Abstract
Immunotherapy has emerged as an effective therapeutic strategy for various cancers, including colorectal cancer (CRC), but only a subset of MSI-H patients can benefit from such therapy. Patched1 (PTCH1) is a frequently altered gene in CRCs and its mutations contribute to unregulated Hedgehog (Hh) signaling. In the study, we evaluated the association of PTCH1 mutations with CRC immunity based on our single-center cohort and multiple cancer genomic datasets. Among 21 enrolled patients, six (28.6%) harbored a PTCH1 mutation based on WES analyses. In CRC patients, the PTCH1 mutation subgroup experienced a higher durable clinical benefit rate than the PTCH1 wild-type subgroup (100% vs. 40%, P = 0.017). In addition, patients with the PTCH1 mutation experienced greater progression-free survival (PFS, P = 0.037; HR, 0.208) and overall survival (OS, P = 0.045; HR, 0.185). A validation cohort from the MSKCC also confirmed the correlation between PTCH1 mutation and better prognosis (P = 0.022; HR, 0.290). Mechanically, diverse antitumor immune signatures were more highly enriched in PTCH1-mutated tumors than in PTCH1 wild-type tumors. Furthermore, PTCH1-mutated tumors had higher proportions of CD8 + T cells, activated NK cells, and M1 type macrophage infiltration, as well as elevated gene signatures of several steps in the cancer-immunity cycle. Notably, the PTCH1 mutation was correlated with tumor mutational burden (TMB), loss of heterozygosity score, and copy number variation burden. Our results show that the mutation of PTCH1 is a potential biomarker for predicting the response of CRC patients to immunotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-02966-9.
Keywords: PTCH1, Colorectal cancer, Immune checkpoint inhibitors, Biomarker
Introduction
Colorectal cancer (CRC) is the third-most common cancer and the second leading cause of cancer-related death in the world [1]. Although early detection and systematic treatment have improved the survival rate of localized CRC, approximately 25% of patients still present with stage IV disease with a 5-year survival of only 14% [2]. Thus, the development of more effective treatments guided by actionable biomarkers for patients with this disease is urgently needed. In recent decades, the development of immune checkpoint inhibitors (ICIs) has revolutionized oncology, including CRC. In 2017, ICIs were approved for clinical use in patients with microsatellite instability-high (MSI-H) or deficient mismatch repair (dMMR) CRC. However, the MSI-H or dMMR subtype is present in approximately 5% of metastatic CRCs [3, 4], and only 30–40% of MSI-H/dMMR patients respond to ICIs [5–7], indicating an unmet clinical need for precision immunotherapy in CRC. Therefore, identifying additional biomarkers to precisely predict the response to ICIs in CRC patients is important.
The Hedgehog (Hh) signaling pathway, which regulates proliferation, angiogenesis, matrix remodeling, and stem cell renewal, plays an important role during tumorigenesis in CRC [8]. Upon secretion by cells, Hh ligands, such as sonic hedgehog (Shh), bind to Patched1 (PTCH1), thereby releasing the suppression of Smoothened (Smo), ultimately activating glioma-associated oncogene (Gli) transcription factors and promoting the transcription of Hh target genes. Recently, Hh signaling was reported to modulate the tumor microenvironment (TME) [9] by increasing immune checkpoint expression and promoting an inflammatory environment [10–12], thus indicating a potential association between Hh signaling and the response to ICIs. PTCH1 is the most frequently altered Hh signaling regulator in CRC [13, 14], however the potential association of PTCH1 with clinical outcomes for CRC patients receiving ICIs is unclear. Therefore, whether the mutation of PTCH1 can predict the clinical outcome of ICI treatment was analyzed using data from our cohort and a publicly available cohort. Subsequently, we assessed immunogenic features based on PTCH1 status to explore the possible underlying mechanism.
Materials and methods
Clinical cohorts
A total of 21 CRC patients treated with a PD-1/PD-L1 inhibitor combined with a CTLA-4 inhibitor between April 2011 and January 2017 at the Beijing Cancer Hospital were retrospectively analyzed. The tumor response was assessed by physicians using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 criteria. Efficacy was defined as a durable clinical benefit (DCB, complete response [CR], partial response [PR], or stable disease [SD] lasting ≥ 6 months) or no durable benefit (NDB, progressive disease [PD] or SD lasting < 6 months) [15]. Another independent cohort consisting of 109 CRC patients treated with ICIs at Memorial Sloan Kettering Cancer Center (MSKCC) was also analyzed to validate the findings in our cohort. Clinical and genetic data were downloaded from the cBioPortal for Cancer Genomics at http://www.cbioportal.org/study?id=tmb_mskcc_2018. In addition, the Cancer Genome Atlas (TCGA) colorectal adenocarcinoma (COADREAD) database (www.cbioportal.org) including clinical and genomic data, as well as expression profile data, was further explored to determine the underlying mechanism.
Genomic data analysis
DNA were extracted from formalin-fixed paraffin-embedded (FFPE) tissue specimens and matched white blood cell (WBC) samples of patients in our cohort. All specimens were collected from patients with written informed consent for their samples to be used in future translational researches including genomics, proteomics. Genetic alterations were subsequently analyzed according to standard principles. The tumor mutational burden (TMB) was measured in mutations per megabase (Mb) and was stratified into two groups, TMB-low (2–36 mutations/Mb) and TMB-high (> 37 mutations/Mb), as reported previously [16]. TMB level, loss of heterozygosity (LOH) score, and copy number variation (CNV) data were obtained from the TCGA database portal and analyzed according to a previously published study [17].
MMR/MSI status detection
The MMR/MSI status was assessed according to the previous study [18]. In brief, the MMR status was identified using immunohistochemistry (IHC) for MMR protein such as monoclonal anti-mutL homolog 1, anti-mutS homolog 2, anti-mutS homolog 6, and PMS1 homolog 2. Deficient MMR tumors were defined as instability at two or more of these markers. Polymerase chain reaction (PCR) was performed to detect the MSI status, which assesses five microsatellite loci comprising BAT-25, BAT-26, D2S123, D5S346, and D17S250. MSI-H tumors were defined as instability at two or more of these markers.
RNA expression profile analysis
Gene expression data were downloaded from the TCGA database portal and transformed into transcript per kilobase million (TPM) values. Subsequently, the log average of gene expression in TPM was used to quantify the enrichment level of three immune signatures, including GEP score (CXCR6, TIGIT, CD27, CD274, PDCD1LG2, LAG3, NKG7, PSMB10, CMKLR1, CD8A, IDO1, CCL5, CXCL9, HLA.DQA1, CD276, HLA-DRB1, STAT1,and HLA-E) [19, 20], immune cytolytic activity (GZMA and PRF1) [21], and IFN-γ signature (IFNG, CXCL10, CXCL9, IDO1, STAT1, and HLA-DRA) [22]. In addition, single-sample gene set enrichment analysis (ssGSEA) [23] and the CIBERSORT algorithm [24] were used to predict infiltration levels of multiple types of immune cells.
The immune response to tumors is triggered by critical steps referred to as the cancer-immunity cycle. In the present study, each step was quantified using immunogram scores (IGSs) [25] as follows: IGS1, T cell immunity; IGS2, tumor antigenicity; IGS3, T cell priming and activation; IGS4, T cell trafficking and infiltration; IGS5, antigen-presenting machinery; IGS6, inhibitory immune cell infiltration; IGS7, immune checkpoint molecules; and IGS8, inhibitory molecules. IGSs were assessed using Gene Set Variation Analysis (GSVA). The distributions of IGSs are presented as radar figures.
Statistical analyses
All statistical analyses were performed using SPSS software version 23.0 for Windows and R 3.6.1. The survival function was estimated using Kaplan–Meier curves and the P value was determined using a log-rank test. Statistical heterogeneity was evaluated using the chi-squared test or Fisher’s exact test. Student’s t test was applied to determine the differences between two groups when data were normally distributed and continuous variables were compared using the Mann–Whitney U test. All reported P values were two-tailed and P values < 0.05 were considered statistically significant.
Results
Patient characteristics
A total of 21 patients from our center were included in the analysis. Baseline characteristics are summarized in Table 1. The median age was 44 years (range, 14–75 years) and 14 (66.7%) patients were male. Among the 21 patients, five experienced CR or PR, eight patients achieved SD, and eight patients had PD, resulting in a DCB rate of 57.1% and NDB rate of 42.9%. The tumors of 18 patients (85.7%) were tested for MSI status using PCR or MMR status using IHC; 16 (88.9%) were MSI-H or dMMR. Of these 16 patients with MSI-H/dMMR tumors, six (37.5%) had PTCH1 mutation. However, PTCH1 mutation was not detected in MSS/pMMR tumors. The median TMB value was 38.1 mutations/Mb (range, 2.3–220.3 mutations/Mb).
Table 1.
Patient’s characteristics
| Characteristics valuea | Number | PTCH1-MUT | PTCH1-WT | P |
|---|---|---|---|---|
| (n = 21) | (n = 6) | (n = 15) | ||
| Age at diagnosis(year) | ||||
| Median age (range) | 44 (14–75) | 28.5 (15–31) | 48 (33–75) | |
| Gender, N (%) | 0.061 | |||
| Male | 14 (66.67%) | 6 (100.00%) | 8 (53.33%) | |
| Female | 7 (33.33%) | 0 | 7 (46.67%) | |
| Histopathology, N (%) | 1.000 | |||
| Adenocarcinoma | 19 (90.48%) | 6 (100.00%) | 13 (86.67%) | |
| Others | 2 (9.52%) | 0 | 2 (13.33%) | |
| Treatment option, N (%) | 1.000 | |||
| Anti-PD-(L)1 | 19 (90.48%) | 6 (100.00%) | 13 (86.67%) | |
| Anti-PD-(L)1 + anti-CTLA-4 | 2 (9.52%) | 0 | 2 (13.33%) | |
| Best response, N (%) | 0.140 | |||
| Complete response | 2 (9.52%) | 1 (16.67%) | 1 (16.67%) | |
| Partial response | 3 (14.28%) | 1 (16.67%) | 2 (13.33%) | |
| Stable disease | 8 (38.10%) | 4 (66.66%) | 4 (26.67%) | |
| Progressive disease | 8 (38.10%) | 0 | 8 (53.33%) | |
| Response group, N (%) | 0.017 | |||
| Durable clinical benefit (DCB) | 12 (57.14%) | 6 (100.00%) | 6 (40.00%) | |
| No durable benefit (NDB) | 9 (42.86%) | 0 | 9 (60.00%) | |
| Tumor MSI/MMR status, N (%) | 0.269 | |||
| MSI-H/dMMR | 16 (76.19%) | 6 (100.00%) | 10 (66.67%) | |
| pMMR | 2 (9.52%) | 0 | 2 (13.33%) | |
| Not available | 3 (14.29%) | 0 | 3 (20.00%) | |
aP values derived from Fisher’s exact tests between the two groups
Predictive role of PTCH1 and TMB in the study cohort
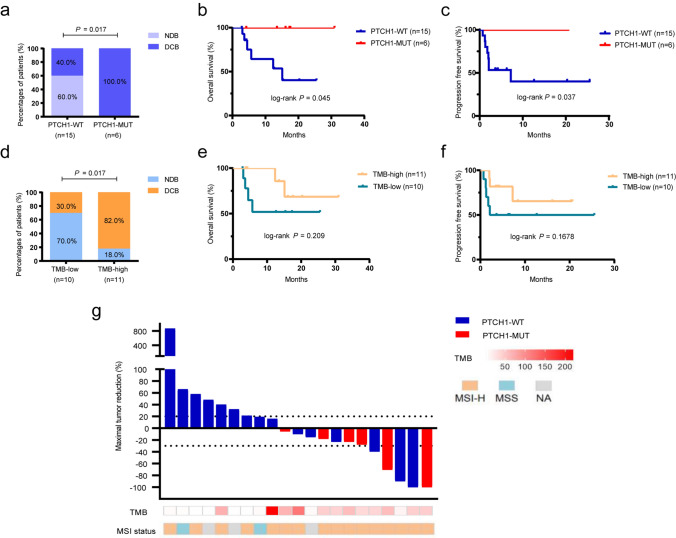
First, whether the PTCH1 mutation was associated with the clinical benefit from ICIs in CRC patients was analyzed. The DCB rate was higher in patients with the PTCH1 mutation than in PTCH1 wild-type patients (100% vs. 40%, P = 0.017; Fig. 1a), this is mainly due to higher proportion of SD lasting ≥ 6 months in PTCH1 mutation group. In addition, a favorable prognosis (better OS/PFS) was identified in the population with the PTCH1 mutation (OS, P = 0.045, HR, 0.185, 95% CI, 0.035–0.965; PFS, P = 0.037 HR, 0.208, 95% CI, 0.048–0.911; Fig. 1b, c). Among 16 MSI-H/dMMR patients, the PTCH1 mutation was also associated with improved OS (P = 0.0607, HR, 0.149, 95% CI, 0.020–1.090) and PFS (P = 0.057, HR, 0.178, 95% CI, 0.030–1.055) compared with PTCH1 wild-type patients, although with marginal significance (Supplementary Fig. S1a and b). Next, the predictive value of TMB was evaluated in our cohort because it is considered a predictive biomarker in multiple cancers [26]. Unfortunately, a statistically significant difference was not observed in the DCB rates or survival outcomes between TMB-high and TMB-low patients when using the cutoff value of 37 as previously suggested (Fig. 1d–f). Waterfall plots depicting the tumor change from baseline in individual patients based on their PTCH1 or TMB status are shown in Fig. 1g.
Fig. 1.
PTCH1 and TMB status correlate with the response to ICI treatment in our cohort a Comparison of the DCB rates in the PTCH1 mutation and PTCH1 wild-type groups. b and c Kaplan–Meier curves comparing OS (b) and PFS (c) in patients with the PTCH1 mutation and wild-type PTCH1 in our cohort. d Comparison of the DCB rates in the TMB-low and TMB-high groups. e and f Kaplan–Meier curves comparing OS (e) and PFS (f) of the TMB-high and TMB-low patients. The cutoff value for TMB-high and TMB-low was defined as 37 mutations/Mb. g Waterfall plot representing the best change from baseline in sum of longest target lesion diameters per patient based on PTCH1, TMB, and MSI status. PTCH1, Patched1 gene; DCB, durable clinical benefit; OS, overall survival; PFS, progression-free survival; TMB, tumor mutational burden; MSI, microsatellite instability
Validation of PTCH1 and TMB as biomarkers in the MSKCC CRC ICI cohort
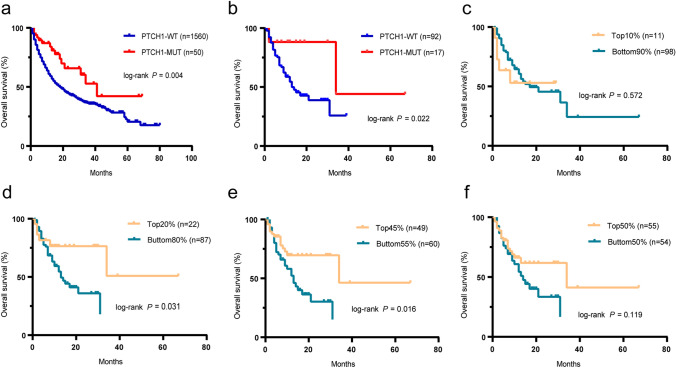
To validate the above results, another cohort (MSKCC ICI) consisting of 1,610 patients treated with ICIs, including 109 CRC patients, was used. In both the pan-cancer cohort and a subset of CRC patients, those harboring the PTCH1 mutation also had better survival outcomes (P = 0.004, HR, 0.485, 95% CI, 0.337–0.697; P = 0.022, HR, 0.29, 95% CI, 0.139–0.607; Fig. 2a, b). Consistent with a previous study [26], a higher TMB (highest 25%) was associated with better OS. However, statistical significance was not always reached when using cutoff values outside of appropriate ranges (Fig. 2c–f, cutoff values of 10%, 20%, 45%, and 50%). Together with the findings from our cohort, these results indicate the robustness of TMB as a predictive biomarker for predicting benefit from ICIs.
Fig. 2.
Survival analysis of patients stratified based on PTCH1 status or TMB in the MSKCC cohort a and b OS in patients with and without the PTCH1 mutation in the MSKCC pan-cancer (a) and MSKCC CRC cohort (b). c-f OS curves were plotted for the patients stratified based on the TMB level in the MSKCC cohort. The cutoffs used in this cohort were the top 10% (c), 20% (d), 45% (e), and 50% (f).
Correlation between the PTCH1 mutation and immune-related signatures
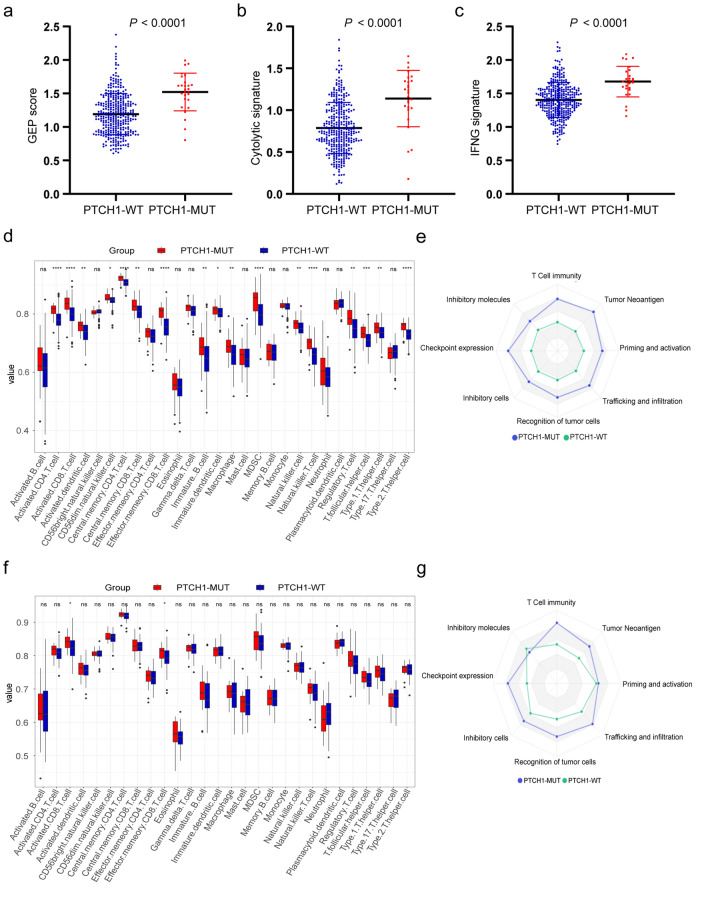
Based on the above observations, we hypothesized that PTCH1 status influenced the shaping of the TME. To verify this hypothesis, the T cell-inflamed gene expression signature (GEP) consisting of 18 genes indicative of a T cell activated tumor, which was shown to predict the response to ICIs [27], was analyzed. Based on the expression profile from the TCGA cohort, PTCH1-mutated tumors displayed a higher GEP signature (Fig. 3a). Furthermore, greater enrichment of IFN-γ and cytolytic signatures in the PTCH1 mutation subgroup were observed (Fig. 3b, c).
Fig. 3.
Correlation between PTCH1 mutation and immune-related signatures in the TCGA cohort a–c Quantitative analysis of GEP scores (a), Cytolytic (b) and IFNG (c) signatures in the PTCH1 mutation and PTCH1 wild-type patients based on the TCGA COADREAD database. d and f Boxplots show the abundance of ssGSEA-derived immune cells in multiple cell subsets based on PTCH1 mutation status in the TCGA COADREAD cohort (d) and its MSI-H subgroup (f). e and g Eight axes of an immunogram involving the cancer-immunity cycle were plotted based on the PTCH1 mutation status in the TCGA COADREAD cohort (e) and its MSI-H subgroup (g). The value of the immunogram score (IGS) was calculated using GSVA. GEP, gene expression signature; GSVA, Gene Set Variation Analysis
To characterize the immune infiltration landscape of the PTCH1 mutation and wild-type subgroups, ssGSEA scores from the 28 immune-related signatures were used to quantify the relative abundance of 28 immune cell types. PTCH1-mutated tumors had higher proportions of CD8+ T cells and activated NK cells, indicating the induction of an antitumor response (Fig. 3d). Similar results were obtained when using the CIBERSORT algorithm (Supplementary Fig. S2). Specifically, in the MSI-H population, CD8+ T cells and effector memory CD8+ T cells were enriched in PTCH1-mutated tumors (Fig. 3f). To obtain a more comprehensive understanding of the relationship between PTCH1 mutation and tumor-immunity interactions, an immunogram was used to visualize the general cancer immunity status in the PTCH1 mutation and wild-type patients. The steps in the cancer-immunity cycle were enhanced in PTCH1-mutated tumors in the entire TCGA COADREAD cohort and its MSI-H subgroup (Fig. 3e, g), indicating that the mutation of PTCH1 in CRC potentially facilitates immune cell infiltration, partially accounting for the better response to ICIs in the PTCH1 mutation subgroup.
Correlations between PTCH1 mutation and genomic parameters
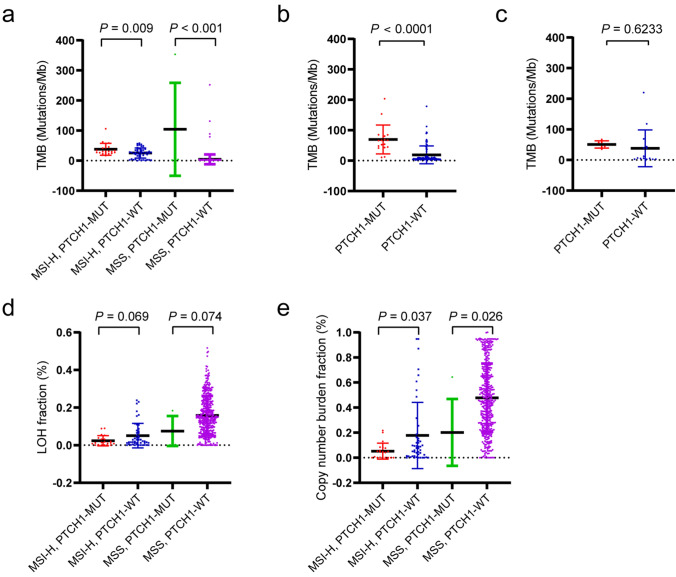
In a previous study, PTCH1-driven skin basal cell carcinoma showed a high level of TMB [28], thus, the TMB levels in the PTCH1 mutation and wild-type groups from our cohort and database were evaluated. The analysis confirmed that the PTCH1 mutation subgroup had a higher TMB than the PTCH1 wild-type subgroup in both MSI-H and MSS populations in the TCGA database (Fig. 4a) and MSKCC cohort (Fig. 4b). However, patients with the PTCH1 mutation in our cohort did not show a higher TMB level; this may be explained by the relatively small sample size (Fig. 4c). Notably, a lower LOH and CNV burden were identified in patients with the PTCH1 mutation regardless of MSI status (Fig. 4d, e). The above results imply that the mutation of PTCH1 may be indicative of genomic tumor features.
Fig. 4.
Correlations between PTCH1 mutation and genomic parameters a Comparison between the PTCH1 mutation and wild-type subgroups in MSI-H and MSS populations based on the TCGA COADREAD database. b Analysis of TMB level in the PTCH1-mutated and wild-type tumors in the MSKCC cohort. c Analysis of TMB level in the PTCH1-mutated and wild-type tumors in our cohort. d and e Comparison of LOH (d) and CNV burden (e) between the PTCH1 mutation and wild-type subgroups in the MSI-H and MSS populations based on the TCGA COADREAD database. LOH, loss of heterozygosity; CNV, copy number variation
Discussion
Our study based on this small cohort of patients shows the PTCH1 mutation was associated with better survival outcomes (OS/PFS) and DCB rates among patients who received ICIs, which is partially attributable to a higher proportion of SD lasting ≥ 6 months in PTCH1 mutation group. Potentially, the PTCH1 mutation established a favorable immune contexture of CRC, thus sensitizing tumors to immunotherapy.
The main challenge of ICI treatment in CRC patients is that only 30–40% of MSI-H/dMMR CRCs respond to ICIs [5–7], and the underlying molecular mechanisms of this clinical observation are not well known. Although quantifying TMB and MSI density may precisely stratify MSI-H patients for ICI treatment [16, 29], the present technical and practical barriers limit the use of these strategies in clinical practice. Furthermore, mixed results of the predictive role of TMB based on different cutoff values were elicited in our and MSKCC cohorts, indicating that clarifying the TMB cutoff values specific to CRC and its MSI-H subtype to identify who will receive optimal benefit is important. Other than TMB, two independent cohorts showed prolonged OS in patients with the PTCH1 mutation. The clinical benefit analysis also indicated that patients with a PTCH1 mutation experienced a higher DCB rate than PTCH1 wild-type patients. The results of the present study suggest that PTCH1 status could also function as a predictive biomarker for response to ICI treatment, in addition to TMB and MSI density. Moreover, The detection of PTCH1 mutations may provide a more accessible way to stratify MSI-H/dMMR CRC patients for immunotherapy, though a study using a larger cohort would be necessarty to validate these findings.
PTCH1 plays a pivotal role in regulating Hh signaling, inactivating mutations that can activate the expression of Hh target genes in an unregulated manner [8]. The involvement of Hh signaling has been associated with tumor development, recurrence, metastasis, and TME regulation in CRC. In recent studies, Hh-driven skin basal cell carcinoma had a significantly high TMB level with an average of 65 mutations per Mb [28, 30], indicating that Hh-driven tumors likely represent an immunogenic entity. Similarly, tumors harboring the PTCH1 mutation had a higher level of TMB in the present study. Recently, Lin A et al. [14] has found that PTCH1 deficiency leads to the increased secretion of cytokines that promote tumor-antigen presentation, facilitate T lymphocytes infiltration. These notions, further support our findings that tumors with PTCH1 mutation display a “hot” TME phenotype, with a high immune infiltration and TMB/neoantigen level, partially explaining why patients with a PTCH1 mutation respond well to ICIs.
The present study has several limitations. First, this was a retrospective study that included patients treated with both single-agent PD-1/PD-L1 blockade and combinational immunotherapy. Second, because MSI detection was not the standard-of-care at the time when some of these patients were treated, three patients lacked MSI status information and two patients were identified with dMMR. Third, experimental information to determine the impact of PTCH1 on immune contexture was lacking. Future investigations based on in vitro and in vivo experiments and larger clinical cohorts are warranted to confirm these findings. However, the present study based on multiple cohorts was the first to demonstrate the value of PTCH1 for predicting the response to ICIs.
In the present study, ICI treatment was shown to be more effective in CRC patients with a PTCH1 mutation. Although we did not have adequate samples in our cohort, it is an important finding that the PTCH1 mutation is characterized by activation of the immune microenvironment and may be used to predict the response of CRC patients to ICIs. Further verification is needed in additional large cohorts.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank textcheck (http://www.textcheck.com) for providing linguistic assistance during the preparation of this manuscript.
Abbreviations
- CNV
Copy number variation
- CR
Complete response
- CRC
Colorectal cancer
- DCB
Durable clinical benefit
- dMMR
Deficient mismatch repair
- FFPE
Formalin-fixed paraffin-embedded
- GEP
Gene expression signature
- Gli
Glioma-associated oncogene
- Hh
Hedgehog
- ICIs
Immune checkpoint inhibitors
- LOH
Loss of heterozygosity
- MSI-H
Microsatellite instability-high
- OS
Overall survival
- PD
Progressive disease
- PFS
Progression-free survival
- PR
Partial response
- PTCH1
Patched1
- SD
Stable disease
- Shh
Sonic hedgehog
- ssGSEA
Single-sample gene set enrichment analysis
- TMB
Tumor mutational burden
- TME
Tumor microenvironment
- TPM
Transcript per kilobase million
- WBC
White blood cell
Author contributions
L. S., H. Z. and Z. L. designed the study. J. G., J. L., X. Z., X. W., Z. P., C. Q., Z. W., J. L. and Y. L. acquired and analyzed the data. Y. W., L. W., H. C., X. J., Y. Y., J. Z., L. W., C. L., N. Z. and S. L. analyzed and interpreted the data. Y. W., H. C. and X. J. wrote, reviewed, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Key Research and Development Program of China (2017YFC1308900, 2018YFC1313302), the National Key Sci-Tech Special Project of China (2018ZX10302207), Beijing Hospitals Authority Youth Programme (QML20191102), and the Digestive Medical Coordinated Development Center of Beijing Hospitals Authority (No. XXT19).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no potential conflicts of interest to this work.
Consent to participate
All the patients were informed of sample collection for translational researches before they signed the consents.
Ethical approval
The tissue samples were collected and used in accordance with approval by the Clinical Research Ethics Committee of Peking University Cancer Hospital and Institute Consent for publication (2020MS01). The study was performed in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Footnotes
Yanni Wang, Huan Chen and Xi Jiao contributed equally to this work.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhihao Lu, Email: pppeirain@126.com.
Henghui Zhang, Email: zhhbao@ccmu.edu.cn.
Lin Shen, Email: shenlin@bjmu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Loree JM, Pereira AAL, Lam M, Willauer AN, Raghav K, Dasari A, Morris VK, Advani S, Menter DG, Eng C, Shaw K, Broaddus R, Routbort MJ, Liu Y, Morris JS, Luthra R, Meric-Bernstam F, Overman MJ, Maru D, Kopetz S. Classifying colorectal cancer by tumor location rather than sidedness highlights a continuum in mutation profiles and consensus molecular Subtypes. Clin Cancer Res. 2018;24(5):1062–1072. doi: 10.1158/1078-0432.Ccr-17-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koopman M, Kortman GAM, Mekenkamp L, Ligtenberg MJL, Hoogerbrugge N, Antonini NF, Punt CJA, van Krieken JHJM. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100(2):266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA., Jr PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le DT, Kim TW, Van Cutsem E, Geva R, Jager D, Hara H, Burge M, O'Neil B, Kavan P, Yoshino T, Guimbaud R, Taniguchi H, Elez E, Al-Batran SE, Boland PM, Crocenzi T, Atreya CE, Cui Y, Dai T, Marinello P, Diaz LA, Jr, Andre T. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: keynote-164. J Clin Oncol. 2020;38(1):11–19. doi: 10.1200/jco.19.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, Goldberg MV, Cao ZA, Ledeine JM, Maglinte GA, Kopetz S, Andre T. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/s1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raleigh DR, Reiter JF. Misactivation of Hedgehog signaling causes inherited and sporadic cancers. J Clin Invest. 2019;129(2):465–475. doi: 10.1172/JCI120850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grund-Gröschke S, Stockmaier G, Aberger F. Hedgehog/GLI signaling in tumor immunity - new therapeutic opportunities and clinical implications. Cell Commun Signal. 2019;17(1):172–172. doi: 10.1186/s12964-019-0459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti J, Holokai L, Syu L, Steele NG, Chang J, Wang J, Ahmed S, Dlugosz A, Zavros Y. Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget. 2018;9(100):37439–37457. doi: 10.18632/oncotarget.26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills LD, Zhang Y, Marler RJ, Herreros-Villanueva M, Zhang L, Almada LL, Couch F, Wetmore C, Pasca di Magliano M, Fernandez-Zapico ME. Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. J Biol Chem. 2013;288(17):11786–11794. doi: 10.1074/jbc.M112.438846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Jin G, Li Q, Wang Z, Hu W, Li P, Li S, Wu H, Kong X, Gao J, Li Z. Hedgehog signaling non-canonical activated by pro-inflammatory cytokines in pancreatic ductal adenocarcinoma. J Cancer. 2016;7(14):2067–2076. doi: 10.7150/jca.15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung JH, Bunz F. A loss-of-function mutation in PTCH1 suggests a role for autocrine hedgehog signaling in colorectal tumorigenesis. Oncotarget. 2013;4(12):2208–2211. doi: 10.18632/oncotarget.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin A, Zhang H, Hu X, Chen X, Wu G, Luo P, Zhang J. Age, sex, and specific gene mutations affect the effects of immune checkpoint inhibitors in colorectal cancer. Pharmacol Res. 2020;159:105028. doi: 10.1016/j.phrs.2020.105028. [DOI] [PubMed] [Google Scholar]
- 15.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS, Miller VA, Lim D, Amanam I, Chao J, Catenacci D, Cho M, Braiteh F, Klempner SJ, Ali SM, Fakih M. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30(7):1096–1103. doi: 10.1093/annonc/mdz134. [DOI] [PubMed] [Google Scholar]
- 17.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Z, Chen H, Jiao X, Zhou W, Han W, Li S, Liu C, Gong J, Li J, Zhang X, Wang X, Peng Z, Qi C, Wang Z, Li Y, Li J, Li Y, Brock M, Zhang H, Shen L. Prediction of immune checkpoint inhibition with immune oncology-related gene expression in gastrointestinal cancer using a machine learning classifier. J Immunother Cancer. 2020;8(2):e000631. doi: 10.1136/jitc-2020-000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott P, Bang Y, Piha-Paul S, Razak A, Bennouna J, Soria J, Rugo H, Cohen R, O'Neil B, Mehnert J, Lopez J, Doi T, van Brummelen E, Cristescu R, Yang P, Emancipator K, Stein K, Ayers M, Joe A, Lunceford JK. T-Cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: keynote-028. J Clin Oncol. 2019;37(4):318–327. doi: 10.1200/JCO.2018.78.2276. [DOI] [PubMed] [Google Scholar]
- 20.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu X, Lu H, Nebozhyn M, Zhang C, Lunceford J, Joe A, Cheng J, Webber A, Ibrahim N, Plimack E, Ott P, Seiwert T, Ribas A, McClanahan T, Tomassini J, Loboda A, Kaufman D. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411):3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rooney M, Shukla S, Wu C, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman D, Albright A, Cheng J, Kang S, Shankaran V, Piha-Paul S, Yearley J, Seiwert T, Ribas A, McClanahan TK. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, Trajanoski Z. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karasaki T, Nagayama K, Kuwano H, Nitadori JI, Sato M, Anraku M, Hosoi A, Matsushita H, Morishita Y, Kashiwabara K, Takazawa M, Ohara O, Kakimi K, Nakajima J. An immunogram for the cancer-immunity cycle: towards personalized immunotherapy of lung cancer. J Thorac Oncol. 2017;12(5):791–803. doi: 10.1016/j.jtho.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Al-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan CW, Tabar V, Mellinghoff IK, DeAngelis LM, Ariyan CE, Lee N, Tap WD, Gounder MM, D’Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, DeMatteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB, Chan TA, Morris LGT. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019 doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/jci91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, Seplyarskiy VB, Sharpe HJ, McKee T, Letourneau A, Ribaux PG, Popadin K, Basset-Seguin N, Ben Chaabene R, Santoni FA, Andrianova MA, Guipponi M, Garieri M, Verdan C, Grosdemange K, Sumara O, Eilers M, Aifantis I, Michielin O, de Sauvage FJ, Antonarakis SE, Nikolaev SI. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 29.Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, Sabio EY, Makarov V, Kuo F, Blecua P, Ramaswamy AT, Durham JN, Bartlett B, Ma X, Srivastava R, Middha S, Zehir A, Hechtman JF, Morris LG, Weinhold N, Riaz N, Le DT, Diaz LA, Jr, Chan TA. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364(6439):485–491. doi: 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman AM, Kato S, Cohen PR, Boichard A, Frampton G, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Genomic landscape of advanced basal cell carcinoma: implications for precision treatment with targeted and immune therapies. Oncoimmunology. 2017;7(3):e1404217–e1404217. doi: 10.1080/2162402X.2017.1404217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Not applicable.