rescues Disease-Relevant Mutant Cystic Fibrosis Transmembrane Conductance Regulator. Chem. Sci 6:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dagliyan O, Tarnawski M, Chu P-H, Shirvanyants D, Schlichting I, V Dokholyan N, and Hahn KM. 2016. Engineering extrinsic disorder to control protein activity in living cells. Science (80-.) 354:1441 LP – 1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Jain A, McDonald LR, Gambogi C, Lee AL, and V Dokholyan N. 2020. Mapping allosteric communications within individual proteins. Nat. Commun 11:3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei Y, Xu L, Mowrey DD, Mendez Giraldez R, Wang Y, Pasek DA, V Dokholyan N, and Meissner G. 2015. Channel Gating Dependence on Pore Lining Helix Glycine Residues in Skeletal Muscle Ryanodine Receptor. J. Biol. Chem 290:17535–17545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dokholyan NV, Li L, Ding F, and Shakhnovich EI. 2002. Topological determinants of protein folding. Proc. Natl. Acad. Sci 99:8637 LP – 8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen JY 1971. Finding the K Shortest Loopless Paths in a Network. Manage. Sci 17:712–716. [Google Scholar]
- 27.Dijkstra EW 1959. A note on two problems in connexion with graphs. Numer. Math 1:269–271. [Google Scholar]
- 28.Suzuki R, and Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- 29.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. . [Google Scholar]
- 30.Mowrey DD, Xu L, Mei Y, Pasek DA, Meissner G, and V Dokholyan N. 2017. Ion-pulling simulations provide insights into the mechanisms of channel opening of the skeletal muscle ryanodine receptor. J. Biol. Chem 292:12947–12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chirasani VR, Xu L, Addis HG, Pasek DA, V Dokholyan N, Meissner G, and Yamaguchi N. 2019. A central core disease mutation in the Ca(2+)-binding site of skeletal muscle ryanodine receptor impairs single-channel regulation. Am. J. Physiol. Cell Physiol 317:C358–C365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Wang Y, Gillespie D, and Meissner G. 2006. Two Rings of Negative Charges in the Cytosolic Vestibule of Type-1 Ryanodine Receptor Modulate Ion Fluxes. Biophys. J 90:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran S, Chakraborty A, Xu L, Mei Y, Samsó M, V Dokholyan N, and Meissner G. 2013. Structural Determinants of Skeletal Muscle Ryanodine Receptor Gating. J. Biol. Chem . 288:6154–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.