Abstract
Around 30–50% of classical Hodgkin lymphoma (cHL) cases in immunocompetent individuals from industrialized countries are associated with the B-lymphotropic Epstein-Barr virus (EBV). Although natural killer (NK) cells exhibit anti-viral and anti-tumoral functions, virtually nothing is known about quantitative and qualitative differences in NK cells in patients with EBV+ cHL vs. EBV- cHL. Here, we prospectively investigated 36 cHL patients without known immune suppression or overt immunodeficiency at diagnosis. All 10 EBV+ cHL patients and 25 out 26 EBV- cHL were seropositive for EBV antibodies, and EBV+ cHL patients presented with higher plasma EBV DNA levels compared to EBV- cHL patients. We show that the CD56dim CD16+ NK cell subset was decreased in frequency in EBV+ cHL patients compared to EBV- cHL patients. This quantitative deficiency translates into an impaired CD56dim NK cell mediated degranulation toward rituximab-coated HLA class 1 negative lymphoblastoid cells in EBV+ compared to EBV- cHL patients. We finally observed a trend to a decrease in the rituximab-associated degranulation and ADCC of in vitro expanded NK cells of EBV+ cHL compared to healthy controls. Our findings may impact on the design of adjunctive treatment targeting antibody-dependent cellular cytotoxicity in EBV+ cHL.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-02956-x.
Keywords: Epstein-Barr virus, Classical Hodgkin lymphoma, Natural killer cells, Rituximab, Antibody-dependent cellular cytotoxicity
Introduction
Classical Hodgkin lymphoma (cHL) is a solid lymphoid cancer characterized by a very low frequency of neoplastic cells, i.e., the Hodgkin and Reed-Sternberg (HRS) cells, that are surrounded by an inflammatory tumor microenvironment (TME) [1]. The pathognomonic HRS cells are of B cell origin with evidence of somatic hypermutation indicating that they are germinal center experienced [2]. cHL can be subclassified into four histologic subtypes (nodular sclerosis, mixed-cellular, lymphocyte-rich and lymphocyte-depleted) and can be staged I to IV depending on tumoral extension. Treatment of cHL with chemotherapy achieves a 5-year cure in more than 80% of the cases [3]. Patients suffering from treatment-refractory cHL might ultimately benefit from novel antibody-based immunotherapies [4].
Around 30–50% of cHL cases in immunocompetent individuals from industrialized countries are associated with the -herpesvirus Epstein-Barr virus (EBV) [5], while in resource-poor countries this association may be higher than 70% and in patients infected with HIV up to 100% [2]. The EBV status of the HRS cell in newly diagnosed cHL is not routinely determined since it does not impact on the choice of chemotherapy regimen in treatment guidelines. EBV establishes an asymptomatic latent infection in the vast majority of adults and is primarily controlled by T cells and natural killer (NK) cells [6]. EBV exhibits potent B cell growth-transformation properties in vitro and is associated with several B cell cancers in immunocompetent and immunocompromised individuals. With respect to cHL, EBV is preferentially linked to the mixed cellularity and lymphocyte-depleted subtypes and is found as monoclonal viral genome in HRS cells [5]. Epidemiological [7] and genetic [8] studies suggest that EBV-negative (EBV-) cHL and EBV-positive (EBV+) cHL differ in their pathogenesis, EBV latent genes thereby providing survival signals for HRS cells.
NK cells contribute to the immune control of malignant cells [9, 10] and viruses [11]. The blood NK cell compartment is mainly composed of the two well-characterized functional CD56bright CD16- and CD56dim CD16+ subsets [12]. The former subset produces large amounts of cytokines upon stimulation, acquires cytotoxicity only after prolonged activation and is enriched in secondary lymphoid organs. On the other hand, NK cells of the latter subset readily kill susceptible targets, can rapidly secrete IFN-γ upon engagement of activating receptors and are involved in the antibody-dependent cellular cytotoxicity (ADCC) mediated by the low affinity FcγRIIIA receptor CD16 [13]. Several maturation stages can be further delineated within the cytotoxic CD56dim NK cell subset based on the expression of NKG2A, Killer-cell Immunoglobulin-like receptors (KIR) [14], the intermediate stage marker CD62L [15] and the terminal differentiation marker CD57 [16].
NK cells in cHL patients at diagnosis are decreased in frequency in the TME [17] as well as in absolute numbers in the peripheral blood [10] compared to in healthy controls (HC). Furthermore, cytotoxicity toward the erythroleukemic K562 [18, 19] and the EBV- cHL L428 [20] cell lines is impaired in NK cells of cHL patients compared to NK cells of HC. Several lines of evidence suggest that NK cells found within the cHL tumor are functionally impaired [21]. Furthermore, exhausted PD-1+ NK cells are expanded in the peripheral blood of cHL patients at diagnosis independently of the EBV status of the tumor [22].
Thus, in the light of increasingly available adjunctive immune treatments for cHL [4] we sought to phenotypically characterize NK cells from patients with EBV+ and EBV- cHL at diagnosis and to functionally test their reactivity toward EBV-infected B cells.
Material and methods
Study design and human samples
Newly diagnosed and treatment naïve adult patients with cHL were prospectively enrolled at the Hematology Department of Gustave-Roussy between October 2015 and July 2018. cHL cases were classified according to the Ann Arbor staging. The cHL histological subtype was defined according to the WHO classification. The presence of EBV in the HRS cells was assessed by EBER-FISH and was not communicated until completion of the phenotypic part of the study. Serologies for HIV and EBV were performed in all cHL patients as part of the baseline evaluation. The EBV-seropositive (i.e., having serological evidence of prior infection with EBV) cHL patients of the study were either designated EBV+ (presence of EBV within the tumor; EBV-seropositive EBV+ cHL) or EBV- (absence of EBV within the tumor; EBV-seropositive EBV- cHL). The EBV-seronegative, i.e., not infected with EBV, cHL patients are EBV- cHL (EBV-seronegative and absence of EBV within the tumor). Five healthy adult donors (HC) living in the same area as the cHL patients (age range 25–49 years, 4 males, all EBV-seropositive) were enrolled secondarily in the study. The institutional ethics committee approved all protocols used and all participants provided informed consent in accordance with the Declaration of Helsinki.
PBMC isolation
Peripheral blood samples of HL patients and HC were drawn at the Hematology Department of Gustave-Roussy, shipped to the Experimental Infectious Disease and Cancer Research laboratory in Zurich and processed for peripheral blood mononuclear cells (PBMC) and plasma isolation within 24 h of blood puncture as stated elsewhere [23].
Quantification of EBV
Quantification of EBV DNA copy number in plasma was performed as previously described [23].
Flow cytometry analysis
Frozen PBMCs were thawed, washed and stained with monoclonal antibodies (Supplementary Table 1) at room temperature for 15 min. LIVE/DEAD Fixable Aqua (Invitrogen) was used for dead cell exclusion. Samples were acquired on a LSR Fortessa (BD Biosciences), and all flow cytometry analyses were performed with FlowJo Version 10 software (Tree star, Inc). The NK cell subset gating strategy is depicted in the Supplementary Fig. 1.
CD107a-based degranulation assay
The HLA class 1 negative lymphoblastoid cell line LCL721.221 (LCL221) was maintained in complete medium (RPMI-1640 medium; Sigma, Life Science) supplemented with 10% heat-inactivated FBS (Sigma), 2 mM Glutamax (Gibco by Life Technologies), 1% Penicillin Streptomycin (Gibco) and tested negative for mycoplasma. LCL221 (Cellosaurus, RRID:CVCL_6263) has been authenticated by Microsynth using STR profiling in February 2020. For the degranulation assay with rIL-2 pre-stimulation, frozen PBMCs from 7 EBV+ and 7 EBV- cHL were thawed and incubated overnight with 100 IU recombinant interleukin-2 (rIL2) per ml. For the antibody-based degranulation assay without rIL-2 pre-stimulation, frozen PBMCs from 5 EBV+ cHL, 5 EBV- cHL and 5 HC were thawed and incubated overnight without any supplementary cytokines. The next day, 1.5×105 PBMC were either resuspended in complete medium (medium control) or co-cultured with LCL221 at an effector to target ratio (E:T ratio) of 10:1 for 5 h with or without the monoclonal antibody anti-CD20 rituximab (1 μg/ml, MabThera). All conditions were performed in duplicates and with addition of the monoclonal antibody anti-CD107a (LAMP-1) pacific blue (Bio Legend) at the beginning of the assay. After 1 h, monensin (1 μg/ml BD Golgi Stop; BD Pharmingen) was added to all samples. At the end of the incubation, cells were stained with mAbs and analyzed by flow cytometry. The gating strategy is depicted in the Supplementary Fig. 2. The spontaneous degranulation (medium control) from NK cell subsets was subtracted in each degranulation analysis.
In vitro NK cell expansion
NK cells were expanded from PBMCs (4 EBV+ HL, 4 EBV- HL, 4 HC) as previously described [24–26] with some modifications. NK cells were cultured in NK cell media consisting of RPMI 1640 (Life Technologies) supplemented with 10% fetal calf serum (FCS; Biochrom), 1% penicillin/streptomycin (Thermo Fisher) and 200 IU/mL IL-2 (Preprotech). PBMCs were thawed, counted and co-cultured with irradiated (130 Gy) K562-mbIL21 feeder cells (kindly provided by Dr. Dean Lee, Nationwide Children’s Hospital, Columbus, United States) at a 1:2 NK:feeder cell ratio at day 0. On day 7 and 14, NK cells were re-stimulated with irradiated (130 Gy) K562-mbIL21 feeder cells (1:1 NK:feeder cell ratio). IL-2 (200 IU/mL) was replenished every 2–3 days. On day 20, cultured cells were CD3-depleted via immunomagnetic depletion according to manufacturer’s instructions (Miltenyi Biotech). Flow cytometric analysis of NK-cell purity and viability was performed on days 0, 14 and 20 of expansion before addition of K562mb-IL21 feeder cells.
CD107a-based degranulation assay and ADCC assay with expanded NK cells
Degranulation assays using expanded NK cells in culture from 4 EBV+ cHL, 4 EBV- cHL and 4 HC were performed as described before but with an E:T ratio of 1:2 with 1 μg/ml rituximab. The ADCC assay was performed according to Lee-MacAry et al. [27]. Briefly, in vitro expanded and T cell depleted NK cells from 4 EBV+ cHL, 4 EBV- cHL and 4 HC were thawed and incubated overnight to rest. The next day LCL221 cells were labeled with PKH-26 (Sigma-Aldrich) and co-cultured with the expanded NK cells at a E:T ratio of 1:3, 1:1 and 3:1 for 4 h in the presence or absence of rituximab (1 μg/ml). Target cell lysis was assessed by flow cytometry on PKH-26-positive cells using TO-PRO-3 iodide (Invitrogen) staining at the end of the co-culture. The baseline target cell death was subtracted from each sample, according to the presence or absence of rituximab.
Statistical analysis
Data were analyzed using Prism software (version 8.41.; GraphPad Software, Inc.). P values < 0.05 were considered significant. For the categorical data, Chi2 statistical test was used. For the numerical data comparing 2 groups, unpaired t test was used. For statistical analysis of 3 groups, we used the one-way ANOVA test with Tukey’s multiple comparisons test. Comparison between paired samples was assessed with the paired t test. Correlation studies were assessed with the Pearson test. Figures were created in Adobe Illustrator. Tables were created in Excel.
Results
One fourth of the cHL are associated with EBV
We prospectively enrolled 36 newly diagnosed and treatment-naïve cHL patients without known immune suppression or overt immunodeficiency. All patients were tested seronegative for HIV. The demographic and cancer characteristics of 26 EBV- cHL patients (72.2%) and 10 EBV+ cHL patients (27.7%) is shown in Supplementary Table 2, with the statistical comparison displayed in Table 1. All 10 EBV+ cHL and 25 out 26 EBV- cHL were tested seropositive for EBV antibodies. The majority (83%) of cHL was diagnosed with the nodular sclerosis subtype. None of the patients suffered from a treatment refractory cHL during the clinical follow-up. The mean age and the frequency of male patients were significantly higher in EBV+ cHL patients compared to their EBV- counterparts. Both groups did not differ in terms of frequency of histological subtypes, Ann Arbor staging, blood lymphocyte counts and EBV-seropositivity.
Table 1.
Comparison of demographic and cancer parameters of patients with EBV-positive versus EBV-negative classical Hodgkin lymphoma. The statistical tests used are stated as footnote
| EBV-positive cHL | EBV-negative cHL | ||
|---|---|---|---|
| (n=10) | (n=26) | P value | |
| Age (years) | 48; 24; 76 | 35; 19; 74 | 0.04 a |
| Male sex (n; %) | 8; 80 | 10; 38.4 | 0.02 b |
| Nodular sclerosis HL (n; %) | 7; 70 | 23; 88 | 0.18 b |
| Ann Arbor stage ≥ III (n; %) | 5; 50 | 12; 46 | 0.83 b |
| Lymphocyte count (G/l) | 1.27; 0.90; 1.70 | 1.26; 0.40; 3.30 | 0.96 a |
| Plasma EBV DNA (copies/ml) | 3414; 0; 11,743 | 26; 0; 143 | 0.0002 a |
| EBV-seropositive (n; %) | 10; 100 | 25; 96 | 0.52 |
aunpaired t test (mean; min; max)
bchi-square test
Reduced frequency of cytotoxic CD56dim CD16+ NK cells in EBV+ cHL
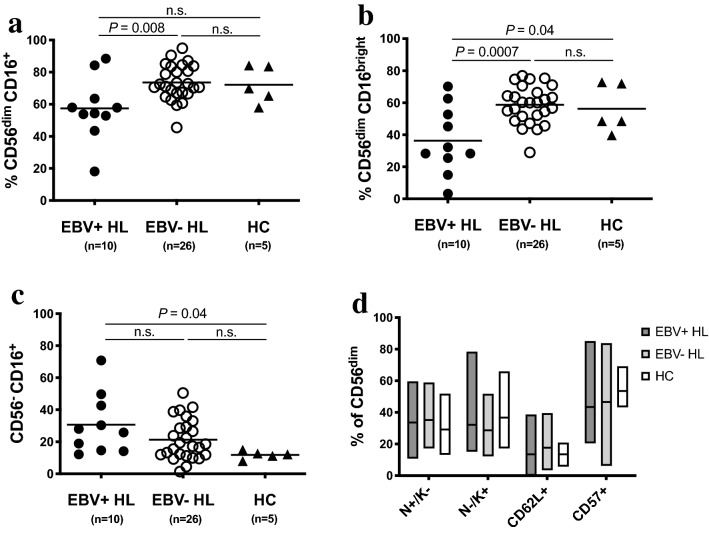
We assessed the frequencies of NK cell subsets in the peripheral blood of cHL patients and HC. The gating strategy is depicted in Supplementary Fig. 1. There were no differences for CD3- CD56+ (Supplementary Fig. 3A) and CD56bright CD16- (Supplementary Fig. 4A) NK cells. However, we found that the CD56dim CD16+ NK cell subset was decreased in frequency in EBV+ compared to EBV- cHL patients (Fig. 1a). CD56dim CD16+ NK cells are composed of two functionally different subset [28], namely CD56dim CD16dim and CD56dim CD16bright. EBV+ cHL exhibited a decreased frequency of CD56dim CD16bright NK cells compared to EBV- cHL and HC (Fig. 1b). On the other hand, we observed in EBV+ cHL an increased frequency of CD56dim CD16- (Supplementary Fig. 4B) and CD56dim CD16dim (supplementary Fig. 4C) NK cells compared to EBV- cHL. The frequency of CD56- CD16+ was higher in EBV+ cHL compared to HC (Fig. 1c). Representative staining of three EBV+ cHL-patients, i.e., one with high frequency of CD56bright CD16- NK cells (left panel) and two with high frequency of CD56dim CD16- NK cells (middle and right panel) is depicted in supplementary Fig. 4D. The level of CD16 expression on monocytes was not reduced in EBV+ cHL patients with low frequencies of CD56dim CD16+ NK cells (supplementary Fig. 4E). The frequencies of CD56dim NK cell maturation stages defined as NKG2A+ KIR-, NKG2A- KIR+, CD62L+ and CD57+ were comparable between the three groups (Fig. 1d). We additionally assessed the frequency of T cell subsets known to be involved in the immune response toward EBV [6]. EBV+ cHL and EBV- cHL patients exhibit similar frequencies of CD4+ T cells, CD8+ T cells, TCR δ-1+ (γδ-1) T cells and TCR δ-2+ (γδ-2) T cells (Supplementary Fig. 3B-E).
Fig. 1.
Decrease of CD56dim CD16+ NK cell subset frequency in EBV+ HL patients compared to EBV- HL patients. Thawed PBMCs from EBV+ and EBV- HL patients and healthy controls (HC) were analyzed by flow cytometry. Frequencies of CD56dim CD16+ a, CD56dim CD16bright b and CD56- CD16+ c NK cells of EBV+ HL (n=10; filled circles), EBV- HL (n=26; open circles) and HC (n=5, triangles). d Frequencies of NKG2A+ KIR- (N+/K-), NKG2A- KIR+ (N-/K+), CD62L+ and CD57+ cells within CD56dim NK cells of EBV+ HL (dark gray, n=10), EBV- HL (light gray, n=26) and HC (white, n=5). Horizontal lines in panels a to c indicate the mean value. Floating bars in panel d indicate mean and minimum and maximum values. Significance was determined by the one-way ANOVA test with Tukey’s multiple comparisons test
Impaired CD56dim NK cell-mediated rituximab-dependent degranulation in EBV+ cHL
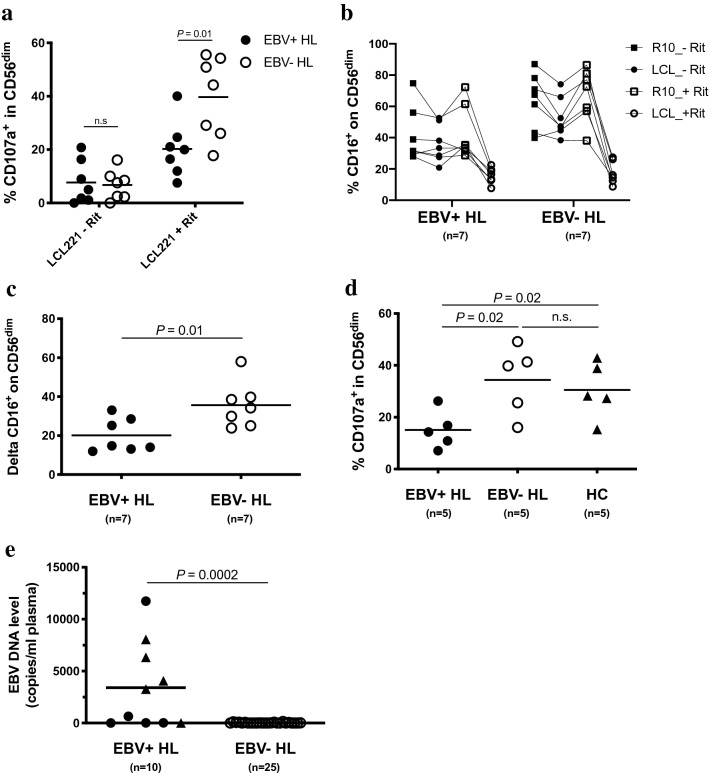
We next assessed the NK cell mediated natural cytotoxicity and CD16-mediated antibody-dependent degranulation upon rIL-2 overnight stimulation and co-culture with the HLA class 1 negative LCL221. Patients from both cHL groups exhibited comparable natural cytotoxicity of the CD56dim NK cell subset (Fig. 2a, left part). The addition of rituximab led in samples of EBV- cHL patients to a sixfold increase of the mean frequency of degranulating CD107a+ CD56dim NK cells (Fig. 2a, right part), but only a 2.5-fold increase in EBV+ cHL patients. Differences in the frequencies of both effector CD56dim CD16+ NK cells and bystander autologous CD20+ B cells might theoretically influence our PBMC-based antibody-dependent degranulation assay. We did confirm the decreased frequency of CD16+ on CD56dim NK cells observed ex vivo (Fig. 1a) after overnight incubation in rIL2-containing medium of EBV+ cHL compared to EBV- cHL samples (Supplementary Fig. S5A). Moreover, we found a positive correlation between the frequency of CD16+ on CD56dim NK cells and the antibody-dependent degranulation of CD56dim upon co-culture with rituximab-coated LCL221 (Supplementary Fig. 5B). The frequency of CD56dim CD16+ within the live PBMC is similar in EBV+ vs. EBV- cHL samples (data not shown), indicating that the effective E:T ratios applied for the 14 samples chosen for this assay are comparable between both groups. On the other hand, EBV+ and EBV- cHL samples did not differ in the frequencies of CD20+ CD19+ B cells with the live PBMC fraction (Supplementary Fig. 5C). We did not find an overall increase in the CD56dim mediated degranulation in the medium control after addition of rituximab (calculated as ratio over medium control without rituximab) nor any correlation with the frequency of CD20+ CD19+ B cells (Supplementary Fig. 5D). As second read-out for antibody-dependent degranulation [29], we quantified the additional CD16 shedding of CD56dim NK cells [30] upon co-culture with LCL221 cells with and without rituximab. CD16 expression on CD56dim NK cells decreased upon addition of rituximab to target cells (Fig. 2b). EBV+ cHL patients exhibited a significantly reduced CD16 shedding upon co-culture with LCL221 with vs. without rituximab compared to their EBV- counterparts (Fig. 2c). We next assessed the rituximab-dependent CD56dim NK cell degranulation without rIL-2 pre-stimulation of PBMC. EBV+ cHL displayed decreased frequencies of CD107a+ CD56dim NK cells compared to EBV- cHL (Fig. 2d). Finally, we observed higher plasma EBV DNA levels in EBV+ compared to EBV- cHL patients (Fig. 2e). Patients with advanced clinical stage (Ann Arbor stage ≥ 3) were overrepresented (4 out of 5) among EBV+ cHL with high plasma EBV DNA level (>1000 copies/ml of plasma). The level of rituximab-dependent degranulation in CD56dim NK cells from the 7 EBV+ cHL patients analyzed did not correlate with plasma EBV DNA levels (data not shown).
Fig. 2.
Impaired rituximab-induced degranulation in CD56dim NK cells from EBV+ HL compared to EBV- HL patients. Overnight rIL-2-pre-stimulated PBMCs from 7 EBV+ HL and 7 EBV- HL patients were co-cultured with LCL721.221 cell line (LCL221) without or with 1 μg/ml of rituximab (Rit) at an effector to target ratio of 10:1 for 5 h. Frequencies of degranulating CD107a+ cells within CD56dim NK cells a of EBV+(filled circles) and EBV-(open circles) HL patients. b Frequencies of CD16+ cells within CD56dim of EBV+ and EBV- HL patients in different experimental conditions (medium control or LCL221; without or with 1 μg/ml rituximab). c CD16 downregulation between the condition LCL221 without rituximab (LCL_-Rit in Fig. 2b) and LCL221 with rituximab (LCL_+Rit) is depicted as delta. d PBMCs of 5 EBV+ HL, 5 EBV- HL patients and 5 healthy controls (HC) were thawed, incubated overnight without rIL2 pre-stimulation and co-cultured the next day with LCL221 in the presence of 1 μg/ml of rituximab at an effector to target ratio of 10:1 for 5 h. Frequencies of degranulating CD107a+ cells within the CD56dim NK cells of EBV+ HL (filled circles), EBV- HL (open circles) and HC (triangles). e Plasma EBV DNA levels (copies/ml of plasma) of EBV+ (filled circles, n=10) and EBV-seropositive EBV- (open circles, n=25) HL patients. EBV+ HL patients were further delineated according to the clinical stage, i.e., early stage I and II (filled circles) vs. advanced stage III and IV (filled triangles). Horizontal lines indicate the mean value in Fig. 2a-e. Displayed P values were determined by unpaired t test for comparison of 2 groups (Fig. 2a, c and e), by the one-way ANOVA test with Tukey’s multiple comparisons test for comparison of 3 groups (Fig. 2d) and by the paired t test for paired samples (Fig. 2b)
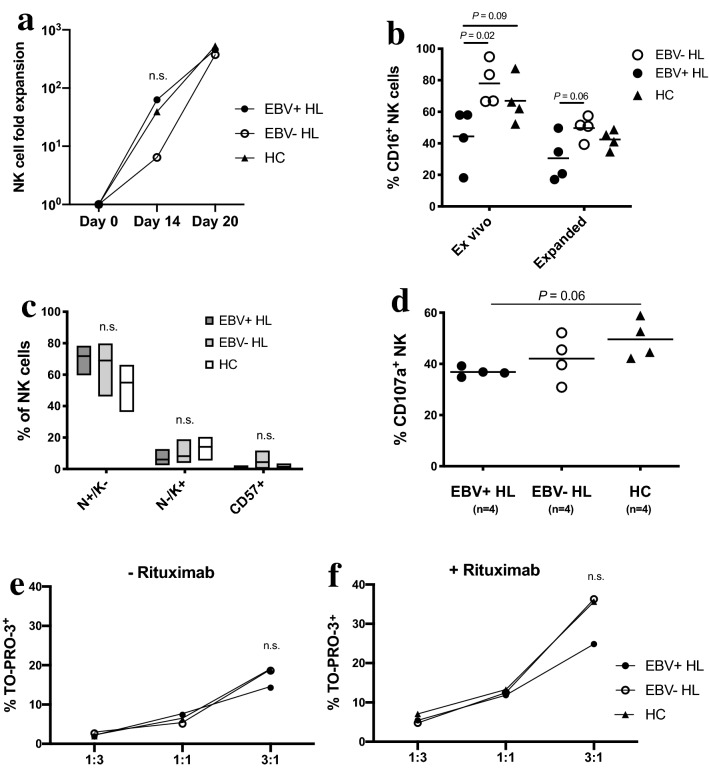
Unaltered in vitro expansion capacity of NK cells and impaired rituximab-dependent degranulation of expanded NK cell from EBV+ cHL
We next assessed if prolonged in vitro expansion of NK cells might rescue the reduced rituximab-dependent degranulation observed in EBV+ cHL. We expanded NK cells from 4 EBV+ cHL, 4 EBV- cHL and 4 HC for 20 days using irradiated K562 expressing IL-21 (K562-mbIL21) and rIL-2 supplementation [24]. The 3 groups did not differ in term of NK cell expansion from day 0 to day 20 (370–530-fold expansion; Fig. 3a). The mean frequency of NK cells at day 20 of expansion was comparable in each group (data not shown) and further increased between 65 and 95% after T cell depletion, except in one EBV+ cHL (15% NK cell) sample with too low cell numbers to undergo additional T cell depletion (E:T ratio in co-culture assays was maintained for this sample). The K562-mbIL21 mediated expansion led to an upregulation of CD56 (data not shown) and downregulation of CD16 expression on NK cells compared to baseline. However, only samples from EBV- cHL and HC exhibited a significant decrease in the frequency of CD16+ NK cells (Fig. 3b). The frequencies of NKG2A+KIR-, NKG2A- KIR+ and CD57+ NK cells (Fig. 3c) did not differ between the three groups. CD62L was not detected on expanded NK cells (data not shown). We performed a degranulation assay with LCL221 coated with rituximab using a lower E:T ratio of 1:2 due to the K562-mbIL21-mediated activation of expanded NK cells. We did observe a decreased frequency of CD107a+ NK cells in EBV+ cHL compared to HC close to the statistical significance (mean 36% vs. 49%, one way ANOVA with Tukey’s multiple comparisons test P=0.06; unpaired t test P=0.01; Fig. 3d). We finally performed with thawed expanded NK cells a flow cytometry based-ADCC assay using PKH26 labeling of LCL221 and the To-Pro-3 iodide dead cell marker. The gating strategy is depicted in the Supplementary Fig. 6. We could not observe between the three groups any significant difference in natural cytotoxicity (Fig. 3e) nor in ADCC (Fig. 3f) with the selected E:T ratios, although a trend toward a decreased ADCC in EBV+ cHL was seen with the highest E:T ratio.
Fig. 3.
Impaired rituximab-induced degranulation in in vitro expanded NK cells from EBV+HL compared to healthy controls (HC). NK cells were expanded from PBMC of 4 EBV+HL (depicted as filled circles or dark gray floating bars), 4 EBV- HL (depicted as open circles or light gray floating bars) and 4 HC (depicted as filled triangles or white floating bars) for 3 weeks. a Fold expansion of CD3- CD56+ NK cells relative to day 0 assessed by NK cell counts at day 0, 14 and 20. b Frequencies of CD16+ cells in CD3- CD56+ were assessed at the end of expansion procedure (expanded) and compared to baseline ex vivo values (thawed PBMC) in 4 EBV+ HL, 4 EBV- HL patients and 4 HC. c Frequencies of NKG2A+ KIR- (N+/K-), NKG2A- KIR+ (N-/K+) and CD57+ cells within expanded NK cells in 4 EBV+ HL, 4 EBV- HL patients and 4 HC. d Expanded NK cells from 4 EBV+ HL, 4 EBV- HL and 4 HC were co-cultured with LCL721.221 cell line (LCL221) with 1 μg/ml of rituximab at an effector to target ratio of 1:2 for 5 h. Frequencies of degranulating CD107a+ cells within CD3- CD56+ NK cells of EBV+ HL, EBV- HL and HC. (e and f) Thawed expanded NK cells of 4 EBV+ HL, 4 EBV- HL and 4 HC were co-cultured with PKH-26-labeled LCL221 either without e or with f 1 μg/ml of rituximab at different effector to target ratios (1:3, 1:1, 3:1) for 4 h. The frequency of dead cells among PKH-26-labeled LCL221 at the end of the co-culture was analyzed by flow cytometry using TO-PRO-3 iodide. The NK cell mediated specific killing of LCL221 was assessed by subtracting the background dead cell level in each sample. Horizontal lines indicate mean values in Fig. 3b-d. Significance between groups was determined by the one-way ANOVA test with Tukey’s multiple comparisons test in Fig. 3a-f and between paired samples by the paired t test in Fig. 3b
Discussion
Here, we analyzed the phenotype and function of peripheral blood NK cells in patients with EBV+ cHL vs. EBV- cHL patients and HC. We found that EBV+ cHL patients exhibited a decreased frequency of the late-differentiated CD56dim CD16+ subset compared to EBV- cHL patients and that this quantitative NK cell subset deficiency translated in an impaired antibody-dependent degranulation toward HLA class 1 negative LCL in EBV+ cHL patients.
The first novel finding of our study is the quantitative deficiency of peripheral blood late-differentiated cytotoxic CD56dim CD16+ NK cells in patients with EBV+ cHL. Vari et al. [22] showed that cHL patients, of whom the EBV status of their HRS cells was not determined, have decreased frequencies and counts of circulating NK cells together with a decreased ratio of CD56dim CD16+/CD56bright CD16- NK cells compared to controls. This finding taken together with our data indicate that EBV+ cHL patients might indeed exhibit a quantitative deficiency of the CD56dim CD16+ NK cells. Furthermore, the decreased frequency of CD56dim CD16+ NK cells cannot be explained by cryo-preservation-induced increase of CD56dim CD16- NK cells [31], since all peripheral blood and PBMC samples of the study were processed according to the same standard operating procedure. The deficiency was more pronounced for the CD56dim CD16bright NK cell subset and does not seem to be related to impaired NK cell maturation as assessed by the surface expression of NKG2A, KIR, CD62L and CD57. We previously reported that early-differentiated blood CD56dim NKG2A+ KIR- NK cells preferentially expand in patients with symptomatic acute EBV infection, i.e., infectious mononucleosis (IM) [23]. This phenotype was not observed in EBV+ cHL patients despite the presence of high levels of EBV DNA in plasma. Interestingly, CD56dim NK cells from both acute IM and EBV+ cHL patients exhibited a decreased surface expression of CD16, which might be due to metalloprotease-mediated shedding triggered by cytokines or ligation of activating receptors [30]. The activation-induced downregulation of CD16 is persistent in long-term in vitro cultured NK cells [29]. Furthermore, the presence of ongoing EBV lytic replication coupled with high level of circulating IgG targeting EBV lytic antigens, as seen in both acute IM and EBV+ cHL, might lead to over-stimulation and ultimately loss of CD16 on CD56dim NK cells. Finally, the quantitative deficiency of CD56dim CD16+ NK cells in the peripheral blood of EBV+ cHL patients might be due to accumulation of this subset within the immunosuppressive TME surrounding EBV-positive HRS cells. We also observed an increased frequency of dysfunctional CD56- CD16+ NK cells in EBV+ cHL compared to HC. Such phenotype has been described in children suffering from endemic Burkitt lymphoma with high plasma EBV DNA levels [32].
The second novel finding is that the quantitative CD56dim CD16+ NK cell deficiency translates in an impaired antibody-dependent degranulation of CD56dim NK cells from EBV+ cHL patients toward HLA class 1 negative LCL. Notably, natural cytotoxicity does not differ according to the EBV status of the cHL, implying that the activating NK cell receptors involved in the recognition of LCL and their signaling pathways are unaltered in both cHL types. The difference in age distribution in EBV+ cHL patients vs. EBV- cHL patients is an unlikely a selection bias, since ADCC function is not influenced by aging [33]. The impaired CD56dim NK cell-mediated rituximab-dependent degranulation of EBV+ cHL could also be confirmed using unstimulated PBMC and was maintained when compared to HC. The frequencies of effector cells, i.e., CD56dim CD16+ NK cells after overnight incubation remains decreased in EBV+ cHL compared to their EBV- counterparts. However, the frequencies of CD56dim CD16+ NK cells within live PBMC are comparable in the tested samples from both groups which makes a bias of effective E:T ratio in favor of EBV- cHL very unlikely. We also did not observe any difference in the frequency of bystander CD20+ B cells, thus ruling out major interferences of PBMC derived B cells in this assay. Unfortunately, we were limited by the low numbers of EBV+ cHL samples and the restricted available PBMC counts in our study which prevented us to functionally test purified unstimulated NK cells. Because rIL-2-pre-stimulation of NK cell did not seem to bias the antibody-dependent degranulation between EBV+ and EBV- cHL, we took advantage of an in vitro NK cell expansion method using K562-mbIL21 and rIL-2. We could show that expanded NK cells from EBV+ cHL exhibited a decreased antibody-dependent degranulation compared to HC. The low magnitude of this decrease was possibly due to the low E:T ratio selected. Furthermore, we did not confirm an impaired ADCC using expanded NK cells from EBV+ cHL. However, the observed trend suggests that a difference might be seen at higher E:T ratios (>10:1) commonly used in many studies on NK cell-based immunotherapy. One limitation of our study is the low numbers of patients in the functional part of the experiments. We could not formally prove that CD56dim CD16+ of EBV+ cHL patients exhibit an intrinsic deficiency in ADCC, since the activation-mediated shedding and the antibody ligation needed for cell sorting would hamper the gating and the functional testing of CD16+ NK cells, respectively [30]. Our study demonstrates that functional autologous NK cells from cHL patients, independently of the EBV status of the tumor, can be efficiently expanded in vitro which is a pre-requisite for the development of a NK cell based-immunotherapy targeting the HRS cells.
Elevated plasma EBV DNA levels is a feature of EBV+ cHL patients in this and other studies [34, 35] and is associated with treatment failure [36]. The origin of the circulating viral DNA is controversial and might arise from tumor-derived release of non-infectious free EBV DNA in the blood or from uncontrolled lytic replication outside of the HRS cells, which characteristically do not express EBV lytic genes. The occurrence of EBV+ cHL is preceded by unusually elevated levels of antibodies to viral capsid and early lytic antigens [37]. EBV+ cHL is associated with a specific anti-EBV antibody pattern [38]. Particularly, the combined expression of IgG specific for the two EBV lytic cycle proteins Zta and VCAp40 allows discriminating between EBV+ cHL and EBV- cHL patients. Early-differentiated CD56dim NKG2A+ KIR- NK cells can restrict EBV-infected B cells expressing lytic genes in vitro [23] and in vivo [39]. Additionally, NK cells can target EBV-infected B cells undergoing lytic replication in vitro via ADCC toward late lytic viral glycoproteins [40]. The essential role of functional CD16 signaling in the immune control of EBV lytic replication was recently demonstrated in patients with complete CD16a deficiency suffering from chronic active EBV disease [41]. Therefore, the reduced frequency of CD56dim CD16+ NK cells and the resulting impaired antibody-dependent NK cell-mediated degranulation of EBV+ cHL described here might lead to an inefficient control of the EBV lytic replication, promoting the progression of lymphomagenesis via a yet unknown mechanism. Nevertheless, the link between the expression of lytic genes and the development of B cell cancers is supported by in vivo studies in mouse models of EBV-associated B cell lymphomas other than cHL [42–44].
Novel immunotherapies using bi-specific monoclonal antibodies targeting the CD30 antigen present on HRS cells are currently evaluated as salvage treatment for patients with refractory or relapsed cHL [4, 20]. Both strategies might rely on the presence of functional NK cells within the TME or in the peripheral blood of cHL patients. The low frequency of blood CD56dim CD16+ NK cells found in some EBV+ cHL patients might impair the clinical efficacy to the bi-specific anti-CD30/CD16A antibody construct AFM13 currently assessed in a phase 2 study of patients with refractory or relapsed cHL (NCT02321592). Notably, AFM13-based immunotherapy in combination with NK cell-activating cytokines has been shown to expand the quantity of tumor-reactive NK cells and to improve NK cell cytotoxicity against tumor cells [45]. Thus, characterization of the EBV status of HRS cells in newly diagnosed cHL patients might be essential for future immunotherapeutic approaches that may include antibodies targeting CD16 and therefore rely on functional NK cell-mediated ADCC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank all the patients for their contribution to this study. We also thank Professor emeritus David Nadal who initiated the study, supervised the project and commented on the manuscript.
Abbreviations
- ADCC
Antibody-dependent cellular cytotoxicity
- cHL
Classical Hodgkin lymphoma
- EBERs
EBV-encoded small RNAs
- EBNA1
Epstein-Barr nuclear antigen 1
- EBV
Epstein-Barr virus
- HC
Healthy controls
- HRS
Hodgkin and Reed/Sternberg
- IM
Infectious mononucleosis
- KIR
Killer-cell Immunoglobulin-like receptors
- LCL
Lymphoblastoid cell line
- LMP1/2a
Latent membrane protein 1/2a
- NK cells
Natural killer cells
- PBMCs
Peripheral blood mononuclear cells
- rIL2
Recombinant interleukin-2
- TME
Tumor microenvironment
Authors’ contributions
TA, EP, AL, PB and DG contributed to the study conception. TA, EP, AL, CM, OC, CB and DG contributed to the study design. Material preparation was performed by TA, EP, SB, MoK, AL, MaK and JM. Patient recruitment was carried out by DG, JL and AD. Data collection was performed by TA, EP, MaK and JM. Data analysis was performed by TA, EP, CB, CM, MaK, JM, OC and DG. The first draft and the revised version of the manuscript was written by TA and EP and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding provided by Universität Zürich. This work was financially supported by the Cancer League of the Canton of Zurich (grant to TA) and Oncosuisse (KFS-3958-08-2016-R).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Consent to participate
All patients provided informed written consent to participate in this research study (CCTIRS N°15.317 and CNIL N° 915369).
Ethical approval
This non-interventional study, which complies with the Declaration of Helsinki, was approved by the appropriate regional ethics committee (“Comités de Protection des Personnes”, Ile-de-France, France) regulated by the government institution “Agence Régionale de Santé” (ARS, Bobigny, France), by the government advisory board for data processing in health care (“Comité Consultatif sur le Traitement de l’Information en matière de Recherche dans le Domaine de la Santé”, Paris, France) (CCTIRS N°15.317), and by the French data protection authority (“Commission Nationale de l’Informatique et des Libertés”, Paris, France) (CNIL N° 915,369).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9(1):15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 2.Murray PG, Young LS. An etiological role for the Epstein-Barr virus in the pathogenesis of classical Hodgkin lymphoma. Blood. 2019;134(7):591–596. doi: 10.1182/blood.2019000568. [DOI] [PubMed] [Google Scholar]
- 3.Vassilakopoulos TP, Angelopoulou MK. Advanced and relapsed/refractory Hodgkin lymphoma: what has been achieved during the last 50 years. Semin Hematol. 2013;50(1):4–14. doi: 10.1053/j.seminhematol.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Michot JM, Lazarovici J, Ghez D, Danu A, Ferme C, Bigorgne A, Ribrag V, Marabelle A, Aspeslagh S. Challenges and perspectives in the immunotherapy of Hodgkin lymphoma. Eur J Cancer. 2017;85:67–77. doi: 10.1016/j.ejca.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Shannon-Lowe C, Rickinson A. The global landscape of EBV-associated tumors. Front Oncol. 2019;9:713. doi: 10.3389/fonc.2019.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rickinson AB, Long HM, Palendira U, Munz C, Hislop AD. Cellular immune controls over Epstein-Barr virus infection: new lessons from the clinic and the laboratory. Trends Immunol. 2014;35(4):159–169. doi: 10.1016/j.it.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Jarrett RF, Krajewski AS, Angus B, Freeland J, Taylor PR, Taylor GM, Alexander FE. The Scotland and Newcastle epidemiological study of Hodgkin’s disease: impact of histopathological review and EBV status on incidence estimates. J Clin Pathol. 2003;56(11):811–816. doi: 10.1136/jcp.56.11.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urayama KY, Jarrett RF, Hjalgrim H, Diepstra A, Kamatani Y, Chabrier A, Gaborieau V, Boland A, Nieters A, Becker N, Foretova L, Benavente Y, Maynadie M, Staines A, Shield L, Lake A, Montgomery D, Taylor M, Smedby KE, Amini RM, Adami HO, Glimelius B, Feenstra B, Nolte IM, Visser L, van Imhoff GW, Lightfoot T, Cocco P, Kiemeney L, Vermeulen SH, Holcatova I, Vatten L, Macfarlane GJ, Thomson P, Conway DI, Benhamou S, Agudo A, Healy CM, Overvad K, Tjonneland A, Melin B, Canzian F, Khaw KT, Travis RC, Peeters PH, Gonzalez CA, Quiros JR, Sanchez MJ, Huerta JM, Ardanaz E, Dorronsoro M, Clavel-Chapelon F, Bueno-de-Mesquita HB, Riboli E, Roman E, Boffetta P, de Sanjose S, Zelenika D, Melbye M, van den Berg A, Lathrop M, Brennan P, McKay JD. Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status-defined subgroups. J Natl Cancer Inst. 2012;104(3):240–253. doi: 10.1093/jnci/djr516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iannello A, Thompson TW, Ardolino M, Marcus A, Raulet DH. Immunosurveillance and immunotherapy of tumors by innate immune cells. Curr Opin Immunol. 2016;38:52–58. doi: 10.1016/j.coi.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai K, Matsuyama S, Miyake S, Suga K, Nakachi K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. The Lancet. 2000;356(9244):1795–1799. doi: 10.1016/s0140-6736(00)03231-1. [DOI] [PubMed] [Google Scholar]
- 11.Hammer Q, Ruckert T, Romagnani C. Natural killer cell specificity for viral infections. Nat Immunol. 2018;19(8):800–808. doi: 10.1038/s41590-018-0163-6. [DOI] [PubMed] [Google Scholar]
- 12.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22(11):633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 13.Lo Nigro C, Macagno M, Sangiolo D, Bertolaccini L, Aglietta M, Merlano MC. NK-mediated antibody-dependent cell-mediated cytotoxicity in solid tumors: biological evidence and clinical perspectives. Ann Transl Med. 2019;7(5):105. doi: 10.21037/atm.2019.01.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116(19):3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 15.Juelke K, Killig M, Luetke-Eversloh M, Parente E, Gruen J, Morandi B, Ferlazzo G, Thiel A, Schmitt-Knosalla I, Romagnani C. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood. 2010;116(8):1299–1307. doi: 10.1182/blood-2009-11-253286. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116(19):3865–3874. doi: 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gattringer G, Greil R, Radaszkiewicz T, Huber H. In situ quantification of T-cell subsets, NK-like cells and macrophages in hodgkin's disease: Quantity and quality of infiltration density depends on histopathological subtypes. Blut. 1986;53(1):49–58. doi: 10.1007/bf00320582. [DOI] [PubMed] [Google Scholar]
- 18.Frydecka I. Natural killer cell activity during the course of disease in patients with Hodgkin’s disease. Cancer. 1985;56(12):2799–2803. doi: 10.1002/1097-0142(19851215)56:12<2799::aid-cncr2820561215>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 19.Konjevic G, Jurisic V, Banicevic B, Spuzic I. The difference in NK-cell activity between patients with non-Hodgkin’s lymphomas and Hodgkin’s disease. Br J Haematol. 1999;104(1):144–151. doi: 10.1046/j.1365-2141.1999.01129.x. [DOI] [PubMed] [Google Scholar]
- 20.Reiners KS, Kessler J, Sauer M, Rothe A, Hansen HP, Reusch U, Hucke C, Kohl U, Durkop H, Engert A, von Strandmann EP. Rescue of impaired NK cell activity in Hodgkin lymphoma with bispecific antibodies in vitro and in patients. Mol Ther. 2013;21(4):895–903. doi: 10.1038/mt.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu J, Ernst DM, Keating A. Acquired natural killer cell dysfunction in the tumor microenvironment of classic Hodgkin lymphoma. Front Immunol. 2018 doi: 10.3389/fimmu.2018.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vari F, Arpon D, Keane C, Hertzberg MS, Talaulikar D, Jain S, Cui Q, Han E, Tobin J, Bird R, Cross D, Hernandez A, Gould C, Birch S, Gandhi MK. Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL. Blood. 2018;131(16):1809–1819. doi: 10.1182/blood-2017-07-796342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azzi T, Lunemann A, Murer A, Ueda S, Beziat V, Malmberg KJ, Staubli G, Gysin C, Berger C, Munz C, Chijioke O, Nadal D. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood. 2014;124(16):2533–2543. doi: 10.1182/blood-2014-01-553024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dieli F, Denman CJ, Senyukov VV, Somanchi SS, Phatarpekar PV, Kopp LM, Johnson JL, Singh H, Hurton L, Maiti SN, Huls MH, Champlin RE, Cooper LJN, Lee DA. Membrane-bound il-21 promotes sustained ex vivo proliferation of human natural killer cells. PLoS ONE. 2012;7(1):e30264. doi: 10.1371/journal.pone.0030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alici E, Shah N, Martin-Antonio B, Yang H, Ku S, Lee DA, Cooper LJN, Decker WK, Li S, Robinson SN, Sekine T, Parmar S, Gribben J, Wang M, Rezvani K, Yvon E, Najjar A, Burks J, Kaur I, Champlin RE, Bollard CM, Shpall EJ. Antigen presenting cell-mediated expansion of human umbilical cord blood yields log-scale expansion of natural killer cells with anti-myeloma activity. PLoS ONE. 2013;8(10):e76781. doi: 10.1371/journal.pone.0076781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Champlin RE, Lee DA, Shpall EJ, Rezvani K, Ahmed S, Gulbis A, Kaur I, Soebbing D, Chen J, Rondon G, Willis D, Cao K, Denman CJ, Bassett R, Schafer JR, Ciurea SO. Phase 1 clinical trial using mbIL21 ex vivo–expanded donor-derived NK cells after haploidentical transplantation. Blood. 2017;130(16):1857–1868. doi: 10.1182/blood-2017-05-785659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee-MacAry AE, Ross EL, Davies D, Laylor R, Honeychurch J, Glennie MJ, Snary D, Wilkinson RW. Development of a novel flow cytometric cell-mediated cytotoxicity assay using the fluorophores PKH-26 and TO-PRO-3 iodide. J Immunol Methods. 2001;252(1–2):83–92. doi: 10.1016/s0022-1759(01)00336-2. [DOI] [PubMed] [Google Scholar]
- 28.Amand M, Iserentant G, Poli A, Sleiman M, Fievez V, Sanchez IP, Sauvageot N, Michel T, Aouali N, Janji B, Trujillo-Vargas CM, Seguin-Devaux C, Zimmer J. Human CD56dimCD16dim cells as an individualized natural killer cell subset. Front Immunol. 2017 doi: 10.3389/fimmu.2017.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodier MR, Lusa C, Sherratt S, Rodriguez-Galan A, Behrens R, Riley EM. Sustained immune complex-mediated reduction in CD16 expression after vaccination regulates NK cell function. Front Immunol. 2016 doi: 10.3389/fimmu.2016.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, Luo X, Cooley S, Verneris M, Walcheck B, Miller J. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17) Blood. 2013;121(18):3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lugthart G, van Ostaijen-ten Dam MM, van Tol MJ, Lankester AC, Schilham MW. CD56(dim)CD16(-) NK cell phenotype can be induced by cryopreservation. Blood. 2015;125(11):1842–1843. doi: 10.1182/blood-2014-11-610311. [DOI] [PubMed] [Google Scholar]
- 32.Forconi CS, Cosgrove CP, Saikumar-Lakshmi P, Nixon CE, Foley J, Ongécha JM, Otieno JA, Alter G, Münz C, Moormann AM. Poorly cytotoxic terminally differentiated CD56negCD16pos NK cells accumulate in Kenyan children with Burkitt lymphomas. Blood Adv. 2018;2(10):1101–1114. doi: 10.1182/bloodadvances.2017015404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazeldine J, Lord JM. The impact of ageing on natural killer cell function and potential consequences for health in older adults. Ageing Res Rev. 2013;12(4):1069–1078. doi: 10.1016/j.arr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hohaus S, Santangelo R, Giachelia M, Vannata B, Massini G, Cuccaro A, Martini M, Cesarini V, Cenci T, D'Alo F, Voso MT, Fadda G, Leone G, Larocca LM. The viral load of Epstein-Barr virus (EBV) DNA in peripheral blood predicts for biological and clinical characteristics in hodgkin lymphoma. Clin Cancer Res. 2011;17(9):2885–2892. doi: 10.1158/1078-0432.CCR-10-3327. [DOI] [PubMed] [Google Scholar]
- 35.Drouet E, Brousset P, Fares F, Icart J, Verniol C, Meggetto F, Schlaifer D, Desmorat-Coat H, Rigal-Huguet F, Niveleau A, Delsol G. High Epstein-Barr virus serum load and elevated titers of anti-ZEBRA antibodies in patients with EBV-harboring tumor cells of Hodgkin's disease. J Med Virol. 1999;57(4):383–389. doi: 10.1002/(sici)1096-9071(199904)57:4<383::aid-jmv10>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Kanakry JA, Li H, Gellert LL, Lemas MV, Hsieh WS, Hong F, Tan KL, Gascoyne RD, Gordon LI, Fisher RI, Bartlett NL, Stiff P, Cheson BD, Advani R, Miller TP, Kahl BS, Horning SJ, Ambinder RF. Plasma Epstein-Barr virus DNA predicts outcome in advanced Hodgkin lymphoma: correlative analysis from a large North American cooperative group trial. Blood. 2013;121(18):3547–3553. doi: 10.1182/blood-2012-09-454694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller N, Evans A, Harris NL, Comstock GW, Jellum E, Magnus K, Orentreich N, Polk BF, Vogelman J. Hodgkin’s disease and Epstein-Barr virus altered antibody pattern before diagnosis. N Engl J Med. 1989;320(11):689–695. doi: 10.1056/NEJM198903163201103. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Jarrett RF, Hjalgrim H, Proietti C, Chang ET, Smedby KE, Yu KJ, Lake A, Troy S, McAulay KA, Pfeiffer RM, Adami HO, Glimelius B, Melbye M, Hildesheim A, Doolan D, Coghill AE. Evaluation of the antibody response to the EBV proteome in EBV-associated classic Hodgkin lymphoma. Int J Cancer. 2019 doi: 10.1002/ijc.32741. [DOI] [PubMed] [Google Scholar]
- 39.Chijioke O, Muller A, Feederle R, Barros MH, Krieg C, Emmel V, Marcenaro E, Leung CS, Antsiferova O, Landtwing V, Bossart W, Moretta A, Hassan R, Boyman O, Niedobitek G, Delecluse HJ, Capaul R, Munz C. Human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein-Barr virus infection. Cell Rep. 2013;5(6):1489–1498. doi: 10.1016/j.celrep.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López-Montañés M, Alari-Pahissa E, Sintes J, Martínez-Rodríguez JE, Muntasell A, López-Botet M. Antibody-dependent NK cell activation differentially targets EBV-infected cells in lytic cycle and bystander B lymphocytes bound to viral antigen-containing particles. J Immunol. 2017;199(2):656–665. doi: 10.4049/jimmunol.1601574. [DOI] [PubMed] [Google Scholar]
- 41.Portilla AP, Moraru M, Moreno AB, Kolb P, Garcia-Morato MB, Ranganath T, Esteso G, Gianelli C, Rodriguez-Pena R, Rodriguez RL, Torres-Canizales JM, Blish CA, Vales-Gomez M, Hengel H, Vilches C, Granados EL, Reyburn HT. Identification of the first cases of complete CD16A deficiency: association with persistent EBV infection. J Allergy Clin Immunol. 2020 doi: 10.1016/j.jaci.2019.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong GK, Gulley ML, Feng WH, Delecluse HJ, Holley-Guthrie E, Kenney SC. Epstein-barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J Virol. 2005;79(22):13993–14003. doi: 10.1128/jvi.79.22.13993-14003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McHugh D, Caduff N, Barros MHM, Rämer PC, Raykova A, Murer A, Landtwing V, Quast I, Styles CT, Spohn M, Fowotade A, Delecluse H-J, Papoudou-Bai A, Lee Y-M, Kim J-M, Middeldorp J, Schulz TF, Cesarman E, Zbinden A, Capaul R, White RE, Allday MJ, Niedobitek G, Blackbourn DJ, Grundhoff A, Münz C. persistent KSHV infection increases ebv-associated tumor formation in vivo via enhanced EBV lytic gene expression. Cell Host Microbe. 2017;22(1):61–73.e67. doi: 10.1016/j.chom.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Shumilov A, Tsai M-H, Schlosser YT, Kratz A-S, Bernhardt K, Fink S, Mizani T, Lin X, Jauch A, Mautner J, Kopp-Schneider A, Feederle R, Hoffmann I, Delecluse H-J. Epstein-Barr virus particles induce centrosome amplification and chromosomal instability. Nat Commun. 2017 doi: 10.1038/ncomms14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pahl JHW, Koch J, Gotz JJ, Arnold A, Reusch U, Gantke T, Rajkovic E, Treder M, Cerwenka A. CD16A activation of NK cells promotes NK cell proliferation and memory-like cytotoxicity against cancer cells. Cancer Immunol Res. 2018;6(5):517–527. doi: 10.1158/2326-6066.CIR-17-0550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.