Abstract
In medulloblastoma, p53 expression has been associated with chemoresistance and radiation resistance and with poor long-term outcomes in the p53-mutated sonic hedgehog, MYC-p53, and p53-positive medulloblastoma subgroups. We previously established a direct role for p53 in supporting drug resistance in medulloblastoma cells with high basal protein expression levels (D556 and DAOY). We now show that p53 genetic suppression in medulloblastoma cells with low basal p53 protein expression levels (D283 and UW228) significantly reduced drug responsiveness, suggesting opposing roles for low p53 protein expression levels. Mechanistically, the enhanced cell death by p53 knockdown in high-p53 cells was associated with an induction of mTOR/PI3K signaling. Both mTOR inhibition and p110α/PIK3CA induction confirmed these findings, which abrogated or accentuated the enhanced chemosensitivity response in D556 cells respectively while converse was seen in D283 cells. Co-treatment with G-actin–sequestering peptide, thymosin β4 (Tβ4), induced p-AKTS473 in both p53-high and p53-low cells, enhancing chemosensitivity in D556 cells while enhancing chemoresistance in D283 and UW228 cells.
Implications:
Collectively, we identified an unexpected role for the PI3K signaling in enhancing cell death in medulloblastoma cells with high basal p53 expression. These studies indicate that levels of p53 immunopositivity may serve as a diagnostic marker of chemotherapy resistance and for defining therapeutic targeting.
Introduction
Medulloblastoma, a primitive neuroectodermal tumor (PNET), arises from granule neuron precursors in the cerebellum or from rhombic lip neural stem cells. Medulloblastoma is the most frequently diagnosed malignant brain tumor in children, accounting for approximately 25% of all childhood brain tumors. Current medulloblastoma therapies (chemotherapy and radiation) are nontargeted, and many survivors suffer from long-term, treatment-related sequelae. Medulloblastoma has been classified into a scheme that incorporates molecular, anatomic, and staging data in a prognostic fashion, namely those associated with SHH and WNT constitutive signaling and so-called groups 3 and 4. While it is generally known that certain groups have a more favorable prognosis, such as the WNT group, further dissection within groups reveal subgroups that have a poorer outcome. Identification of the molecular basis of these differences, therefore, is critical for design of logical therapeutic intensification. The possible role of the p53 tumor suppressor gene in medulloblastoma has been investigated at the genomic and genetic level within these medulloblastoma subgroups (1–4). Defects in the p53 pathway, including p53 nuclear accumulation, MDM2 amplification and CDKN2A dysfunction, are established molecular features of medulloblastoma with validated relationships to disease molecular pathology (5, 6) and are associated with aggressive variants of MB (3). SHH-activated medulloblastoma were reclassified as p53 wildtype (WT) and p53 mutant based on the substantial differences in the long-term survival of the two classes (7). In addition, relatively high expression of p53 in SHH and genes associated with PI3K/AKT and MAPK/ERK pathways in group 3 medulloblastoma are associated with poor prognosis and survival outcome (8).
Studies have shown that strong p53 medulloblastoma tumor immunopositivity correlates with poor clinical prognosis, regardless of p53 genetic status (9–12). Increased p53 expression has been reported to be associated with chemoresistance and radiation resistance, and the long-term outcomes in the p53-mutated SHH (13), MYC-p53 (14), and p53-positive (wild or mutant) medulloblastoma subgroups are poor (11, 15). p53 mutations occur most often in the SHH (15%) and WNT (12%) subgroups of medulloblastoma, and patients with concomitant SHH or Myc amplification have the worst survival outcomes (4).
In recurrent medulloblastoma, p53 pathways defects, p53 mutations, and p53 nuclear staining in patients screened at diagnosis to relapse were found in 100%, 79%, and 24% of the cases, respectively (14). Interestingly, the accumulation of p53 protein, which in the majority of cases was WT (p53, n = 101; p53 mutant, n = 3), conferred a significantly worse survival in central nervous system PNETs (P < 0.0001; ref. 16). In primary medulloblastoma samples, higher expression of both WT and mutant p53 was correlated with overall poor survival, with the p53-positive tumors having protein levels nearly equivalent to DAOY cells (6). Collectively, these data raise the possibility that the normal regulation and function of WT p53 may be compromised in medulloblastoma.
Typically, oncogenic p53 mutations lead to significant structural changes, resulting in altered interactions at the gene and protein levels (17, 18). However, partial or complete inactivation of WT p53 can influence the p53 network, p53 protein levels, p53 aggregation, p53 differential transactivation, and/or the loss or gain of p53 function leading to alterations in its normal tumor-suppressive role (19–21). Using the NeuroD2:SmoA1 mouse model, inhibition of p53 regulatory mechanisms was found to suppress WT p53 function (22). Alterations in normal p53 function may therefore be important in the pathogenesis of medulloblastoma, as studies in other genetically engineered mouse models have shown that Myc expression alone fails to induce medulloblastoma, and either a mutant p53 gene or p53 ablation is required to induce medulloblastoma (13, 23, 24). Given the critical role of p53 in tumor suppression, there is a need to better understand the functional regulation of p53 in medulloblastoma pathogenesis and to better define the impacts of both p53-WT and p53-mutated medulloblastoma to identify new markers and therapeutics to improve clinical outcomes.
Our previous studies established that the high basal p53 protein expression of either WT (D556) or p53 mutant (DAOY) in medulloblastoma cells was involved in chemoresistance, and p53 suppression increased drug sensitivity in both cell types (25). We now show that p53 knockdown induced chemoresistance in medulloblastoma cells with low basal WT p53 protein levels (e.g., D283, UW228). In addition, the enhanced sensitivity to cisplatin and vincristine in p53-high cells following p53 knockdown was associated with an induction of AKT/mTOR signaling. Finally, we established that thymosin β4 (Tβ4), an FDA-approved, naturally occurring peptide, induced the PI3K/AKT pathway in medulloblastoma cells, enhancing chemosensitivity to first-line drugs in p53-high medulloblastoma cells while suppressing drug responses in p53-low medulloblastoma cells.
Materials and Methods
Cell lines and reagents
The human medulloblastoma cell lines D556 (RRID:CVCL_1165), DAOY (RRID:CVCL_1165), and D283 (RRID:CVL_1155) were maintained in complete RPMI containing 10% FBS, l-glutamine, Na-Pyruvate, and 100 U/mL penicillin-streptomycin and incubated at 37°C, in a humidified atmosphere with 5% CO2.
The cell lines UW228 (RRID:CVCL_8585), D425(RRID:CVCL_1165), and D458 (RRID:CVCL_1165) were maintained in complete DMEM containing 10% FBS, 2% l-glutamine, and 100 U/mL penicillin-streptomycin.
Short tandem repeat DNA fingertpinting was performed by PCR on the DOAY, D556, UW228, D425, and D458 cells in the Georgtown-Lombardi Tissue Culture Shared Resource as described previously (25). No cross-contamination was observed. All cell lines were split twice weekly or as needed and were screened for Mycoplasma contamination by PCR. After roughly 50 passages the cells were replenished with fresh cultures established from frozen stocks.
A total of 10 mmol/L RAD001 (Sigma-Aldrich SML2282) stock solution for cell culture was prepared in DMSO (Sigma-Aldrich), was stored at −20°C and diluted with fresh culture medium immediately before use. Cisplatin (PHR1624), vincristine (V8388), and doxorubicin (PHR1404) were purchased from Sigma-Aldrich and vincristine were dissolved with 0.9% NaCl at a concentration of 1 mg/mL, doxorubicin and cisplatin was dissolved in DMSO and stored at 4°C. VMY-1-103, a dansylated analog of purvalanol B developed at Georgetown University, Washington, DC, was dissolved in DMSO to a concentration of 10 mg/mL and further diluted in DMSO for drug treatments as described previously (25).
Cell viability and growth
Cell viability was determined using trypan blue dye exclusion staining. Cell counting was performed using a hemocytometer as described previously (25, 26). Cellular apoptosis was also assessed by Annexin V antibody (BioLegend) staining immediately after treatment with VMY (30 μmol/L), vincristine (0.5 μmol/L), cisplatin (12.5 μmol/L), and doxorubicin (4.8 μmol/L) and analyzed using FACStar Plus dual laser FACSort system (Becton Dickson) as previously described by us (27–29).
Immunoblotting
Protein extracts were separated on 4% to 20% Tris-glycine gels and electroblotted onto polyvinylidene difluoride membranes as described previously (29, 30). Antibodies against p53 (Millipore, #05-224), p-H2A.X (Ser139; Cell Signaling Technology, #9718), mTOR (Cell Signaling Technology, #2983), p-mTORS2448 (Cell Signaling Technology, #2971), 4E-BP1 (Cell Signaling Technology, #9644), PTEN (Cell Signaling Technology, #9559), S6K [Abcam (E175) ab32359], PI3 Kinase p110α (Cell Signaling Technology, #4249), RIP (Cell Signaling Technology, #4926), p-AKTS473 (Cell Signaling Technology, 9271), PARP (Cell Signaling Technology, #9542), BCL2 (Cell Signaling Technology, #15071), BCL-xl (Cell Signaling Technology, #2764), FAS (Santa Cruz Biotechnology, #sc-8009), FOXL1 (Santa Cruz Biotechnology, #sc-130373), β-actin (Cell Signaling Technology, #4967), and GAPDH (Cell Signaling Technology, #2983) were purchased from indicated companies. Densitometry was performed using ImageJ analysis software (RRID:SCR003070) as described previously (29, 30).
Immunofluorescence imaging
Cells were seeded on glass coverslips and treated with DMSO or VMY for 18 hours. Cells were washed with PBS and fixed in 10% formalin for 10 minutes. Cells were then washed three times with PBS, permeabilized with 0.1% Triton X-100, and washed an additional three times with PBS. Cells were then blocked with 1% BSA for 20 minutes and washed with PBS. Cells were then incubated with anti-p53 (1:150, Millipore #05-224) for 1 hour at room temperature. Slides were then washed with PBS (3×) and stained with the secondary antibodies Alexa Fluor goat 488 anti-mouse (1:150, Life Technologies, A-10667) for 30 minutes at room temperature. Slides were then counter stained with DAPI for 5 minutes. Confocal microscopy was performed on a Zeiss LSM510 Meta microscope using a 40× lens.
RNAi
siRNA's for FAS (sc-29311), FASL (sc-29313), and FOXL1 (sc-106746) were obtained from Santa Cruz Biotechnology. A total of 20 nmol of siRNA with lipofectamine 2000 was transfected into D556 cells. The same concentration of nonspecific siRNA (sc-37007) was used as control. Cells were cultured for 3 days and targeted gene expression was examined by immunoblotting.
Production of lentivirus short hairpin RNA vectors for mTOR and p53 knockdown
For adenovirus knockdown experiments, Adp53shRNA and empty pLKO vector control plasmids were purchased commercially (Vector Biolabs, #1854) and used as described by the manufacturer as described previously (25). Briefly, a total of 20 μg of lentiviral vector carrying short hairpin RNA (shRNA; mTOR or p53), 15 μg of packaging vectors pHR′8.2ΔR (Addgene), 2 μg of pCMV-VSV-G (RRID:Addgene8454), and 50 μL Lipofectamine2000 (Invitrogen) were mixed and incubated with 293T cells at 37°C. After 24 hours, the media was changed, and the cell supernatants were collected after an additional 24 hours and recombinant virus was stored at −80°C until use. The cells were seeded at 30% confluency and viral infections were performed for 72 hours prior to treatment with drugs. Efficiency of the knockdown was monitored by p53 and mTOR immunoblotting as described previously (26).
Expression of p110α
Transient transfections with the p110α plasmid (Addgene, #16643) were performed as recommended by the manufacturer's protocol. Briefly, cells were seeded in 6-well plates and allowed to attach overnight. The p110α expression plasmid was transfected using Lipofectamine LTX reagent (Thermo Fisher Scientific, #15338100) and 24 hours after transfection, the cells were treated with aforementioned drugs. After 18 hours of drug treatment, cells were collected and analyzed for cell death or protein expression using cell viability assays and immunoblot.
Reactive oxygen species measurement
For reactive oxygen species (ROS) measurement experiments, the cell permeant reagent 2′,7′-dichlorofluorescin diacetate (DCFDA; Abcam, ab113851) and CellROX (Cell Signaling Technology, #C10422) were used. DCFDA was dissolved in DMSO and stored at −20°C until use. DCFDA final concentration of DCFDA was set at 3 mmol/L in all experiments. D556 cells were stained with CellROX to detect ROS according to the manufacturer's recommendations.
Tβ4
Medulloblastoma cells were pretreated with Tβ4 (Regenex Pharmaceuticals) for 24 hours before adding drugs. The dose curve for Tβ4 was performed with 4.5 μmol/L doxorubicin. To assess the activation of p110α by Tβ4, we performed dose–response studies of Tβ4 (5, 10, 25, 50, 75, 150 nmol/L) in D556 cells and found 5 nmol/L Tβ4 efficiently induced cell death and, therefore, 5 nmol/L was used subsequently in all experiments.
Reverse-phase protein array analysis
Cell pellets were isolated from the p110α and control transfected D556 cells (as described above) and washed 3× with PBS. Cells were collected by scrapping and protein lysates were collected. Reverse-phase protein arrays (RPPA) were performed as described previously (31, 32).
RNA sequencing and pathway analyses
RNA was extracted from D556 and DAOY treated with cisplatin and cisplatin plus Tβ4 as described above using the Zymo research RNA mini-prep (Zymo Research Corp., R2050). RNA quality was analyzed using bioanalyzer (Bioanalyzer 2100, Agilent) and used at RNA index numbers >9.6. Extracted RNA was submitted to Otogenetics Corporation for Illumina Hi-seq RNA sequencing (RNA-seq) from total RNA (PE100 and HiSeq2000/2500 sequencing; 20 million reads). Data were processed as described (25, 33) and submitted to the NCBI https://www.ncbi.nlm.nih.gov/sra/PRJNA766838.
Ingenuity Pathway Analysis
Gene interaction networks, biofunctions, and pathway analysis were generated by Ingenuity Pathway Analysis (IPA) (Qiagen, RRID:SCR_008653), with microarray data interpretation via mapping of differentially expressed genes to known molecular functions, pathways, and networks available in the Ingenuity database as described previously (33). The significance was set at a P value and adjusted P value of 0.05 and a fold change of 1.5. The significance of the association between the input dataset and the functions or pathways was determined on z-score. The z-score is a statistical measure of the match between expected relationship direction and observed gene expression as predicted by IPA. The calculated z-score predicts activation or inhibition of a pathway based on positive or negative z-score, respectively.
Statistical analyses
Data were presented as the mean ± SD. Comparisons between two groups were made using two-tailed t test, and the proportion of Annexin V–positive apoptotic cells was determined using the χ2 test. Analyses were performed using the GraphPad Prism 5 software (GraphPad Software, Inc., RRID:SCR002798). P < 0.05 was considered as statistically significant. Statistical differences were marked by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, not significant.
Results
p53 expression in molecular subtypes of medulloblastoma
We have previously shown that the high levels of basal p53 protein played a significant role in supporting chemoresistance in DAOY (p53C242F mutant, SHH group) and D556 (p53 WT, group 3) medulloblastoma cells. Specifically, we established that p53 suppression enhanced the sensitivity of DAOY and D556 cells to the experimental drug VMY [VMY-1–103, a CDK1/CDK2 inhibitor (25)] or to doxorubicin, as measured by colony-forming assays, DNA degradation assays, and Annexin V staining, suggesting a surprising commonality in the p53 signaling in both cell lines despite the differences in their p53 gene status (25).
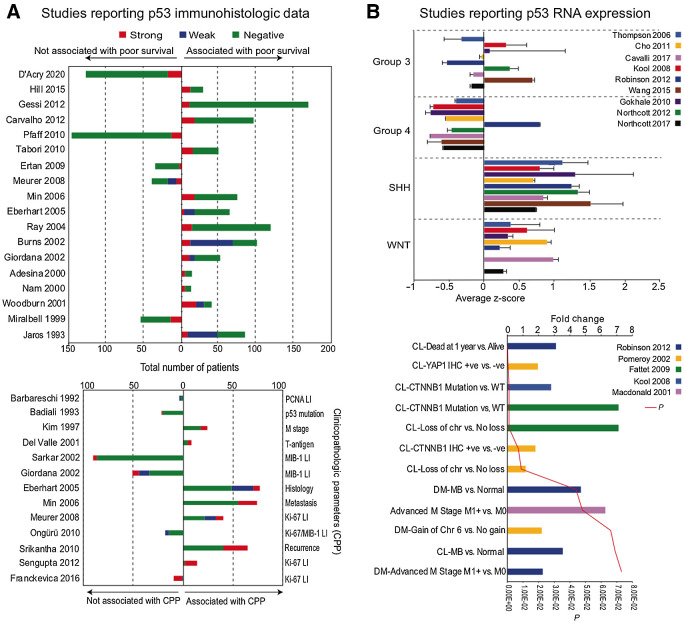
To better understand the possible clinical significance of high basal p53 protein expression levels in medulloblastoma, we have now performed an extensive literature and database search for studies reporting p53 protein and RNA expression levels in medulloblastoma. To the best of our knowledge, a total of 18 studies directly assessed p53 protein expression in primary medulloblastoma samples by IHC staining to predict survival outcomes in medulloblastoma, and 13 studies reported a significant association between intense p53 staining with poor overall survival (refs. 9–11, 14–16, 34–40; Fig. 1A). No correlation of prognosis or survival with p53 immunopositivity was noted in other studies (refs. 2, 34, 41–45; Fig. 1A). p53 immunopositive was unrelated to genetic status of p53 in most of the studies (11, 16, 41, 46). Intense p53 expression was correlated with high apoptotic/proliferative index (45, 47), advanced metastatic stage (48), early recurrence (49), T-antigen of human neurotropic polyomavirus JC (50), and clinical histology (10); however, no survival data were available (Fig. 1A, bottom), while some studies lacked association of p53 immunopositivity with features such as proliferation and apoptosis (Fig. 1A, bottom; refs. 51–55). p53 protein expression is not detected in the normal cerebellum (https://www.proteinatlas.org; ref. 50).
Figure 1.
A, Previous studies reporting p53 protein levels in medulloblastoma samples. Top, Previous studies reporting p53 immunostaining in human medulloblastoma samples and association with survival outcomes. Bottom, Previous studies with p53 immunostaining in human medulloblastoma samples and association with clinicopathologic parameters (CPP). B, Previous studies reporting p53 RNA levels in medulloblastoma samples. Top, Average z-score distribution of p53 mRNA levels in medulloblastoma subgroups. Bottom, Fold change in p53 mRNA expression in different pathologic medulloblastoma conditions. PCNA, proliferating cell nuclear antigen; LI, labeling index; CL, classic; DM, desmoplastic; Chr, chromosome; CTNNB1, β-catenin; M0, no evidence of distant metastases; M1, distant metastases present.
The distribution of p53 RNA levels across the established molecular classification of medulloblastoma (56) was assessed using transcriptomic data from medulloblastoma datasets for which molecular classification was available, extracted from the R2 platform (http://r2.amc.nl; refs. 13, 57–64). The p53 expression levels in SHH were significantly higher than group 3, group 4, and WNT (P < 0.01; Fig. 1B) and reanalysis of the exome sequencing data of Northcott (65) further displayed the distribution of expression of all p53 exons in the medulloblastoma subgroups (Supplementary Fig. S1A). Next the association of p53 RNA expression levels with clinical outcomes was assessed using Oncomine (www.oncomine.org). The p53 expression levels were significantly higher in classic and desmoplastic medulloblastoma versus normal cerebellum (66), as well as in advanced M stage (67) and in medulloblastoma samples with CTNNB1 muation (Fig. 1B, bottom; refs. 13, 68). In addition, p53 RNA expression levels were substantially higher in patients who died within 1 year versus those who survived (Fig. 1B, bottom; ref. 66), although different outcomes have been reported previosuly (58).
p53 protein levels impact chemosensitivity in medulloblastoma cells
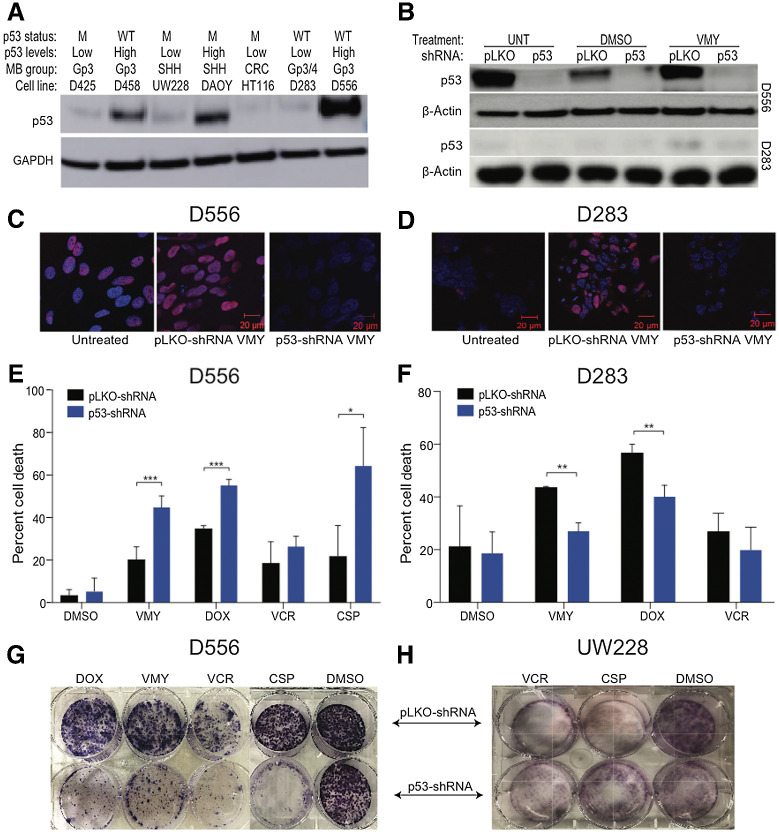
High basal p53 protein expression is seen in D556 and DAOY (25). We next assessed the levels of p53 protein expression in D283, D425, D458, and UW228. As shown in Fig. 2A, D458, D556, and DAOY cells express distinctly higher p53 protein levels versus the D283, UW228, and D425 cells, allowing us to classify these medulloblastoma cell lines as p53-high (D556, DAOY, and D458) and p53-low (D283, UW228, and D425). The status of the p53 gene is also shown in Fig. 2A.
Figure 2.
A, Immunoblot analyses of p53 protein expression and genetic status of six different human medulloblastoma cell lines. GAPDH was used as a loading control. B, Cells were infected with either p53 shRNA or control pLKO lentiviral followed by 18 hours of drug treatment and Western blots were run. C–F, Effect of p53 silencing of in p53 high (D556) and p53 low (D283) cell lines. Cells were infected with either p53-shRNA or control pLKO lentivirus, followed by 18 hours of drug treatment. Immunofluorescence images of p53 (red) subcellular localization and DAPI (blue) nuclear staining. Merged images are shown for D556 (C) and D283 (D) cells. The effects of p53 knockdown on cell viability in D556 (E) and D283 (F) cells following treatment with VMY (30 μmol/L), doxorubicin (DOX, 4.5 μmol/L), vincristine (VCR, 0.5 μmol/L), and cisplatin (CSP, 12.5 μmol/L). Data are mean ± SD of at least three independent experiments. P: *, <0.05; **, <0.01; ***, <0.001. G and H, Colony-forming assays of D556 and UW228 cells following p53 knockdown and treatment with VCR (0.05 μmol/L), DOX (0.05 μmol/L), or CSP (0.5 μmol/L). M; mutant p53, WT; wildtype, Gp; group, CRC; colorectal cancer.
p53 was genetically silenced in both D556 and D283 cells using the previously validated pLKO-p53-shRNsA (Fig. 2B; ref. 25). Treatment of D556 cells with VMY resulted in an increase in p53 nuclear positivity, which was blocked by p53 shRNA knockdown as previously shown (Fig. 2C; ref. 25). p53 immunopositivity in D283 cells was also increased by VMY, albeit to a lesser extent compared with D556 cells, and this increase was effectively inhibited by p53 shRNA knockdown (Fig. 2D).
Next, the pLKO-shRNA control and p53-shRNA knockdown cells were treated with VMY, doxorubicin, vincristine, or cisplatin. Similar to our previous study, p53-shRNA knockdown increased the chemosensitivity of D556 cells while increasing drug resistance in D283 cells (Fig. 2E and F, respectively). Dose-escalation experiments performed in D556 cells in the presence and absence of p53-shRNA or the p53 chemical inhibitor, pifithrin-α (25), established that the heightened chemosensitivity to doxorubicin and VMY was consistent across a broad range of concentrations (Supplementary Fig. S1B and S1C, respectively). Suppression of p53 by pifithrin-α in D556 cells resulted in increased cell aneuploidy (Supplementary Fig. S2A), while p53 ablation by CRISP-R was genotoxic (Supplementary Fig. S2B), suggesting that p53 may contribute to medulloblastoma tumorigenesis by enabling mitosis in the context of a destabilized cell genome (69). Colony-forming assays were next performed. In D556 cells, the genetic silencing of p53 led to significant decreases in colony formation following treatment with VMY, doxorubicin, vincristine, or cisplatin versus shRNA control (Fig. 2G) while in UW228 cells, silencing of p53 led to significant increases (Fig. 2H).
Identification of actionable targets to enhance chemosensitivity
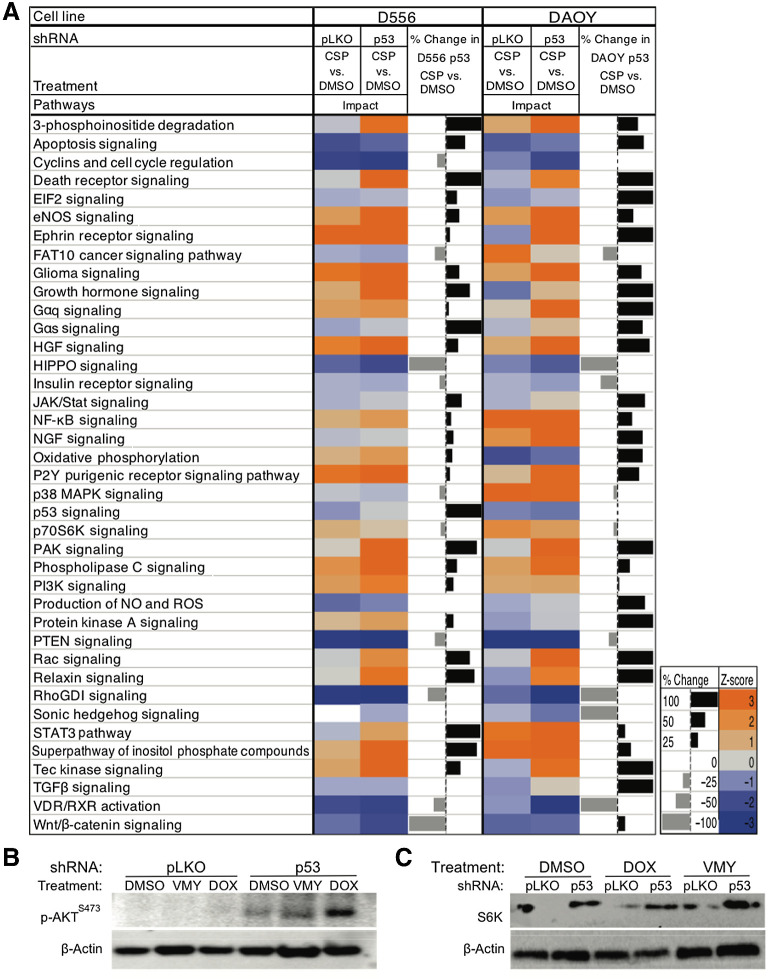
Despite differing in medulloblastoma subgroup and in p53 status, pathway analysis of the RNA-seq data from D556 and DAOY cells treated with VMY displayed noticeable similarities in transcriptomic signatures (Supplementary Fig. S2C). In addition, the RNA-seq data from the D556 and DAOY cells treated with cisplatin in the presence and absence of p53 identified additional shared transcriptomic signatures (Fig. 3A). The complete list of differentially expressed genes [P < 0.05, fold change (FC) 1.5] is presented in Supplementary Data S1. These data further suggest that p53 protein levels and subcellular localization, not simply its genetic status, in part influence response to drugs.
Figure 3.
Pathway analysis following p53 knockdown. A, Gene ontology and pathway analysis was performed on D556 or DAOY cells and either p53 shRNA or control pLKO lentiviral infection, followed by 18 hours of drug treatment. The calculated z-scores predicting activation (orange) or inhibition (blue) of the pathways shown. B and C, The effects of the genetic inhibition of p53 on p-AKTS473 and S6K protein levels in D556 cells, treated with VMY (30 μmol/L) or doxorubicin (DOX, 4.5 μmol/L) versus DMSO control.
As silencing of p53 in humans is not a viable approach, we sought to identify secondary regulators associated with the enhanced chemosensitivity observed with p53 suppression. IPA was performed on the transcriptomic data from VMY or cisplatin-treated D556 cells in the presence or absence p53-shRNA genetic inhibition (Fig. 3A). IPA identified targets included the induction of family members of TNF-receptor superfamily (FasR) and various transcription regulators (FOXL1). However, rescuing the effects of p53 inhibition by siRNA knockdown of FOXL1 (Supplementary Fig. S3A–S3C) or FasR (Supplementary Fig. S3D–S3F) was unsuccessful.
IPA further revealed an unanticipated induction of genes associated with the PI3K signaling pathway, including mTOR and AKT. The induction of the PI3K/mTOR signaling pathway by p53-shRNA knockdown, in both the presence and absence of doxorubicin, was verified in D556 by the increased levels of both p-AKTS473 and S6Kinase (S6K) by immunoblotting (Fig. 3B and C; Supplementary Fig S3G and S3H). The densitometric valuses are based on the gene expression studies and available literature (70, 71) we further explored the possibility that increasing PI3K-associated signaling may impact components of the translational machinery. 4E-BP1 and ELF2-α expression was modestly repressed at the protein level in the VMY- and doxorubicin-treated cells versus pLKO control (Supplementary Fig. S4A). The RNA-seq data further indicated an induction of the ROS and eNOS pathways upon p53 suppression in D556 cells. However, no differences in ROS production or oxidative stress were observed in either D556 or D283 cells using DCFDA and CellRox assays (data not shown).
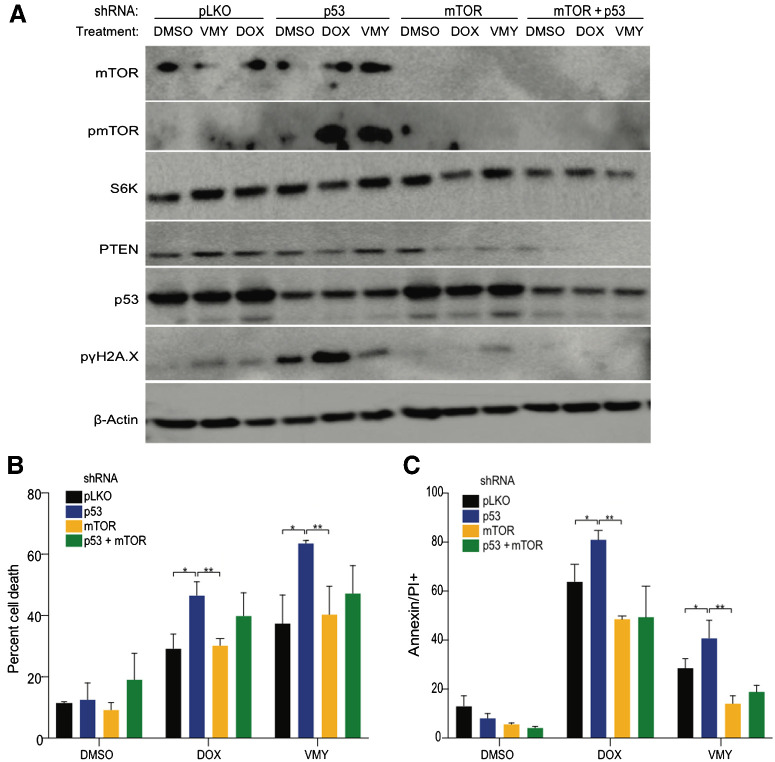
AKT/mTOR regulates p53-mediated chemosensitivity
Experiments were next performed to determine whether AKT/mTOR suppression or overexpression differentially regulated chemosensitivity in p53-high versus p53-low medulloblastoma cells. First, mTOR was genetically or chemically inhibited using mTOR-shRNA lentivirus or RAD001, respectively, in the presence or absence of p53 suppression. D556 cells infected independently and simultaneously with p53-shRNA and mTORC1-shRNA were treated with drugs for 18 hours. Immunoblotting confirmed that mTOR targeting shRNA reduced mTOR protein levels and downstream proteins including phospho-mTOR, S6K, and phospho-γH2A.X (Fig. 4A). While dual mTOR-shRNA and p53-mTOR-shRNA targeting significantly reduced cell death, as determined by trypan blue exclusion and flow cytometry, the addition of the knockdown of p53 did not significantly impact cell survial over the levels of p53-mTOR-shRNA alone (Fig. 4B and C). Similar results were obtained with combinations of RAD001 or rapamycin (RAPA) and p53 knockdown in D556 cells (Supplementary Fig. S4B). Conversely, increased cell death was seen in p53-low D425 and UW228 cells when targeted with mTOR-shRNA and p53-mTOR-shRNA (Supplementary Fig. S4C and S4D). These chemical and genetic approaches each suggest a role for AKT and mTOR activation in accentuating cell death in p53-high medulloblastoma cells and indicate that mTOR suppression is sufficient for enhancing cell survival in the p53-high medulloblastoma cells.
Figure 4.
Effects of mTOR/AKT pathway modulation in medulloblastoma cells. A, Immunoblot of D556 cells infected with p53- and mTORC1-shRNAs or control pLKO-shRNA lentivirus followed by 18 hours exposure to DMSO, VMY (30 μmol/L), or DOX (4.5 μmol/L). Cell viability as a result of infection with p53- and/or mTOR-shRNAs versus pLKO-control shRNA lentivirus in D556 cells treated for 18 hours with DMSO, VMY (30 μmol/L) or doxorubicin (4.5 μmol/L) and assessed using trypan blue (B) or Annexin V (C) and propidium iodide (PI) staining by flow cytometry. Data are shown as percent change in staining versus pLKO-control infected cells. Data are mean ± SD of three independent experiments. P: *, <0.05; **, <0.01.
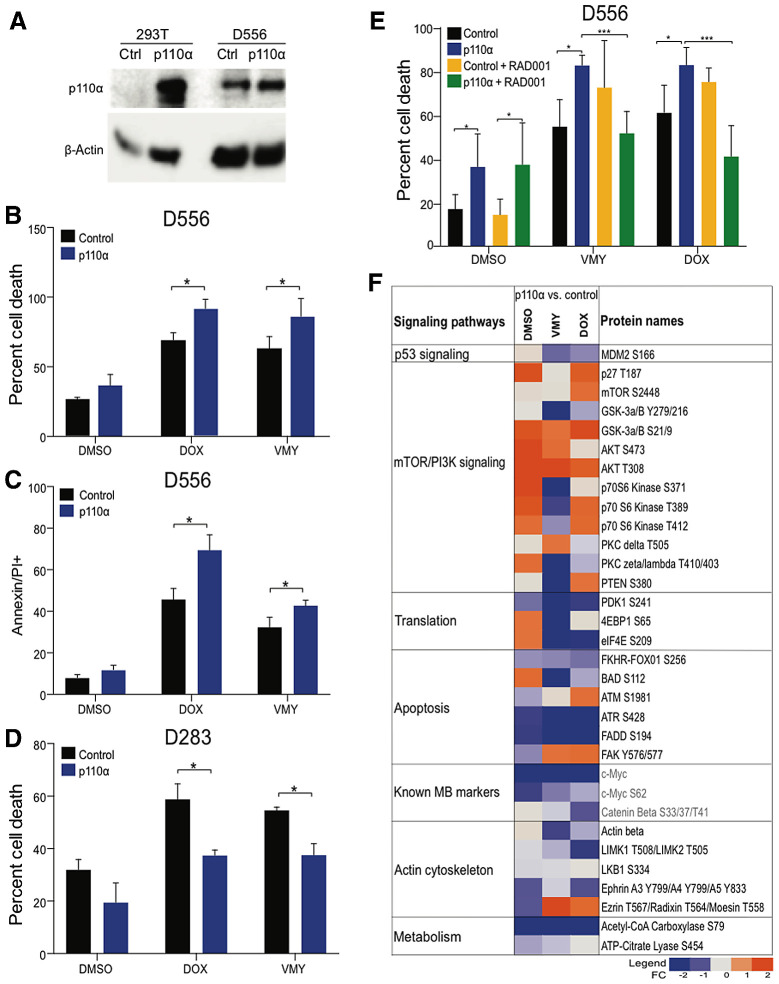
To further define the role of AKT/mTOR signaling in cell death, medulloblastoma cells were transfected with p110α/PIK3CA or control expression vectors (Fig. 5A). p110α significantly enhanced chemosensitivity in D556 cells (Fig. 5B and C), while significantly suppressing chemosensitivity in D283 cells (Fig. 5D). RAD001 abrogated the enhanced chemosensitivity induced by p110α in D556 cells (Fig. 5E).
Figure 5.
Effects of overexpression of p110α on D556 and D283 cell survival. A, p110α expression levels in transfected D556 and 293T cells. The effects of increased p110α expression on D556 cell viability determined by trypan blue (B) and and Annexin V/PI (C) staining. D, The effects of increased p110α expression on cell viability in p53-low D283 cells. E, Suppression of p110α-enhanced chemosensitivity by RAD001 (1 μmol/L). F, RPPA analysis performed on p110α transfected D556 cells and treated with VMY (30 μmol/L) or doxorubicin (DOX, 4.5μmol/L). A panel of 159 total and phosphoproteins was used. Data are represented as fold change in protein levels in p110α versus vector control cells. Key medulloblastoma-related proteins are shown in red. Data are mean ± SD of three independent experiments. P: *, <0.05; **, <0.01; ***, <0.001.
RPPA were performed on D556 cells transfected with p110α or the empty vector control plasmid, following by 18 hours of VMY or doxorubicin treatment. Unsupervised hierarchical cluster analysis verified that the p110α−transfected cells grouped separately versus the empty vector controls (Supplementary Fig. S5A). The RPPA data also verified the induction of AKT/mTOR pathway components by p110α and identified robust reductions in clinically relevant medulloblastoma molecular signatures, including Myc, phospho-Myc, and phospho-β-catenin (Fig. 5F). Despite the induction of AKT/mTOR signaling, downstream components of the pathway involved in translation, including ELF2-α, 4E-BP1, and PDK1, were suppressed, as were tricarboxylic acid cycle-related proteins, including acetyl Co-A carboxylase and ATP citrate lysate (Fig. 5F).
Tβ4 enhanced chemosensitivity and triggered necroptosis in human medulloblastoma cells
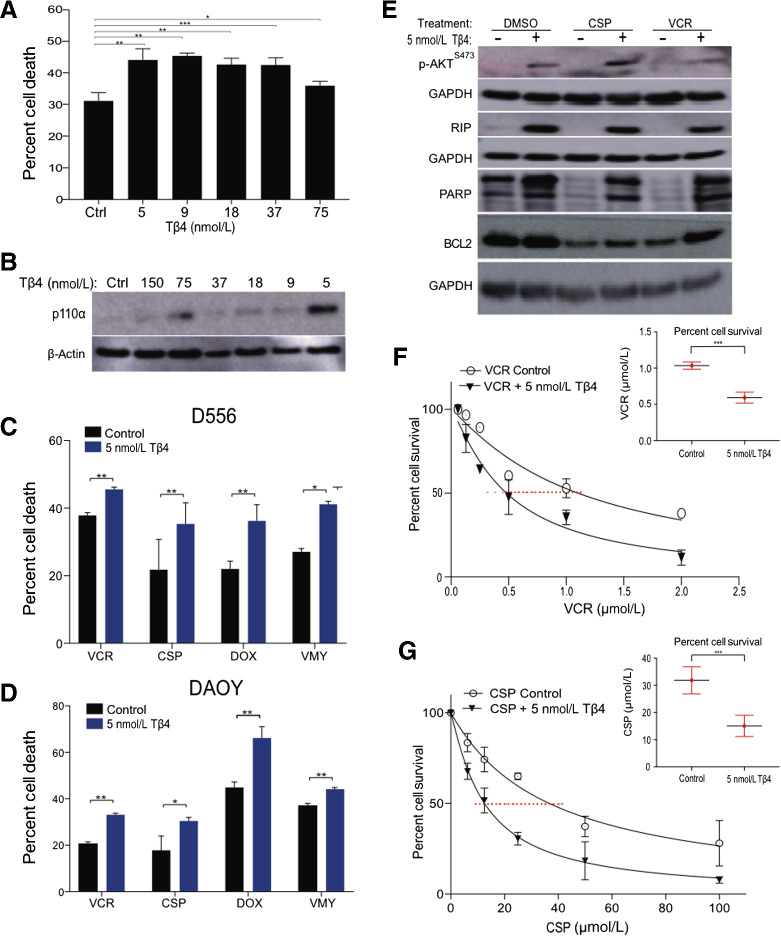
Tβ4 is a multifunctional, regenerative, FDA-approved orphan peptide/drug for treating epidermolysis bullosa. Tβ4 crosses the blood–brain barrier, induces PI3K/AKT signaling and supports recovery from stroke (72, 73). To begin investigating whether Tβ4 induces PI3K/AKT signaling in medulloblastoma cells and impacts chemosensitivity, we performed dose-escalation studies in D556 cells in the presence of doxorubicin. Significant increases in cell death were observed with Tβ4 (Fig. 6A). Immunoblotting confirmed the induction of PIK3CA by Tβ4 down to 5 nmol/L (Fig. 6B).
Figure 6.
Tβ4 enhanced chemosensitivity in medulloblastoma cells. A, Dose–response survival curves as assessed by trypan blue (average ± SD of N = 3 separate experiments) of D556 cells treated with doxorubicin (DOX, 4.5 μmol/L) and Tβ4 at concentrations shown. B, Immunoblots of p110α levels and β-actin in the cells from A. Cell survival of D556 (C) and DAOY (D) in response to the VMY (30 μmol/L), doxorubicin (4.5 μmol/L), vincristine (0.5 μmol/L), and cisplatin (12.5 μmol/L) in presence or absence of 5 nmol/L Tβ4 as measured by trypan blue. E, Immunoblots of p-AKTS437 and RIP levels in D556 cells treated as shown. EC50 dose–response experiments performed on D556 cells treated with vincristine (F) and cisplatin (G) in presence or absence of Tβ4. Insets; EC50 values of D556 cells treated as shown. Data are mean ± SD of three independent experiments. P: *, <0.05; **, <0.01; ***, <0.001.
Experiments using VMY-, doxorubicin-, cisplatin- or vincristine-treated D556 and DAOY cells established that 5 nmol/L Tβ4 consistently increased chemosensitivity (Fig. 6C and D). Similar responses were seen in the p53-high, D458 cells (Supplementary Fig. S5B). Immunoblots indicated an induction of DNA damage-associated proteins inducing RIP, PARP, p-AKTS473 and BCL2, in the presence of Tβ4 (Fig. 6E), Dose–response studies performed in presence or absence of 5 nmol/L Tβ4 with vincristine and cisplatin demonstrated a marked decrease in IC50 of the drugs (Fig. 6F and G).
Conversely, Tβ4 treatment of p53-low D283 and UW228 cells reduced responses to these drugs, consistent with the p53 suppression data (Supplementary Fig. S5C–S5E). Immunoblots established that RIP was induced in UW228 cells, albeit to a lesser extent than in D556 cells, and modest effects on BCL2 and PARP were indicative of reduced cell death in presence of Tβ4 (Supplementary Fig. S5F).
Discussion
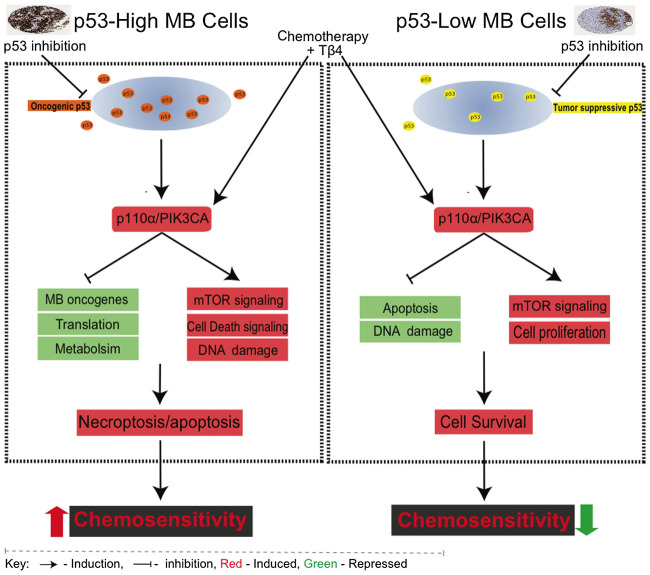
Collectively, the molecular and genetic data presented herein indicate that basal p53 protein levels in medulloblastoma cells influence the effects of AKT/mTOR signaling on key cell death pathways in response to chemotherapy (Fig. 7). The differential influences of these regulatory pathways in p53 high versus low medulloblastoma cells suggest that regulation of cell survival and cell death mechanisms may be predicted by p53 expression levels. p53 protein levels may therefore be a predictive biomarker for enhancing outcomes in patients whose medulloblastoma cells express high levels of p53 protein through the addition of Tβ4 to the chemotherapeutic regimen.
Figure 7.
Schematic model of differential p53 and PI3K function in medulloblastoma cells. Left, p53 acts as a supporting oncogene in medulloblastoma cells with constitutive p53 expression. Suppression of p53 in these cells leads to enhanced PI3K/AKT signaling resulting in enhanced chemosensitivity. Co-treatment with Tβ4 obviates the need for suppressing p53, directly activating the PI3K/AKT pathway to increase chemosensitivity. Right, p53 functions as a classic tumor suppressor protein in cells with low basal p53 expression. Either suppression of p53 or induction of PI3K/AKT signaling reduces chemosensitivity in these cells.
There is a clear precedent for the role of p53 as a tumor suppressor in various cancers (74). Several integrated transcriptomic and genomic studies have revealed significant insights into the prognostic value of p53-mutated medulloblastoma tumors (75). Specifically, detailed studies have been performed to define the prognostic value of p53 mutations within the molecular subgroups of medulloblastoma (7, 65). Additional studies have addressed the role of p53 protein expression in predicting survival outcomes in medulloblastoma (2, 9–11, 14–16, 35, 37–45). Interestingly, many of these studies provide evidence for significantly shorter survival in patients with p53 immunopositive tumors. Studies which reported no statistical significance between p53 IHC expression and survival do show a statistically significant correlation between p53 and Ki-67 (48, 55).
Silencing of p53 also influenced the ploidy of D556 cells, suggesting that p53 may contribute to medulloblastoma tumorigenesis by facilitating mitosis and stabilizing the cancer cell genome (18). p53 protein accumulation in cancer cells is usually attributed to mutations or frame shift deletions, resulting in either loss of tumor suppressor function or gain of oncogenic function (4). However, p53 functional inactivation can be achieved by deregulated co-regulators, for example methylation of p14ARF or deletion of INK4/ARF, in addition to p53 mutation (76) or upstream regulators of p53, such as I2PP2A (inhibitor 2 of phosphatase 2A) in SHH-activated p53-WT medulloblastoma (22). In addition, p53 expression was upregulated by 3.12 FC in patients who died within the first year (Fig. 1B; ref. 66).
Our previous study found that p53 protein accumulation in medulloblastoma cell lines is responsible for enhanced chemoresistance, suggesting a novel role for p53 in regulation of responses to chemotherapy (25). Furthermore, we established that cellular responses to chemotherapy upon p53 suppression comparable in p53-mutant DAOY and the p53-WT D556 cells, suggested a complex molecular basis of p53 functionally dysregulated medulloblastoma types. On the basis of this observation, we examined p53 protein expression levels in six medulloblastoma cell lines. D283 (p53-WT) and UW228 (p53-mutant) express p53 protein at much lower levels than D556 (p53-WT) and DAOY (p53-mutant). As opposed to D556 and DAOY cells, the genetic or chemical inhibition of p53 in D283 and UW228 cells resulted in significant reductions in drug-induced cytotoxicity, as seen previously in other cancers (26). To the best of our knowledge, these data are first to establish this function of p53 in medulloblastoma cells.
The impacts of the PI3K pathway on viability prompted us examine the role of AKT and mTOR signaling in medulloblastoma cells (77, 78). The RPPA data indicated that p110α induction promoted medulloblastoma cell death by compromising key survival and proliferative pathways, and possibly p53 itself via inhibition of MDM2S166 levels in D556 cells (Fig. 5F; refs. 79, 80). p110α/PIK3CA mutations are frequent in cancers but are less common in brain cancer (81). One possible explanation for the increased levels of basal p53 protein levels in D556 and DAOY cells may be a chronic feed-forward suppression of the PI3K pathway by p53. This concept is supported in part by the data showing that mTOR suppression increased cell survival which was not impacted by modulation of p53 levels (Fig 4B and C).
Using a “drug repositioning” approach, we found that Tβ4 induced PI3K/AKT signaling and significantly enhanced the efficacies of first-line chemotherapies in the p53-high D556 and DAOY cells versus reduced responses to these drugs in the p53-low D283 cells (Fig. 7).
The enhanced chemosensitivity by Tβ4 in the p53-high cells occurred through an induction of PI3K/AKT and necroptosis signaling (RIP) and the suppression of Myc (Fig. 5F). This unique interplay of p53 and the PI3K pathway in p53-WT and p53-mutated medulloblastoma warrants additional investigation to evaluate the therapeutic value of modulating p53/AKT/mTOR in vivo or in clinical trials. p53 protein expression and subcellular localization can be assessed in resected patient medulloblastoma samples by standard IHC or cytospin quantification approaches. Post-treatment quality of life of patients with medulloblastoma plays an increasingly important role in the choice of therapies. Current medullobalstoma therapy classifies pattients into four groups, based on histologic, genetic, anatomic, and staging considerations. Even though each category has broad predictive outcomes, significant variability of these outcomes exists within each group, suggestive of the need for further dissection of pathways that impact on survival as well as morbidity. While additional experiments are required, given the observation that targeting the PI3K pathway can uncouple its regulation by p53, it is possible that Tβ4 may reduce the post-treatment morbidity and sequelae by enabling the dose reduction of chemotherapeutics.
Authors' Disclosures
A. Naeem reports a patent for US 2021/0244797 A1 pending. L.F. Kromer reports grants from NIH during the conduct of the study. E.F. Petricoin reports personal fees from Perthera, Inc., Theralink Technologies, Inc., and Ceres Nanosciences, Inc., outside the submitted work; in addition, E.F. Petricoin has a patent for Reverse Phase Protein Array and associated markers issued, licensed, and with royalties paid from Theralink Technologies. M. Pierobon reports personal fees from Theralink Technologies outside the submitted work. S. Fricke reports grants from NIH during the conduct of the study. C. Albanese reports grants from NIH, DOE, and non-financial support from RegeneRx Biopharmaceuticals during the conduct of the study; in addition, C. Albanese has a patent for US 2021/0244797 A1 issued to Georgetown University. No disclosures were reported by the other authors.
Acknowledgments
Support was provided by P30 CA051008 (Weiner) and the Partners in Research (Albanese). C. Albanese, and E.F. Parasido were funded in part by the Department of Energy/Lawrence Livermore National Laboratory - DE-AC52-07NA27344. C. Ihemelandu was funded in part by the NIH CTSA; KL2TR001432. The following Georgetown-Lombardi resources were used: The Genomics and Epigenomics Shared Resource (GESR), the Microscopy and Imaging Shared Resource (MISR), the Flow Cytometry Shared Resource (TCSR), the Tissue Culture Shared Resource (TCSR) and the Histology and Tissue Shared Resource HTSR). We thank Dr. Patricia Ribeiro for help with the fingerprinting.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Contributions
A. Naeem: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. V. Harish: Formal analysis, investigation. S. Coste: Data curation, writing–original draft, writing–review and editing. E.M. Parasido: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. M.U. Choudhry: Conceptualization, data curation. L.F. Kromer: Data curation, investigation, writing–original draft. C. Ihemelandu: Conceptualization, writing–original draft, writing–review and editing. E.F. Petricoin: Formal analysis, investigation, writing–original draft, writing–review and editing. M. Pierobon: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. M.S. Noon: Formal analysis, investigation. V.M. Yenugonda: Resources. M. Avantaggiati: Conceptualization, writing–original draft, writing–review and editing. G.M. Kupfer: Conceptualization, writing–original draft, writing–review and editing. S. Fricke: Conceptualization, writing–original draft, writing–review and editing, involved with this study since the beginning and throughout. O. Rodriguez: Conceptualization, formal analysis, writing–original draft, writing–review and editing. C. Albanese: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Philipova T, Baryawno N, Hartmann W, Pietsch T, Druid H, Johnsen JI, et al. Differential forms of p53 in medulloblastoma primary tumors, cell lines and xenografts. Int J Oncol 2011;38:843–9. [DOI] [PubMed] [Google Scholar]
- 2. Pfaff E, Remke M, Sturm D, Benner A, Witt H, Milde T, et al. TP53 mutation is frequently associated with CTNNB1 mutation or MYCN amplification and is compatible with long-term survival in medulloblastoma. J Clin Oncol 2010;28:5188–96. [DOI] [PubMed] [Google Scholar]
- 3. Zhukova N, Ramaswamy V, Remke M, Pfaff E, Shih DJH, Martin DC, et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol 2013;31:2927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramaswamy V, Nör C, Taylor MD. p53 and meduloblastoma. Cold Spring Harb Perspect Med 2016;6:a026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carr-Wilkinson J, O'Toole K, Wood KM, Challen CC, Baker AG, Board JR, et al. High frequency of p53/MDM2/p14ARF pathway abnormalities in relapsed neuroblastoma. Clin Cancer Res 2010;16:1108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castellino RC, De Bortoli M, Lin LL, Skapura DG, Rajan JA, Adesina AM, et al. Overexpressed TP73 induces apoptosis in medulloblastoma. BMC Cancer 2007;7:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016;131:803–20. [DOI] [PubMed] [Google Scholar]
- 8. Park AK, Lee JY, Cheong H, Ramaswamy V, Park SH, Kool M, et al. Subgroup-specific prognostic signaling and metabolic pathways in pediatric medulloblastoma. BMC Cancer 2019;19:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woodburn RT, Azzarelli B, Montebello JF, Goss IE. Intense p53 staining is a valuable prognostic indicator for poor prognosis in medulloblastoma/central nervous system primitive neuroectodermal tumors. J Neurooncol 2001;52:57–62. [DOI] [PubMed] [Google Scholar]
- 10. Eberhart CG, Chaudhry A, Daniel RW, Khaki L, Shah KV, Gravitt PE. Increased p53 immunopositivity in anaplastic medulloblastoma and supratentorial PNET is not caused by JC virus. BMC Cancer 2005;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tabori U, Baskin B, Shago M, Alon N, Taylor MD, Ray PN, et al. Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations. J Clin Oncol 2010;28:1345–50. [DOI] [PubMed] [Google Scholar]
- 12. Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 2011;7:285–95. [DOI] [PubMed] [Google Scholar]
- 13. Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 2008;3:e3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hill RM, Kuijper S, Lindsey JC, Petrie K, Schwalbe EC, Barker K, et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015;27:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gessi M, Von Bueren AO, Rutkowski S, Pietsch T. P53 expression predicts dismal outcome for medulloblastoma patients with metastatic disease. J Neurooncol 2012;106:135–41. [DOI] [PubMed] [Google Scholar]
- 16. Burns ASYW, Jaros E, Cole M, Perry R, Pearson AJ. The molecular pathology of p53 in primitive neuroectodermal tumours of the central nervous system. Br J Cancer 2002;86:1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev 2012;26:1268–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muller PAJ, Vousden KH. P53 mutations in cancer. Nat Cell Biol 2013;15:2–8. [DOI] [PubMed] [Google Scholar]
- 19. Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem 2008;77:557–82. [DOI] [PubMed] [Google Scholar]
- 20. Billant O, Friocourt G, Roux P, Voisset C. P53, a victim of the prion fashion. Cancers 2021;13:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol 2019;20:199–210. [DOI] [PubMed] [Google Scholar]
- 22. Wei Y, Maximov V, Morrissy SA, Taylor MD, Pallas DC, Kenney AM. P53 function is compromised by inhibitor 2 of phosphatase 2A in sonic hedgehog medulloblastoma. Mol Cancer Res 2019;17:186–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solomon H, Buganim Y, Kogan-Sakin I, Pomeraniec L, Assia Y, Madar S, et al. Various p53 mutant proteins differently regulate the ras circuit to induce a cancer-related gene signature. J Cell Sci 2012;25:3144–52. [DOI] [PubMed] [Google Scholar]
- 24. Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell 2012;21:155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waye S, Naeem A, Choudhry MU, Parasido E, Tricoli L, Sivakumar A, et al. The p53 tumor suppressor protein protects against chemotherapeutic stress and apoptosis in human medulloblastoma cells. Aging 2015;7:854–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ringer L, Sirajuddin P, Tricoli L, Waye S, Choudhry MU, Parasido E, et al. The induction of the p53 tumor suppressor protein bridges the apoptotic and autophagic signaling pathways to regulate cell death in prostate cancer cells. Oncotarget 2014;5:10678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ringer L, Sirajuddin P, Heckler M, Ghosh A, Suprynowicz F, Yenugonda VM, et al. VMY-1–103 is a novel CDK inhibitor that disrupts chromosome organization and delays metaphase progression in medulloblastoma cells. Cancer Biol Ther 2011;12:818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Albanese C, Wu K, D'Amico M, Jarrett C, Joyce D, Hughes J, et al. IKKα regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell 2003;14:585–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ringer L, Sirajuddin P, Yenugonda VM, Ghosh A, Divito K, Trabosh V, et al. VMY-1–103, a dansylated analog of purvalanol B, induces caspase-3-dependent apoptosis in LNCaP prostate cancer cells. Cancer Biol Ther 2010;10:320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodriguez OC, Choudhury S, Kolukula V, Vietsch EE, Catania J, Preet A, et al. Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy. Cell Cycle 2012;11:4436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parasido EM, Silvestri A, Canzonieri V, Belluco C, Diodoro MG, Milione M, et al. Protein drug target activation homogeneity in the face of intratumor heterogeneity: implications for precision medicine. Oncotarget 2017;8:48534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baldelli E, Calvert V, Hodge A, VanMeter A, Petricoin EF, Pierobon M. Reverse phase protein microarrays. Methods Mol Biol 2017;1606:149–60. [DOI] [PubMed] [Google Scholar]
- 33. Naeem A, Dakshanamurthy S, Walthieu H, Parasido E, Avantaggiati M, Tricoli L, et al. Predicting new drug indications for prostate cancer: the integration of an in silico proteochemometric network pharmacology platform with patient-derived primary prostate cells. Prostate 2020;80:1233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giordana MT, Duó D, Gasverde S, Trevisan E, Boghi A, Morra I, et al. MDM2 overexpression is associated with short survival in adults with medulloblastoma. Neuro Oncol 2002;4:115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Min HS, Lee YJ, Park K, Cho B-K, Park S-H. Medulloblastoma: histopathologic and molecular markers of anaplasia and biologic behavior. Acta Neuropathol 2006;112:13–20. [DOI] [PubMed] [Google Scholar]
- 36. Adesina AM, Dunn ST, Moore WE, Nalbantoglu J. Expression of p27kip1 and p53 in medulloblastoma: relationship with cell proliferation and survival. Pathol Res Pract 2000;196:243–50. [DOI] [PubMed] [Google Scholar]
- 37. Carvalho RM, Pinto GR, Yoshioka FKN, Lima PDL, Souza CRT, Guimarães AC, et al. Prognostic value of the TP53 Arg72Pro single-nucleotide polymorphism and susceptibility to medulloblastoma in a cohort of Brazilian patients. J Neurooncol 2012;110:49–57. [DOI] [PubMed] [Google Scholar]
- 38. Ray A, Ho M, Ma J, Parkes RK, Mainprize TG, Ueda S, et al. A clinicobiological model predicting survival in medulloblastoma. Clin Cancer Res 2004;10:7613–20. [DOI] [PubMed] [Google Scholar]
- 39. Nam DH, Wang KC, Kim YM, Chi JG, Kim SK, Cho BK. The effect of isochromosome 17q presence, proliferative and apoptotic indices, expression of c-erbB-2, bcl-2 and p53 proteins on the prognosis of medulloblastoma. J Korean Med Sci 2000;25:452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jaros E, Lunec J, Perry RH, Kelly PJ, Pearson ADJ. p53 protein overexpression identifies a group of central primitive neuroectodermal tumours with poor prognosis. Br J Cancer 1993;68:801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. D'Arcy CE, Nobre LF, Arnaldo A, Ramaswamy V, Taylor MD, Naz-Hazrati L, et al. Immunohistochemical and nanoString-Based Subgrouping of Clinical Medulloblastoma Samples. J Neuropathol Exp Neurol 2020;79:437–47. [DOI] [PubMed] [Google Scholar]
- 42. Narayan V, Jaiswal J, Sugur H, SD S, Rao S, Chatterjee A, et al. Proteomic profiling of medulloblastoma reveals novel proteins differentially expressed within each molecular subgroup. Clin Neurol Neurosurg 2020;196:106028. [DOI] [PubMed] [Google Scholar]
- 43. Ertan Y, Sezak M, Demirağ B, Kantar M, Çetingül N, Turhan T, et al. Medulloblastoma: Clinicopathologic evaluation of 42 pediatric cases. Childs Nerv Syst 2009;21:353–6. [DOI] [PubMed] [Google Scholar]
- 44. Miralbell R, Tolnay M, Bieri S, Probst A, Sappino AP, Berchtold W, et al. Pediatric medulloblastoma: Prognostic value of p53, bcl-2, Mib-1, and microvessel density. J Neurooncol 1999;45:103–10. [DOI] [PubMed] [Google Scholar]
- 45. Meurer RT, Martins DT, Hilbig A, Ribeiro MDC, Roehe AV, Barbosa-Coutinho LM, et al. Immunohistochemical expression of markers KI-67, NeuN, synaptophysin, p53 and HER2 in medulloblastoma and its correlation with clinicopathological parameters. Arq Neuropsiquiatr 2008;66:385–90. [DOI] [PubMed] [Google Scholar]
- 46. Gessi M, Von Bueren A, Rutkowski S, Pietsch T. P53 expression predicts poor outcome in patients with metastatic medulloblastoma. Clin Neuropathol 2012;106:135–41. [DOI] [PubMed] [Google Scholar]
- 47. Sengupta S, Chatterjee U, Banerjee U, Ghosh S, Chatterjee S, Ghosh AK. A study of histopathological spectrum and expression of Ki-67, TP53 in primary brain tumors of pediatric age group. Indian J Med Paediatr Oncol 2012;33:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim EJ, Park SS, Lee YH, Choi AH, Rha SH, Lee SY, et al. Immunohistochemical study on expression of the p53 protein in medulloblastoma/PNET. J Korean Cancer Assoc 1997;29:867–73. [Google Scholar]
- 49. Srikantha U, Balasubramaniam A, Santosh V, Somanna S, Bhagavatula ID, Ashwathnarayana CB. Recurrence in medulloblastoma - influence of clinical, histological and immunohistochemical factors. Br J Neurosurg 2010;24:280–8. [DOI] [PubMed] [Google Scholar]
- 50. Del Valle L, Baehring J, Lorenzana C, Giordano A, Khalili K, Croul S. Expression of a human polyomavirus oncoprotein tumour suppressor proteins in medulloblastomas. Mol Pathol 2001;54:331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Öngürü Ö, Karslioglu Y, Özcan A, Çelik E. Anti-apoptotic and growth-promoting markers in adult medulloblastomas. Clin Neuropathol 2010;29:384–9. [PubMed] [Google Scholar]
- 52. Badiali M, Iolascon A, Loda M, Scheithauer BW, Basso G, Trentini GP, et al. P53 gene mutations in medulloblastoma immunohistochemistry gel shift analysis, and sequencing. Diagn Mol Pathol 1993;2:23–8. [PubMed] [Google Scholar]
- 53. Sarkar C, Pramanik P, Karak AK, Mukhopadhyay P, Sharma MC, Singh VP, et al. Are childhood and adult medulloblastomas different? A comparative study of clinicopathological features, proliferation index and apoptotic index. J Neurooncol 2002;59:49–61. [DOI] [PubMed] [Google Scholar]
- 54. Barbareschi M, Iuzzolino P, Pennella A, Allegranza A, Arrigoni G, Dalla Palma P, et al. p53 Protein expression in central nervous system neoplasms. J Clin Pathol 1992;45:583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Franckevica I, Kirsakmens G, Kleina R. Childhood medulloblastoma in Latvia: morphologic and molecular implications for diagnostics and personalised treatment. Virchows Arch 2016;16:9–15. [Google Scholar]
- 56. Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol 2012;8:340–51. [DOI] [PubMed] [Google Scholar]
- 57. Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012;488:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Northcott PA, Shih DJH, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012;488:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang X, Dubuc AM, Ramaswamy V, Mack S, Gendoo DMA, Remke M, et al. Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol 2015;129:449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gokhale A, Kunder R, Goel A, Sarin R, Moiyadi A, Shenoy A, et al. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J Cancer Res Ther 2010;6:521–9. [DOI] [PubMed] [Google Scholar]
- 63. Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol 2006;24:1924–31. [DOI] [PubMed] [Google Scholar]
- 64. Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 2011;29:1424–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 2011;29:1408–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002;415:436–42. [DOI] [PubMed] [Google Scholar]
- 67. MacDonald TJ, Brown KM, Lafleur B, Peterson K, Lawlor C, Chen Y, et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet 2001;29:143–52. [DOI] [PubMed] [Google Scholar]
- 68. Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S, et al. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol 2009;218:86–94. [DOI] [PubMed] [Google Scholar]
- 69. Gualberto A, Aldape K, Kozakiewicz K, Tlsty TD. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci U S A 1998;95:5166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Meng Y, Wang W, Kang J, Wang X, Sun L. Role of the PI3K/AKT signaling pathway in apoptotic cell death in the cerebral cortex of streptozotocin-induced diabetic rats. Exp Ther Med 2017;13:2417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu YT, Tan HL, Huang Q, Ong CN, Shen HM. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy 2009;5:824–34. [DOI] [PubMed] [Google Scholar]
- 72. Morris DC, Chopp M, Zhang L, Lu M, Zhang ZG. Thymosin β4 improves functional neurological outcome in a rat model of embolic stroke. Neuroscience 2010;169:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xiong Y, Mahmood A, Meng Y, Zhang Y, Zhang ZG, Morris DC, et al. Neuroprotective and neurorestorative effects of thymosin β4 treatment following experimental traumatic brain injury. Ann N Y Acad Sci 2012;1270:51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017;170:1062–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thornton C, Hagberg H. Mechanisms of cell death in the developing brain. In: Polin RA, Abman SH, Rowitch DH, Benitz WE, editors. Fetal and neonatal physiology. 5th ed. Elsevier; 2017. p. 76–85. [Google Scholar]
- 76. Frank AJ, Hernan R, Hollander A, Lindsey JC, Lusher ME, Fuller CE, et al. The TP53-ARF tumor suppressor pathway is frequently disrupted in large/cell anaplastic medulloblastoma. Brain Res Mol Brain Res 2004;121:137–40. [DOI] [PubMed] [Google Scholar]
- 77. Gao R, Lv G, Zhang C, Wang X, Chen L. TRIM59 induces epithelial-to-mesenchymal transition and promotes migration and invasion by PI3K/AKT signaling pathway in medulloblastoma. Oncol Lett 2018;15:8253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Akgül S, Li Y, Zheng S, Kool M, Treisman DM, Li C, et al. Opposing tumor-promoting and -suppressive functions of rictor/mTORC2 signaling in adult glioma and pediatric SHH medulloblastoma. Cell Rep 2018;24:463–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Feng J, Tamaskovic R, Yang Z, Brazil DP, Merlo A, Hess D, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem 2004;34:35510–7. [DOI] [PubMed] [Google Scholar]
- 80. Gottlieb TM, Fernando J, Leal M, Seger R, Taya Y, Oren M. Cross-talk between Akt, p53 and Mdm2: possible implications for the regulation of apoptosis. Oncogene 2002;8:1299–303. [DOI] [PubMed] [Google Scholar]
- 81. Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol 2010;3:1221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]