Abstract
The validity of taxa around Trichophyton rubrum was evaluated by a combination of phenetic and molecular methods. Morphological and physiological features were compared to results of sequencing of the internal transcribed spacer region of the ribosomal operon, PCR fingerprinting, and amplified fragment length polymorphism analysis. The 15 species and varieties investigated (Trichophyton circonvolutum, Trichophyton fischeri, Trichophyton fluviomuniense, Trichophyton glabrum, Trichophyton gourvilii, Trichophyton kanei, Trichophyton kuryangei, Trichophyton megninii, Trichophyton pedis, Trichophyton raubitschekii, Trichophyton rodhaini, Trichophyton rubrum var. nigricans, Trichophyton soudanense, Trichophyton violaceum var. indicum, and Trichophyton yaoundei) were reclassified or synonymized as T. rubrum or T. violaceum.
The members of the Trichophyton rubrum complex are the most common agents of dermatomycoses, primarily causing tinea pedis, onychomycosis, tinea corporis, and tinea capitis. Trichophyton megninii Blanchard (5), described in 1896, is the oldest identified taxon in the group. In the 1920s and 1930s, the species was common in Western Europe (6) as an etiological agent of tinea barbae. Now it is endemic in the Mediterranean countries, mainly causing tinea corporis (25).
The most prevalent species of the complex worldwide is T. rubrum (Castellani) Semon. It was described by Castellani (8) in 1910, when all other main dermatophytes had already been known for several decades. The species was suggested to have evolved in the late nineteenth century as a cause of chronic tinea corporis. It has since spread throughout the world as the etiological agent of onychomycosis and tinea pedis (34).
Another currently predominant species, Trichophyton violaceum, was described by Sabouraud (7) in 1902, 6 years after T. megninii. This species mostly causes tinea capitis and is distributed particularly in North Africa and the Middle East. The remaining species of the T. rubrum complex (Trichophyton circonvolutum, Trichophyton fischeri, Trichophyton fluviomuniense, Trichophyton glabrum, Trichophyton gourvilii, Trichophyton kanei, Trichophyton kuryangei, Trichophyton pedis, Trichophyton raubitschekii, Trichophyton rodhainii, Trichophyton soudanense, and Trichophyton yaoundei) are extremely rarely isolated as agents of dermatomycosis, and most of them were described much later, between 1960 and 1990.
Pleomorphism and cultural variability still make the dermatophytes notoriously difficult to identify. Georg (17) and Shadomy and Philpot (39) introduced a physiological identification system, which has been elaborated particularly by Canadian mycologists (25). However, in practice, test results are often difficult to read, and the system is insufficiently discriminative. Using molecular tools, Gräser et al. (21) analyzed 100 strains of T. rubrum, including phenotypic varieties such as var. nigricans. They did not detect any DNA variability among the strains studied. The diagnostic potential of molecular diagnosis thus seems to be significantly higher. The present study aimed to reveal the taxonomic structure of all species closely related to T. rubrum. This was done by concurrent application of several molecular methods (internal transcribed spacer [ITS] sequencing, PCR fingerprinting, and amplified fragment length polymorphism [AFLP] analysis) and by comparison of the results with conventional physiological, morphological, clinical, and geographical data for the same strains.
MATERIALS AND METHODS
Fungal strains.
The strains used and their origins are listed in Table 1. Names applied are according to current taxonomy as used in the Centraalbureau voor Schimmelcultures (CBS) list of cultures (http://www.cbs.knaw.nl), with the exception of Trichophyton pervesii Catanei, which had been reduced to synonymy with T. rubrum on morphological grounds. When available, type or authentic strains were used. Of the oldest species, no type strains are available because they were not cultured. All strains were grown at 27°C for 3 weeks.
TABLE 1.
Trichophyton species and strains analyzed in this studya
| Trichophyton species strain | CBS no. | Status | Source |
|---|---|---|---|
| T. abissinicum | 126.34 | T of B. abissinica | Human/tinea capitis |
| T. balcaneum | 359.62 | T | Human |
| T. circonvolutum | 286.30 | ||
| T. fischeri | 288.86 | Contaminant | |
| T. fischeri | 100081 | T | Contaminant |
| T. fluviomuniense | 592.68 | T | Human/skin |
| T. fluviomuniense | 191.69 | Human/skin | |
| T. glabrum | 499.48 | Human/skin | |
| T. gourvilii | 360.62 | Onychomycosis | |
| T. kanei | 289.86 | T | Human/buttock |
| T. kuryangei | 517.63 | T | Child/tinea capitis |
| T. kuryangei | 422.67 | Human/tinea capitis | |
| T. megninii | 735.88 | Human/skin of chin | |
| T. megninii | 734.88 | Human/skin of chin | |
| T. pedis | 189.69 | Onychomycosis | |
| T. raubitschekii | 100084 | T | Human/skin |
| T. raubitschekii | 287.86 | Human/skin | |
| T. raubitschekii | 202.88 | Human/tinea pedis | |
| T. raubitschekii | IFM 45885 | ||
| T. rodhainii | 376.49 | T | Human/tinea cruris |
| T. rubrum | 392.58 | Human/tinea pedis | |
| T. rubrum | 303.38 | AUT of T. pervesii | Child/tinea capitis |
| T. rubrum | 304.60 | T of T. cerebriforme | Human/skin |
| T. rubrum var. nigricans | ATU TR9 | ||
| T. rubrum var. nigricans | ATU TR16 | ||
| T. soudanense | 452.61 | Human | |
| T. violaceum var. indicum | 319.31 | T | Folliculitis of hair |
| T. violaceum var. violaceum | 374.92 | Human/skin | |
| T. yaoundei | 730.88 | Human | |
| T. yaoundei | 305.60 | AUT | Child/tinea capitis |
T, type strain; AUT, authentic strain; IFM, Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University, Japan; ATU, Faculty of Agriculture, University of Tokyo, Japan.
Conventional identification.
Microscopic morphology was studied on malt extract agar and colony morphology on Sabouraud's glucose agar. Physiological testing included the following features. (i) Production of urease was studied in urea broth (Oxoid B.V., Haarlem, The Netherlands) by the method of Christensen (12). Tests were read with intervals of up to 7 days at 27°C. (ii) In vitro hair perforation was tested at 27°C in sterile water containing 2 drops of 10% yeast extract and sterilized prepuberal human hair (33). Results were judged microscopically with up to 4-week intervals. (iii) The requirements for vitamins and amino acids were examined at 27°C on commercially available Trichophyton agars (Difco, Brunschwig Chemie B.V., Amsterdam, The Netherlands) (15): T1, vitamin-free Casamino Acids (CAS) agar; T2, CAS plus inositol; T3, CAS plus inositol plus thiamine; T4, CAS plus thiamine; T5, CAS plus nicotinic acid; T6, vitamin-free ammonium nitrate agar; and T7, vitamin-free ammonium nitrate agar plus l-histidine. Tests were read periodically for a maximum of 4 weeks.
DNA extraction.
A minipreparation method for DNA from fungi was used as described previously (18).
PCR fingerprinting.
The following oligonucleotides were used as single primers in the PCR experiments: the simple repeat sequence (AC)10 (30) and T3B, which is derived from the ITS region of the tRNA (5′-AGG TCG CGG GTT CGA ATC C) (27). Amplification reactions were performed in volumes of 50 μl containing 25 ng of template DNA, reaction buffer (10 mM Tris-HCl [pH 8.0]–50 mM KCl–1.5 mM MgCl2; additionally 3 mM magnesium acetate was added for T3B and 4 mM MgCl2 was added for (AC)10), 200 μM (each) deoxynucleoside triphosphates (Pharmacia LKB Biotechnology Inc., Piscataway, N.J.), and 2.5 U of Taq DNA polymerase (Perkin Elmer, Roche Molecular Systems, Inc., Branchburg, N.Y.). The primers T3B and (AC)10 were added at final concentrations of 25 pmol/50 μl of assay mixture and 10 pmol/50 μl of assay mixture, respectively. Samples were overlaid with sterile, light mineral oil (Sigma, Deisenhofen, Germany) and amplified through 32 cycles in a thermocycler (Perkin Elmer 9600) as follows: initial denaturation for 5 min at 95°C, denaturation for 15 s at 95°C, annealing for 30 s at 52°C for T3B and 54°C for (AC)10, and extension for 1.2 min at 72°C. This was followed by a final extension step of 6 min at 72°C.
Approximately 20 μl of each reaction was loaded on 1.2% NA-agarose gels (Pharmacia Biotech AB, Uppsala, Sweden) and electrophoresed for 5 h at 3 V cm−1 in 0.5 × buffer (89 mM Tris-borate–2.5 mM EDTA, pH 8.3). The gels were stained with ethidium bromide and photographed.
AFLP analysis.
AFLP was based on selective amplification of a subset of genomic restriction fragments using PCR (37, 45). Briefly, restriction fragments for amplification were generated in 40-μl reaction volumes. Genomic DNA (0.5 to 1 μg) was digested with 5 U each of EcoRI and MseI at 37°C for 3 h. Then 10 μl of a solution containing 5 pM EcoRI and 50 pM MseI adapters, 1 U of T4 DNA ligase, and 1× ligase buffer (50 mM Tris-HCl [pH 7.6]–10 mM MgCl2– 1 mM ATP–1 mM dithiothreitol–5% [wt/vol] polyethylene glycol 8000) was added. The ligation reaction was incubated at room temperature for 3 h. After ligation the reaction mixture was diluted 1:10. The following combinations of AFLP primer pairs with three selective nucleotides each (bold letters) were used for the amplification of the EcoRI-MseI fragments: (i) EcoRI-ATG (5′-GAC TGC GTA CCA ATT CAT G) and MseI-TGC (5′-GAT GAG TCC TGA GTA ATG C), (ii) EcoRI-TGC (5′-GAC TGC GTA CCA ATT CTG C) and MseI-CTA (5′-GAT GAG TCC TGA GTA ACT A), and (iii) EcoRI-TGC (5′-GAC TGC GTA CCA ATT CTG C) and MseI-TGC (5′-GAT GAG TCC TGA GTA ATG C).
The amplification reactions were performed in 25-μl volumes containing 8 μl of the 1:10 diluted ligation mixture as the template, 1× reaction buffer (10 mM Tris-HCl [pH 8.0]–50 mM KCl–1.5 mM MgCl2), 200 μM (each) dNTPs, 1.25 U of Taq DNA, and 25 pmol of AFLP primers/25 μl. Samples were overlaid with sterile, light mineral oil (Sigma) and amplified through 36 cycles in a thermocycler (Perkin-Elmer 9600) as follows: denaturation for 30 s at 94°C, annealing for 30 s (see below), and extension for 1 min at 72°C. The annealing temperature of 65°C in the first cycle was subsequently reduced by 0.7°C for each of the next 12 cycles and was kept at 56°C for the remaining 23 cycles. Electrophoresis in 1% agarose gels on 5-μl aliquots of the PCR products confirmed successful amplification. Samples of 20 μl were mixed with 10 μl of loading buffer (98% formamide–10 mM EDTA–0.15% bromophenol blue–0.15% xylene cyanol) denatured at 94°C for 5 min, followed immediately by chilling on ice. Fifteen to twenty microliters was loaded on 6% MDE Hydrolink acrylamide gels (FMC Bioproducts, Rockland, Maine) and run in 0.5× Tris-borate-EDTA buffer for 5 to 6 h at constant power (40 W). The gels were silver stained as follows: fixation in 1% HNO3 for 10 min and incubation in 0.2% AgNO3 for 20 min and then in 0.28 M NaCO3 plus 1 ml of formaldehyde (in 4 liters) for 10 to 30 min. The reaction was stopped by incubation in 10% CH3COOH. Between each step the gels were washed in Aqua-bidest, for 5 to 10 min and finally dried on Whatman 3MM chromatographic paper using a gel drier.
DNA fragment analysis.
Binary matrices were assessed for each of the five AFLP and PCR fingerprinting data sets by using polymorphism analysis of the computer program GelCompar (Applied Maths, Kortrijk, Belgium). Only dominant bands were included, and differences in band intensity were not taken into account. After using automatic search options, the bands were edited manually. The five data sets were analyzed separately and then by combining all binary matrices. Pairwise distances and phenograms were computed using total character differences and the unweighted pair group method with arithmetic means in the program PAUP 4.0 (42).
Sequence determination.
The ribosomal ITS region was amplified using universal primers LR1 (5′-GGT TGG TTT CTT TTC CT) and SR6R (5′-AAG TAA AAG TCG TAA CAA GG), corresponding to positions 73 to 57 of the 25S and positions 1744 to 1763 of the 18S nuclear rDNA genes of Saccharomyces cerevisiae, respectively. For sequencing both strands, one of the two primers was biotinylated in two reciprocal PCRs. Single-stranded DNA was obtained for direct sequencing by using streptavidin-coated magnetic beads (Dynabeads M 280; Dynal, Oldendorf, Germany). Each of the strands was sequenced using the same infrared-labeled primers in the sequencing reaction (SequiTherm ExelTM Long Read Cycle Sequencing Kit; Biozym Technologies, Oldendorf, Germany), combined with a LI-COR automatic DNA sequencer.
Sequence alignment and phylogenetic analysis.
Sequence alignment and phylogenetic analysis were performed using CLUSTAL V (Deutsches Krebsforschungszentrum, Heidelberg, Germany) and PAUP 4.0 (42). Parsimony analysis was conducted with unambiguously aligned sequences by using stepwise addition of sequences of the heuristic search option of PAUP. Gaps were treated as fifth-character states. The robustness of branches was assessed by bootstrap analysis with 100 replicates.
RESULTS
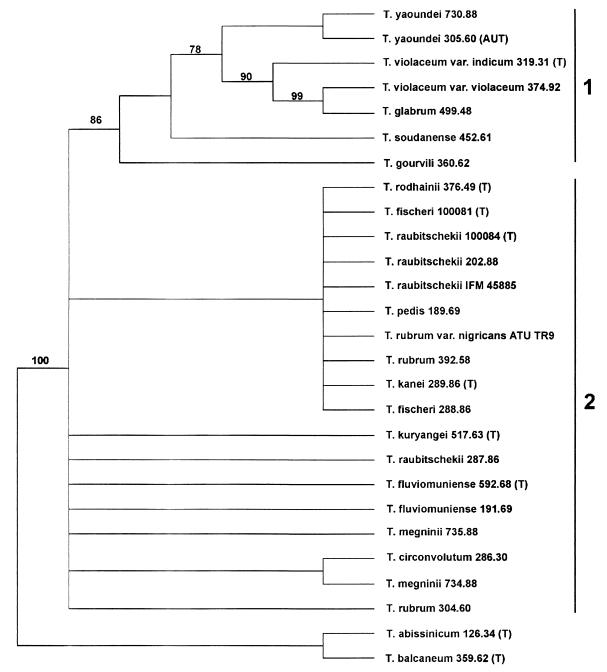
Figure 1 displays the bootstrap consensus tree inferred from 93 informative sites out of 630 characters, including the ITS 1, 2, and 5.8S sequences of the T. rubrum complex. Within the complex, two main clades can be distinguished. The species and strains forming clade 1 (T. glabrum Sabouraud, T. gourvilii Catanei, T. soudanense Joyeux, T. violaceum Bodin var. indicum Acton & McGuire, and two strains of T. yaoundei [Cochet et Doby-Dubois]) are supported by a high bootstrap value of 86% (Fig. 1). Clade 1 is separated from the species of clade 2 by seven substitutions. Clade 2 comprises all remaining type strains, including T. rodhainii Vanbreuseghem, T. fischeri Kane (2 strains), T. raubitschekii Kane et al. (4 strains), T. pedis Ota, T. rubrum (Castellani) Sabouraud (3 strains, including var. nigricans), T. kanei Summerbell, T. kuryangei Vanbreuseghem et Rosenthal, T. fluviomuniense Miguens (2 strains), T. circonvolutum Sabouraud, and T. megninii Blanchard (2 strains). Subclustering of the taxa within clade 2 is not supported by bootstrap analysis and appears to be insignificant since strains of the same species (e.g., T. rubrum 392.58 versus 304.60, T. raubitschekii 100084 versus 287.86, or T. megninii 735.88 versus 734.88) are found in different subclusters. Sequence comparison of Trichophyton abissinicum (Agostini) Nannizzi with the species of the Trichophyton mentagrophytes-Trichophyton tonsurans complex (18) resulted in similarity with Trichophyton immergens Milochevitch, Trichophyton balcaneum Castellani, and Trichophyton radicosum Catanei (data not shown). Therefore, T. abissinicum and T. balcaneum were used as outgroup species.
FIG. 1.
Bootstrap consensus tree obtained for ITS sequences of dermatophytes listed in Table 1. The tree was generated by using stepwise addition of sequences from the heuristic search option of PAUP (version 4.0b2). Ninety-three of the 630 selected sites were informative, and the tree length is 170 steps. Bootstrap values are shown above 70%. T. abissinicum and T. balcaneum were used as outgroup strains. EMBL accession numbers for the sequences are as follows: T. abissinicum, AJ270790; T. circonvolutum, AJ270791; T. fischeri, AJ270792-3; T. fluviomuniense, AJ270794-5; T. glabrum, AJ270796; T. gourvilli, AJ270797; T. kanei, AJ270798; T. kuryangei, AJ270799; T. megninii, AJ270800, Z97994; T. pedis, AJ270801; T. raubitscheckii, AJ270802-5; T. rodhaini, AJ270806; T. rubrum, AJ270807-8, Z97993; T. soudanense, AJ270809; T. violaceum, AJ270810-11; T. yaoundei, AJ270812-13. AUT, authentic strain; T, type strain.
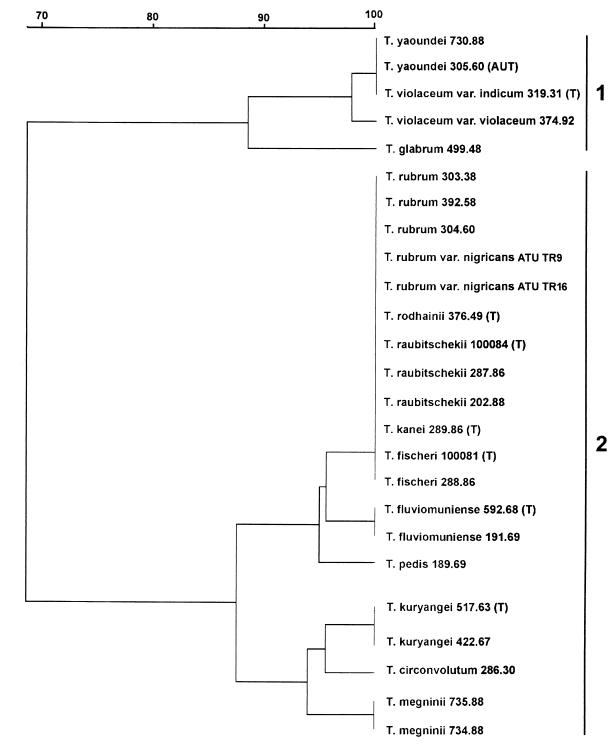
Results of the two fingerprinting and the three AFLP data sets are summarized in Table 2 and in the phenogram shown in Fig. 2. With all primers and primer pairs, a total of 65 fragments was generated. With the exception of the T3B, groupings within the T. rubrum complex were similar, irrespective of the primer used. The similarity levels ranged between 100 and 43% with an average level of 69% when all fragments were combined (Fig. 2). Nine genotypes were produced, with a minimum of one with fingerprint primer T3B and a maximum of four with AFLP primer pair EcoRI-TGC–MseI-CTA and primer (AC)10 (Fig. 3; Table 2).
TABLE 2.
Summary of PCR fingerprinting and AFLP analysesa
| Primer or primer pair | No. of band | % Similarity | No. of genotypes |
|---|---|---|---|
| T3B | 17 | 100 | 1 |
| (AC)10 | 14 | 73 | 4 |
| EcoRI-ATG–MseI-TGC | 7 | 43 | 3 |
| EcoRI-TGC–MseI-CTA | 12 | 58 | 4 |
| EcoRI-TGC–MseI-TGC | 15 | 53 | 3 |
| Total | 65 | 69 | 9 |
Similarity values were computed using total character differences between strains.
FIG. 2.
Phenogram computed from the DNA fragment profiles obtained with all PCR fingerprinting and AFLP primer sets. The bar scale shows the similarity (in percent) computed by total character differences using PAUP 4.0b2. AUT, authentic strain; T, type strain.
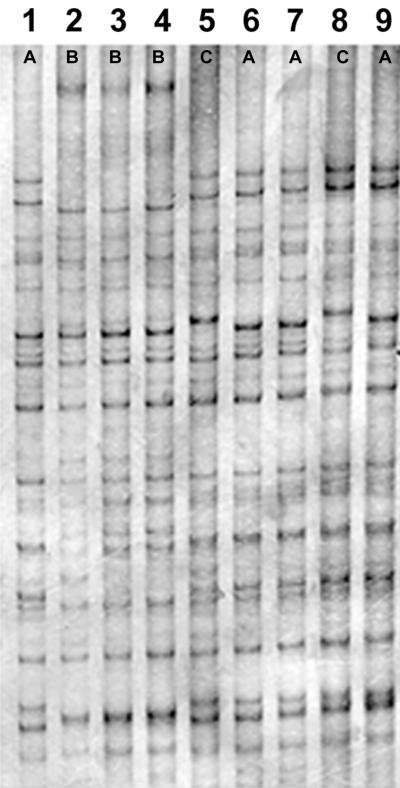
FIG. 3.
PCR fingerprinting pattern obtained with primer T3B from representatives of the Trichophyton rubrum complex. Lane 1, kb ladder; lane 2, T. rubrum 392.58; lane 3, T. glabrum 499.48; lane 4, T. violaceum var. indicum 319.31; lane 5, T. yaoundei 305.60, lane 6, T. kuryangei 517.63; lane 7, T. kanei 289.86; lane 8, T. fischeri 100081; lane 9, T. megninii 735.88; lane 10, T. raubitschekii 100084.
The T. rubrum complex was divided in two main groups, i.e., clusters 1 and 2. At the 88% similarity level, cluster 2 was subdivided (Fig. 2). Figure 4 shows representative genotypes (A, B, and C) for these three clusters. From the AFLP patterns it is obvious that genotypes A and C (both cluster 2) were more closely related when compared to B (cluster 1). Unfortunately we were unable to generate any AFLP or PCR fingerprinting data for T. soudanense and T. gourvilii. We know from our earlier studies that fragment analyses and sequencing data generally support each other. Therefore, taxonomic inferences for both species are drawn on the basis of the sequencing results only.
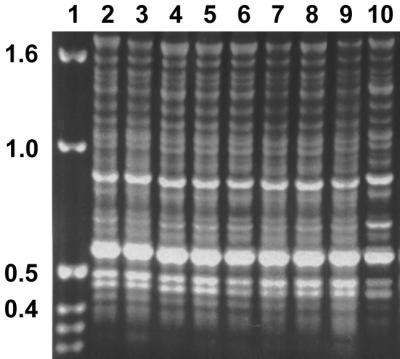
FIG. 4.
AFLP patterns (A, B, C) obtained with primer pair EcoRI-TGC–MseI-TGC from representatives of the T. rubrum complex. Lane 1, T. rubrum 392.58; lane 2, T. glabrum 499.48; lane 3, T. violaceum var. indicum 319.31; lane 4, T. yaoundei 305.60, lane 5, T. kuryangei 517.63; lane 6, T. kanei 289.86; lane 7, T. fischeri 100081; lane 8, T. megninii 735.88; lane 9, T. raubitschekii 100084.
Results of physiological and morphological studies using the same strains as studied with ITS sequencing, AFLP, and fingerprinting are presented in Table 3. No significant differences were found between the strains regarding the physiological features.
TABLE 3.
Physiological data of the strains used in this study
| Species | CBS no. | Ua | Hb | TIc | T2c | T3c | T4c | T5c | T6c | T7c | MiCd | MaCd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade 1 | ||||||||||||
| T. glabrum | 499.48 | W | − | + | + | ++ | ++ | + | + | + | + | − |
| T. gourvilii | 360.62 | − | − | + | + | + | + | + | + | + | − | − |
| T. soudanense | 452.61 | − | − | + | + | + | + | ++ | + | + | ++ | − |
| T. violaceum var. indicum | 319.31 | W | ND | ND | ND | ND | ND | ND | ND | ND | − | − |
| T. violaceum var. violaceum | 374.92 | + | − | + | + | ++ | ++ | + | + | + | − | − |
| T. yaoundei | 730.88 | D | − | + | + | + | + | + | + | + | − | − |
| T. yaoundei | 305.60 | + | − | + | + | + | + | + | + | + | + | − |
| Clade 2 | ||||||||||||
| T. fischeri | 288.86 | W | − | + | + | + | + | + | + | + | ++ | + |
| T. fischeri | 100081 | W | − | ++ | ++ | ++ | ++ | ++ | + | + | ++ | − |
| T. fluviomuniense | 592.68 | D | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| T. fluviomuniense | 191.69 | D | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| T. kanei | 289.86 | − | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | + |
| T. kuryangei | 517.63 | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | ++ | − |
| T. kuryangei | 422.67 | D | − | ++ | ++ | ++ | ++ | ++ | − | ++ | ++ | − |
| T. megninii | 735.88 | D | − | ++ | ++ | ++ | ++ | ++ | + | + | ++ | − |
| T. megninii | 734.88 | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | ++ | − |
| T. pedis | 189.69 | − | − | + | + | + | + | + | + | + | ++ | − |
| T. raubitschekii | 100084 | W | − | ++ | ++ | ++ | ++ | ++ | + | + | ++ | ++ |
| T. raubitschekii | 287.86 | + | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| T. rodhainii | 376.49 | D | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − |
| T. rubrum | 392.58 | − | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | − |
| T. rubrum | 303.38 | − | − | ++ | ++ | + | ++ | ++ | + | + | ++ | + |
| T. rubrum | 304.60 | D | − | ++ | ++ | ++ | ++ | ++ | + | + | ++ | ++ |
| T. rubrum var. nigricans | TR 16 | + | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − |
| T. rubrum var. nigricans | TR 9 | W | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − |
U, urease test; +, positive; W, weak; D, delayed; −, negative after 7 days of incubation.
H, hair perforation test; +, positive; −, negative after 4 weeks of incubation. ND, not done.
T, Trichophyton agar; T2 through T5, growth relative to T1; T7, growth relative to T6, where T1 and T6 are basis test media without supplement (see Materials and Methods); ++, good growth; +, poor growth; −, no growth after 4 weeks of incubation. ND, not done.
MiC, microconidia; MaC, macroconidia; ++, abundant; +, sparse; −, absent.
DISCUSSION
All species of the complex appear to be obligately anthropophilic, nearly exclusively transmitted from human to human. Animal infections have rarely been reported, and experimental transmission of T. rubrum from cultures to animals was problematic (11). None of the species of the T. rubrum complex is known to have a teleomorph; they are not even distantly related to any known Arthroderma species (19). In an extended search for teleomorphs, Young (47) found only five strains producing sterile cleistothecia out of 600 isolates analyzed. In contrast, other anthropophilic dermatophytes, such as Trichophyton interdigitale, Microsporum audouinii, and Microsporum ferrugineum, are phylogenetically close to described teleomorphs of the genus Arthroderma (18, 20). The loss of sexuality within the T. rubrum complex is a unique phenomenon among the dermatophytes. Together with the low virulence of the species concerned, this suggests a fine-tuned adaptation to the human host. Occasional human infections by otherwise zoophilic species such as Trichophyton verrucosum should be regarded as “spill-over” infections (E. Holmes, personal communication), which are evolutionary dead ends because they are not transmitted any further, whereas low-virulence T. rubrum strains can continue as chronic cutaneous infections, since they have changed their route of transmission. The latter had been promoted during the 19th century by the wide use of closed footwear, providing a moist and warm environment around the skin.
Within the species complex analyzed, two monophyletic clades (1 and 2) can be distinguished based on ITS sequence data. The clear separation of the clade 1 species is due to the varying length of a TA motif. This sequence motif is located at the end of ITS2 and is up to fourfold longer in clade 1 taxa than in those of clade 2. T. violaceum is the oldest identified taxon in clade 1. Clade 2 contains strains identified with the ancient names T. rubrum and T. megninii, although no type or authentic material is known to be preserved from either of these species.
Although the combined AFLP and PCR fingerprint data display three groups, only two of them are differentiated by a fairly high distance value of 31%. Thus, in main traits, these data are in concordance with the sequencing results. We therefore conclude that only two species can be distinguished within the T. rubrum complex, i.e., clade 1 (T. violaceum) and clade 2 (T. rubrum and T. megninii). For reasons explained below we maintain T. rubrum for strains classified in clade 2.
The clinical pictures of species in clades 1 and 2 show clear differences. Clade 1 species primarily cause tinea capitis as endothrix infection, whereas clade 2 species predominantly are agents of tinea pedis, onychomycosis, and tinea corporis. They have occasionally been described to cause tinea capitis, but then (e.g., “T. megninii”) only in connection with ectothrix infections. Distribution of species around T. violaceum is mostly restricted to Africa, whereas those around T. rubrum are distributed worldwide.
In their phenetic characters, the two clades are clearly different. The main characteristics are summarized as follows.
T. violaceum.
Colonies of T. violaceum strains grow slowly and are glabrous, leathery, wrinkled, and purple-red, yellow, or apricot-red; reverse is dark yellow, red-brown, purple, or violet. Macroconidia are absent. Microconidia, when present, are tear shaped. Hyphae are highly distorted; they may have reflexive branching (T. soudanense), and chlamydospores (T. yaoundei) may be present. Urease is mostly positive. In vitro hair perforation tests are negative.
T. rubrum.
Colonies of T. rubrum grow quickly and are fluffy to downy, white, sometimes becoming rose colored when aging; reverse is wine-red to olive, or sometimes yellow. Macroconidia are sparse or abundant, variable in size, and cyclindrical to cigar shaped, with a tendency to disarticulate. Microconidia are mostly present and pyriform to clavate. Urease is mostly positive. In vitro hair perforation tests are negative.
Below, the identity of anamorph strains and typification of anamorph species are discussed. For full nomenclature of each taxon, the reader is referred to the work of de Hoog et al. (14).
(i) T. abissinicum (Agostini) Nannizzi.
T. abissinicum (Agostini) Nannizzi (2, 29) was originally described as Bodinia abissinica and was isolated from a case of tinea capitis of a native in Eritrea by Agostini (2). CBS 126.34 is probably Agostini's original isolate, because it was sent to the CBS by G. Pollacci, who was Agostini's director at that time. A molecular analysis (data not shown) indicated that this species took an intermediate position between T. tonsurans and T. interdigitale, together with T. balcaneum, T. immergens, and T. radicosum. As is the case with those three species, the taxonomic position of this species remains as yet unresolved.
(ii) T. circonvolutum Sabouraud.
No authentic material from T. circonvolutum Sabouraud (36) is known to be preserved. CBS 186.30 is a secondary strain and was sent by G. Pollacci in 1930. Unfortunately the first description is poor. On the basis of morphology, Dodge (15) supposed it was perhaps referable to the genus Favotrichophyton along with Favotrichophyton violaceum (later T. violaceum), but our data indicate that CBS 186.30 is T. rubrum. This is in agreement with the clinical picture in the protologue, because the type specimen was isolated from a case of tinea corporis (buttock lesion).
(iii) T. fischeri Kane.
CBS 100081 is the type strain of T. fischeri Kane (24). Based on phenotypic characters, Kane separated this species from T. rubrum by more abundant sporulation (not in agreement with our data [see Table 3]) and its inability to form a red pigment on CAS-erythritol-albumin agar. The species is described as nonpathogenic since it was isolated as a contaminant of blood plates and from sputum of a patient with pneumonia caused by Pneumocystis carinii (35). The molecular and physiological data indicate that T. fischeri and T. rubrum are conspecific. In addition, it is known that arthroconidia of T. rubrum can survive up to 18 months in the environment (4), and thus airborne contamination appears not to be unusual.
(iv) T. fluviomuniense Miguens.
CBS 592.68 is the type strain of T. fluviomuniense Miguens (28). It was isolated from a case of tinea corporis. The taxon is conspecific with T. rubrum.
(v) T. glabrum Sabouraud.
Probably no type material has been preserved for T. glabrum Sabouraud (36). Strain CBS 499.48 is a secondary isolate of strain E. Rivalier originating from a tinea capitis in France. Colonies of T. violaceum lacking the ability to form the pigment were previously called T. glabrum. Our data confirm the opinion of Rippon (34), who treated the name as a synonym of T. violaceum.
(vi) T. gourvilii Catanei.
No authentic material is known to be preserved from T. gourvilii Catanei (9). CBS 360.62 is a secondary strain which was isolated from a native of Togo suffering from a nail mycosis. This species is considered by some authors to be conspecific with T. violaceum (32). Our data support their observation.
(vii) T. kanei Summerbell.
CBS 289.86, originating from a tinea corporis, is the type strain of T. kanei Summerbell (40). The species resembles T. rubrum morphologically, but differs by its inability to produce microconidia. PCR fingerprinting and AFLP patterns along with ITS sequencing data show 100% homology to T. rubrum.
(viii) T. kuryangei Vanbreuseghem & Rosenthal.
CBS 517.63 is the type strain of T. kuryangei Vanbreuseghem & Rosenthal (44). In agreement with our molecular data, Varsavsky and Ajello (45) suggested that the species is conspecific with T. megninii on morphological grounds, the latter being a synonym of T. rubrum.
(ix) T. megninii Blanchard.
T. megninii Blanchard (5) dates back to 1896, and it is the oldest name available in the T. rubrum complex. Cultures were not made at that time, and authentic material hence is not available. CBS 734.88 and 735.88 are secondary strains, which were isolated by M. Pereiro from cases of human tinea corporis in Spain. The micromorphology resembles that of T. rubrum, suggesting that it might be a variant of T. rubrum. T. megninii is physiologically distinct from T. rubrum in its requirement of l-histidine. Since the strains with this characteristic have close molecular resemblance to T. rubrum, probably a metabolic mutant is concerned. Thus, the strains with this characteristic are regarded as synonyms of T. rubrum. However, the protologue is insufficiently clear to be certain whether Blanchard's (5) taxon is identical to this mutant; we therefore treat the name T. megninii as of doubtful identity. Bodin (7) further listed a Trichophyton roseum Sabouraud. Authentic material for this taxon is lost. In the protologue, strains from humans and chickens are listed, and hence it is difficult to guess which species was concerned. T. roseum is therefore regarded as a doubtful species.
(x) T. pedis Ota.
No authentic material is known to be preserved for T. pedis Ota (31). CBS 189.69 is a secondary strain isolated from a nail mycosis. This strain was identified as T. rubrum.
(xi) T. pervesii Catanei.
T. pervesii Catanei (10) strain CBS 303.38 is authentic for the species. The author failed to provide a Latin description as required by the International Code of Botanical Nomenclature (ICBN) and hence the name is invalid (Article 36, ICBN). In the CBS list of cultures, the species was already synonymized with T. rubrum on morphological grounds. Our molecular results confirm these data.
(xii) T. raubitschekii Kane, Salkin, Weitzman & Smitka.
CBS 100084 is the type strain of T. raubitschekii Kane, Salkin, Weitzman & Smith (26). The morphological and physiological features separating it from T. rubrum are relatively minute (the species [i] is urease positive, [ii] has restricted growth on lactose agar, [iii] shows brown rather than red pigmentation on casein dextrose agar, and [iv] produces abundant macroconidia). Using restriction fragment length polymorphism analysis of the mitochondrial DNA, Ishizaki et al. (22) revealed identical patterns between T. raubitschekii and T. rubrum, suggesting conspecificity. Our data confirm their suggestion. Therefore, this species is reduced to synonymy with T. rubrum.
(xiii) T. rodhainii Vanbreuseghem.
T. rodhainii Vanbreuseghem (43) strain CBS 376.49 is the type strain of this species. The taxon name is illegitimate, as no Latin description was provided by the author. Ajello (3) already equated the species with T. rubrum, which is confirmed by our data.
(xiv) T. rubrum (Castellani) Semon.
No type material is known to be preserved for T. rubrum (Castellani) Semon (8, 38). The authentic strain originated from a case of tinea cruris. CBS 392.58, isolated from tinea pedis, shows all of the morphological characteristics described by Castellani, which are currently maintained as diagnostic for the species (14, 25), and it is therefore indicated as the neotype strain for T. rubrum.
(xv) T. rubrum var. nigricans Frágner.
T. rubrum var. nigricans Frágner (15) is characterized by its melanoid pigmentation. No type material was available; ATU TR9 and 16 are secondary isolates isolated by A. Hasegawa. T. rubrum var. nigricans is reduced to synonymy with T. rubrum.
(xvi) T. soudanense Joyeux.
The authentic material for T. soudanense Joyeux (23) is probably lost. CBS 452.61 (equals R.V. 10184) was isolated by R. Vanbreuseghem from a patient in Zaire in 1959. The species' commonest clinical manifestation is identical to that of T. violaceum, i.e., “black dot,” an endothrix infection of the scalp. The taxon is a synonym of T. violaceum.
(xvii) T. violaceum var. indicum Acton & McGuire.
The type strain of T. violaceum var. indicum Acton & McGuire (1) is CBS 319.31. According to the authors, it differed only in cultural characteristics, but not in morphology. The taxon is synonymous with T. violaceum.
(xviii) T. violaceum Sabouraud.
No type material is known to be preserved for T. violaceum Sabouraud (7). The protologue (7) describes an isolate from a tinea corporis in Paris, which is in accordance with the cultural and morphological criteria maintained in recent treatments of the genus Trichophyton (14). Therefore CBS 374.92, from a human tinea corporis in The Netherlands, has been selected as the neotype strain.
(xix) T. yaoundei Cochet & Doby-Dubois.
T. yaoundei Cochet & Doby-Dubois (13) strain 305.60 is authentic and was isolated from a patient with tinea capitis. The species name is invalidly described as it lacked a Latin diagnosis (Article 36, ICBN). It was considered by some authors (32) to be conspecific with T. violaceum; our data support this observation. A recent ITS sequence study by Summerbell et al. (41) has placed T. yaoundei near Trichophyton simii, a monkey dermatophyte. This placement is based on a misidentified isolate deposited in the University of Alberta Microfungus Collection and Herbarium.
ACKNOWLEDGMENTS
We thank Jirko Kühnisch for excellent technical assistance. Parts of the sequences were kindly provided by M. El Fari.
Funding was provided by the Deutsche Forschungsgemeinschaft, GR 1147/1-1 and GR 1147/1-2, to H.-J. Tietz and Y. Gräser. Part of the work was carried out at the CBS by the senior author and was financially supported by this institute.
REFERENCES
- 1.Acton H W, McGuire C. “Cooly itch.” a purulent folliculitis due to the Trichophyton violaceum variety indicum. Indian Med Gaz. 1929;65:241–246. [PMC free article] [PubMed] [Google Scholar]
- 2.Agostini A. Una nuova specie di Bodinia causa di tigna umana nell'Eritrea. Atti Ist Bot Univ Pavia. 1930;II:118–125. [Google Scholar]
- 3.Ajello L. A taxonomic review of the dermatophytes and related species. Sabouraudia. 1968;6:147–159. doi: 10.1080/00362176885190271. [DOI] [PubMed] [Google Scholar]
- 4.Baer R L, Rosenthal S A, Furnari D. Survival of dermatophytes applied on the feet. J Invest Dermatol. 1955;24:619–662. doi: 10.1038/jid.1955.83. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard R. Parasites vegetaux à l'exclusion des bacteries. In C. Bouchard, Traité de pathologie génerale vol. 2. Paris, France: Masson et Cie; 1896. pp. 811–926. [Google Scholar]
- 6.Blank F. Dermatophytes of animal origin transmissible to man. Prog Med Sci. 1955;229:302–316. [PubMed] [Google Scholar]
- 7.Bodin E. Les champignons parasites de l'homme. Paris, France: Masson et Cie; 1902. [Google Scholar]
- 8.Castellani A. Observation on a new species found in tinea cruris. Br J Dermatol. 1910;5:148–150. [Google Scholar]
- 9.Catanei A. Description de Trichophyton gourvili n. sp., agent d'une teigne de l'homme. Bull Soc Pathol Exot. 1933;26:377–381. [Google Scholar]
- 10.Catanei A. Description de deux nouvelles espèces et d'une variété nouvelle de champignons provoquant des teignes chez l'homme. Arch Inst Pasteur Alger. 1937;15:265–270. [Google Scholar]
- 11.Chakraborty A N, Ghosh S, Blank F. Isolation of Trichophyton rubrum (Castellani) Sabouraud, 1911, from animals. Can J Comp Med Vet Sci. 1954;18:436. [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen W B. Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella. J Bacteriol. 1946;52:461–466. doi: 10.1128/jb.52.4.461-466.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochet G, Doby-Dubois M. Contribution à la connaissance des teignes infantiles du Cameroun (note préliminaire) Semaine Hop Paris. 1957;33:2980–2986. [PubMed] [Google Scholar]
- 14.de Hoog, G. S., J. Guarro, M. J. Figueras, and J. Gené. Atlas of clinical fungi, 2nd ed., in press. Centraalbureau voor Schimmelcultures/Universitat Rovira i Virgili, Baarn/Reus, The Netherlands.
- 15.Dodge C W. Medical mycology. St. Louis, Mo: C.V. Mosby Co.; 1935. p. 533. [Google Scholar]
- 16.Frágner P. Trichophyton rubrum (Cast.) Sabouraud var. nigricans var. nova. Ceska Mykol. 1966;20:27–28. [Google Scholar]
- 17.Georg L K. Routine nutritional test for the identification of dermatophytes. J Bacteriol. 1957;74:113–121. doi: 10.1128/jb.74.2.113-121.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gräser Y, Kuijpers A, Presber W, de Hoog G S. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol. 1999;37:315–330. doi: 10.1046/j.1365-280x.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 19.Gräser Y, El Fari M, Vilgalys R, Kuijpers A F A, de Hoog G S, Presber W, Tietz H-J. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ITS region. Med Mycol. 1999;37:105–114. [PubMed] [Google Scholar]
- 20.Gräser, Y., A. F. A. Kuijpers, M. El Fari, W. Presber, and G. S. de Hoog. Molecular and conventional taxonomy of the Microsporum canis complex. Med. Mycol., in press. [DOI] [PubMed]
- 21.Gräser Y, Kühnisch J, Presber W. Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum. J Clin Microbiol. 1999;37:3713–3717. doi: 10.1128/jcm.37.11.3713-3717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishizaki H. Fungal taxonomy based on mitochondrial DNA analysis. Jpn J Med Mycol. 1993;34:243–251. [Google Scholar]
- 23.Joyeux C. Sur le Trichophyton soudanense, sp. nov. C R Soc Biol Paris. 1912;72:15–16. [Google Scholar]
- 24.Kane J. Trichophyton fischeri sp. nov.: a saprophyte resembling Trichophyton rubrum. Sabouraudia. 1977;15:231–241. [PubMed] [Google Scholar]
- 25.Kane J, Summerbell R, Sigler L, Krajden S, Land G. Laboratory handbook of dermatophytes. Belmont, Calif: Star Publishing Company; 1997. [Google Scholar]
- 26.Kane J, Salkin I F, Weitzman I, Smitka C. Trichophyton raubitschekii, sp. nov. Mycotaxon. 1981;13:259–266. [Google Scholar]
- 27.McClealland M, Peterson M, Welsh J. Length polymorphisms in tRNA intergenic spacer detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J Clin Microbiol. 1992;30:1499–1504. doi: 10.1128/jcm.30.6.1499-1504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miguens M P. Un nuevo dermatofito de origin tropical; Trichophyton fluviomuniense sp. nov. Sabouraudia. 1968;6:312–317. [PubMed] [Google Scholar]
- 29.Nannizzi A. Repertorio sistematico del wiceti dell' uomo e degli animali. Trattato di Micopatol Um. 1934;4:174–175. [Google Scholar]
- 30.Niesters H G M, Goessens W H F, Meis J M F G, Quint W G V. Rapid polymerase chain reaction-based identification assay for Candida species. J Clin Microbiol. 1993;31:904–910. doi: 10.1128/jcm.31.4.904-910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ota M. Sur deux espèces nouvelles de dermatophytes en Mandchourie: Microsporum ferrugineum et Trichophyton pedis n. sp. Bull Soc Pathol Exot. 1992;15:588–596. [Google Scholar]
- 32.Otçenásek M, Dvorák J. Ecological classification of dermatophytes. Mykosen. 1975;18:425–434. [PubMed] [Google Scholar]
- 33.Padhye A A, Young C N, Ajello L. Hair perforation as a diagnostic criterium in the identification of Epidermophyton, Microsporum and Trichophyton species. Pan Am Sci Publ. 1980;396:115–120. [Google Scholar]
- 34.Rippon J W. Medical mycology: the pathogenic fungi and pathogenic Actinomycetes. Philadelphia, Pa: The W. B. Saunders Co.; 1988. p. 178. [Google Scholar]
- 35.Rosenthal S A, Scott J S, Summerbell R C, Kane J. First isolation of Trichophyton fischeri in the United States. J Clin Microbiol. 1998;36:3389–3391. doi: 10.1128/jcm.36.11.3389-3391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabouraud R. Maladies du cuir chevelu. Paris, France: Masson et Cie; 1910. [Google Scholar]
- 37.Savelkoul P H M, Aarts H J M, De Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker J L W, Schouls L, Lenstra J A. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999;37:3083–3091. doi: 10.1128/jcm.37.10.3083-3091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semon H C. Tinea unguium. Br J Derm Syph. 1922;34:397–400. [Google Scholar]
- 39.Shadomy H J, Philpot C M. Utilization of standard laboratory methods in the laboratory diagnosis of problem dermatophytes. Am J Clin Pathol. 1980;74:197–201. doi: 10.1093/ajcp/74.2.197. [DOI] [PubMed] [Google Scholar]
- 40.Summerbell R C. Trichophyton kanei, sp. nov., a new anthropophilic dermatophyte. Mycotaxon. 1987;28:509–523. [Google Scholar]
- 41.Summerbell R C, Haugland R A, Li A, Gupta A K. rRNA gene internal transcribed spacer 1 and 2 sequences of asexual, anthropophilic dermatophytes related to Trichophyton rubrum. J Clin Microbiol. 1999;37:4005–4011. doi: 10.1128/jcm.37.12.4005-4011.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swofford D L. PAUP. Phylogenetic analysis using parsimony. Version 4. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 43.Vanbreuseghem R. Contribution à l'étude des dermatophytes du Congo Belge. Description du Trichophyton mégaspore T. rodhaini n. sp. Ann Parasitol Hum Comp. 1949;24:243–251. [Google Scholar]
- 44.Vanbreuseghem R, Rosenthal S A. Trichophyton kuryangei n. sp. nouveau dermatophyte Africain. Ann Parasitol Hum Comp. 1961;36:797–803. doi: 10.1051/parasite/1961365797. [DOI] [PubMed] [Google Scholar]
- 45.Varsavsky E, Ajello L. The perfect and imperfect forms of a new keratinophilic fungus Arthroderma ciferrii sp. nov.: Trichophyton georgii sp. nov. Riv Patol Veg. 1964;4:351–364. [Google Scholar]
- 46.Vos P, Hogers R, Bleeker M, Reijans A N, de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;21:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young C N. –1968. Pseudocleistothecia in Trichophyton rubrum. Sabouraudia. 1967;6:160–162. [PubMed] [Google Scholar]