Abstract
ITSN1 plays an important role in brain development. Recent studies in large cohorts of subjects with neurodevelopmental disorders have identified de novo variants in ITSN1 gene thereby suggesting that this gene is involved in the development of such disorders. The aim of this study is to provide further proof of such a link. We performed trio exome sequencing in a patient presenting autism, intellectual disability, and severe behavioral difficulties. Additional affected patients with a neurodevelopmental disorder harboring a heterozygous variant in ITSN1 (NM_003024.2) were collected through a worldwide collaboration. All patients underwent detailed phenotypic and genetic assessment and data was collected and shared by healthcare givers. We identified ten novel patients from eight families with heterozygous truncating or missense variants in ITSN1 gene. In addition, four previously published patients from large meta-analysis studies were included. In total, 7/14 patients presented a de novo variant in ITSN1. All patients showed neurodevelopmental disorders from autism spectrum disorders (90%), intellectual disability (86%), and epilepsy (30%). We demonstrated that truncating variants are in the first half of ITSN1 whereas missense variants are clustered in C-terminal region. We suggest ITSN1 gene is involved in development of an autism spectrum disorder with variable additional neurodevelopmental deficiency, thus confirming the hypothesis that ITSN1 is important for brain development.
Subject terms: ADHD, DNA sequencing
Introduction
Over the past decade, the introduction of next-generation sequencing technologies has enhanced the understanding of many Mendelian neurodevelopmental conditions that affect more than 1–3% of population worldwide. Variants were found at over 1000 genes responsible for such phenotypes [1]. Neurodevelopmental disorders includes intellectual disability (ID) characterized by defects in adaptive function and intellectual abilities [2], and autism spectrum disorders (ASD—HP:0000729) characterized by early dysfunction in social interactions, communication deficits, and the presence of repetitive and restricted behaviors [3]. Exome sequencing (ES) has become a key diagnostic tool for establishing a genetic diagnosis in neurodevelopmental disorders because of their wide genetic heterogeneity. ES offers interesting diagnostic yields ranging from 20% for ASD cohorts to 50% for ID cohorts [4–6]. However, molecular bases remain unknown in at least half of affected individuals.
ITSN1 gene (intersectin 1) is located on chromosome 21q22.11 and encodes a cytoplasmic membrane-associated protein involved in many cellular processes such as exocytosis/endocytosis [7, 8], cytoskeleton rearrangements, apoptosis, and mitogenic signaling [7]. Indeed, ITSN1 has been implicated in various cell pathway including GTPases, Ras/MAPKinase, or reelin signaling [7, 9]. Two isoforms were described: the ubiquitous short isoform (ITSN1-s) and the long isoform (ITSN1-l) expressed predominantly in neurons [10]. Furthermore, several studies in mammals showed high expression of ITSN1-l in neurons and major role in brain biology including development of dendritic spines, cortical midline connectivity, synaptic vesicle recycling, neuronal migration, synaptic plasticity [9], and more recently in learning and memory [11]. Large cohorts of patients with neurodevelopmental disorders, including ASD and ID have identified four isolated patients with a de novo variants in ITSN1 gene [12–15].
Here, we describe neurodevelopmental features identified in eight unrelated families with heterozygous variants in ITSN1 predicted to be deleterious.
Methods
DNA sequencing
DNA was extracted from peripheral whole-blood samples, using the QIAamp DNA Mini Kit (Qiagen) following standard procedures.
For patient 1, exome capture and sequencing were performed at Integragen SA from 1 µg of genomic DNA per individual using the TWIG kit. The resulting libraries were sequenced on a HiSeq 4000 (Illumina) according to the manufacturer’s recommendations for paired-end 76 bp reads. Obtained reads were aligned to the human genome reference sequence (GRCh37/hg19 assembly) using Burrows–Wheeler aligner (version 0.7.15). Duplicate reads were marked using Picard MarkDuplicates (version 2.4.1) (http://broadinstitute.github.io/picard/) and aligned read were then processed using GATK BaseRecalibrator and PrintReads (Genome Analysis Toolkit; version 3.8) to recalibrate base quality scores, according to GATK Best Practices recommendations. Quality control was performed on all BAM files by calculating depth of coverage onto RefSeq database (release 2018-11-11) with GATK DepthOfCoverage. SNPs and indels were identified from BAM files using GATK HaplotypeCaller. All variants identified were annotated using SnpEff (version 4.3). Patient 5 also beneficed of ES with trio analysis (child and parents). For patient 4, ES was provided by Yale University Centers for Mendelian Genomics on an Illumina HiSeq 2000 instrument with blood samples pooled 6 per lane. Libraries (TruSeq DNA v2 Sample Preparation kit; Illumina, San Diego, CA) and exome capture (EZ Exome 2.0, Roche) were performed according to manufacturer protocols. FASTQs were filtered, aligned, and variants were filtered and annotated by Codified Genomics (proprietary algorithm, San Diego, CA). For patients 3 6, 7, 8 using genomic DNA from the proband and parents, the exonic regions and flanking splice junctions of the genome were captured using the IDT xGen Exome Research Panel v1.0 (Integrated DNA Technologies, Coralville, IA) (patient 8 and 9) or the Clinical Research Exome kit (Agilent Technologies, Santa Clara, CA) (Manwaring). Massively parallel (NextGen) sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19, and analyzed for sequence variants using a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol has been previously described and are available on request. The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/).
Sanger sequencing
Genomic DNA was amplified by polymerase chain reaction (PCR) using HotStarTaq PCR kit (Qiagen) according to the manufacturer’s protocol. PCR products were purified by Agencourt CleanSEQ system (Beckman Coulter) and sequenced with the BigDye Terminator Cycle Sequencing kit, v3.1 (Applied Biosystems) in ABI 3730 sequencer (Applied Biosystems).
Datasharing
Datasharing was performed using the Matchmaker Exchange platform GeneMatcher (https://genematcher.org/). All variants are submitted on ClinVAR portal (https://www.ncbi.nlm.nih.gov/clinvar/; submission SCV001806780- SCV001806787)
Results
We report a 27-year-old male patient born from healthy unrelated parents. The family was already known from the genetic clinics because of a type 2 multiple endocrine neoplasia in the father’s side, with the identification of a RET variant (LRG_518t2: p.(Cys634Tyr)). Affected family members have a lender build. He was hospitalized at birth for 10 days for hypotonia and feeding difficulties. He walked independently at age 18 months. He developed hyperactivity, with violent outbursts of anger and anxiety. The diagnosis of autism was made at the age of 4. At about the age of 8, the parents reported a speech arrest until the age of 18, with increased anxiety, self-mutilation, aggressivity, and anorexia. Treatment with Diazepam and Cyamemazine was instituted, with Clonazepam when needed. He developed epilepsy at the age of 12, treated with Valproate, with the addition of Topiramate a few years later. Word associations reappeared from the age of 18. At the age of 20, he was admitted to a psychiatric hospital all week. At the age of 28, bilateral cataract was diagnosed. Cerebral CT scan was normal. He had mild dysmorphic features (right preauricular skin tag, everted lower lip vermilion, prominent nose, prominent maxilla, hollow cheeks, frizzy hair), he had a bent walking attitude with bent back and legs. He also had striae, arachnodactyly, flat feet, long and thin habitus (Table 1). Cardiac investigations not showed malformations suggesting Marfan syndrome (OMIM # 154700). We performed trio-ES (patient and his parents) and identified a de novo nonsense variant in ITSN1 gene (NM_003024.2:p.(Gln654*)). The known missense variant in RET gene (NM_020630.4: p.(Cys634Tyr)) was also found.
Table 1.
Clinical and molecular data of ITSN1 patients (NM_003024.2).
| Present reported cases | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | |
| Country | France | USA | USA | USA | Danmark | Canada | USA | USA | USA | USA |
| Sex | M | M | M | M | M | M | M | M | M | F |
| Age at last examination | 27 years 7months | 18 years | 3 years | 14 years | 10 years 1 month | 7 years | 6 years | 2 years 6 months | 7 years 5 months | 4 years 9 months |
| Weight | −0.5 SD | +0.3 SD | +2 SD | NA | +1 SD | <−2 SD | +2.5 SD | +1.5 SD | −0.2 SD | +0.79 SD |
| Height | +3.62 SD | −2 SD | +1.7 SD | NA | −1.5 SD | <−2 SD | +2 SD | +0.2 SD | 0 SD | +1.73 SD |
| OFC | +1.4 SD | 0 SD | 0 SD | NA | NA | −2 SD | +0.5 SD | +2.5 SD | +2 DS | NA |
| Cranio-facial features | Preauricular skin tag [HP:0000384], everted lower lip vermilion [HP:0000232], prominent nose [HP:0000448], prominent maxilla [HP:0430028] | Brachycephaly [HP:0000248], thickened helices [HP:0000391], epicanthus [HP:0000286], everted lower lip vermilion [HP:0000232] | Mild dolichocephaly [HP:0000268], broad forehead [HP:0000337], epicanthus [HP:0000286] | − | − | Microcephaly [HP:0000252], epicanthus [HP:0000286], prominent nose [HP:0000448], smooth philtrum [HP:0000319], thin upper vermillion [HP:0011339], micrognathia [HP:0000347] | Mild hypertelorism [HP:0000316], epicanthus [HP:0000286], wide mouth [HP:0000154], Widely spaced teeth [HP:0000687] | Macrocephaly [HP:0000256], High forehead [HP:0000348], epicanthus [HP:0000286] | High forehead [HP:0000348] | |
| Ocular anomalies | Cataract [HP:0000518], strabismus [HP:0000486] | Refractive error | NA | − | NA | Hypermetropia [HP:0000540], astigmatism [HP:0000483], | − | − | Hypermetropia [HP:0000540], astigmatism [HP:0000483], amblyopia [HP:0000646], accomodative esotropia [HP:0031764] | − |
| Hearing impairment [HP:0000365] | − | + | NA | − | − | − | − | − | − | − |
| Musculo-skeletal features | pes planus [HP:0001763], joint laxity [HP:0001388], arachnodactyly [HP:0001166], slender build [HP:0001533] | Pes cavus [HP:0001761], bilateral Hammertoe [HP:0001765] | Overlapping toe [HP:0001845] | − | − | Short stature [HP:0004322], brachydactyly [HP:0001156], single transverse palmar crease [HP:0000954] | − | Pes planus [HP:0001763] | Bilateral clinodactyly [HP:0030084] | − |
| Cardiac features | − | Syncopes [HP:0001279], | NA | − | − | − | − | − | − | − |
| Gastro-intestinal anomalies | Constipation [HP:0002019] | Constipation [HP:0002019], gastroesophageal reflux [HP:0002020], | NA | − | − | Gastroesophageal reflux [HP:0002020] | Diarrhea frequent in infancy [HP:0002014] | − | − | − |
| Other features | Striae extensae, Medullary thyroid carcinoma [HP:0002865], Episodic hypokalemia [HP:0012726], potomania | Eczema [HP:0000964], gynecomastia [HP:0000771], asthma [HP:0002099] | inguinal hernia [HP:0000023] Hydrocele [HP:4000037] | Sensitive gag reflex, Hydrocele [HP:4000037], Inguinal hernia [HP:0000023] | − | Eczema [HP:0000964] | - | Right undescended testicle, right inguinal hernia [HP:0000023], burried penis, phimosis [HP:0001741], Feeding difficulties [HP:0011968] | Café-au-lait spots, obstructive sleep apnea [HP:0002870] | Frequent ear infections, feeding difficulties [HP:0011968] |
| Global developmental delay [HP:0001263] | + | + | + | + | + | + | + | + | + | + |
| Intellectual disability (IQ test) [HP:0001249] | Mild (NA) | Mild (69) | NA | Mild (61) | Mild (67) | − | Mild-Moderate | NA | NA | NA |
| Autistic behavior [HP:0000729] | + | + | + | + | + | − | + | + | + | + |
| Delayed speech and language development [HP:0000750] | + | + | + | + | + | + | + | + | + | + |
| Echolalia [HP:0010529] | + | − | NA | + | NA | − | NA | NA | + | + |
| Speech apraxia [HP:0011098] | NA | + | NA | + | + | + | NA | NA | NA | NA |
| Seizure (age at onset) [HP:0001250] | + (12 years) | − | − | − | + (1 year 6 months) | − | − | − | + | − |
| Behavioral abnormality[HP:0000708] | Bulimia, anorexia, self-mutilation, anxiety, ADHD, stereotypies, aggressivity | ADHD | Poor socialization, repetitive behaviors | ADHD, anxiety, motor tics disorder | Stereotypies, social interaction problems | Impulsivity, aggressive behavior, ADHD, social difficulties | NA | − | ADHD | Tantrums |
| Gait disturbance [HP:0001288] | NA | + | − | − | − | − | NA | − | − | − |
| Hypotonia [HP:0001252] | + | + | + | + | − | − | + | − | − | − |
| Sleep disturbance [HP:0002360] | − | − | − | − | NA | − | NA | − | + | − |
| Psychiatric disorder | + | NA | NA | Very early onset psychosis | − | NA | NA | − | − | − |
| Brain MRI anomalies | − | − | NA | − | − | Prominent perivascular spaces, high signal white matter, slow myelination | Delayed myelinization, gliosis | − | NA | NA |
| Variant | chr21:g.35169690 C > T | chr21::g.35190737dupA | chr21:g.35138177 A > G | chr21:g.35154339 G > T | chr21: g.35254559 A > T | chr21:g.35144580 G > T | chr21:g.35147116_35147119delAGAA | chr21:g.35195890 G > A | chr21:g.35195890 G > A | chr21:g.35195890 G > A |
| c.1960C > T | c.2894dupA | c.789-2 A > G | c.1726G > T | c.4354 A > T | c.1258 G > T | c.1389_1392delAGAA | c.3116 G > A | c.3116 G > A | c.3116 G > A | |
| p.(Gln654*) | p.(Tyr965*) | p.? | p.(Glu576*) | p.(Asn1452Tyr) | p.(Glu420*) | p.(Lys463Asn*5) | p.(Trp1039*) | p.(Trp1039*) | p.(Trp1039*) | |
| PolyPhen-2 | NA | NA | NA | NA | 0.952 | NA | NA | NA | NA | NA |
| CADD | 38 | NA | NA | 39 | 29 | NA | NA | 40 | 40 | 40 |
| GnomAD frequency | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Inheritance | de novo | NA (adopted) | de novo | Inherited from father | de novo | NA (adopted) | NA | Inherited from father | Inherited from father | Inherited from father |
NA not available, ADHD attention-deficit hyperactivity disorder, MRI magnetic resonance imaging, OFC occipitofrontal circumference, HPO number are noted [HP].
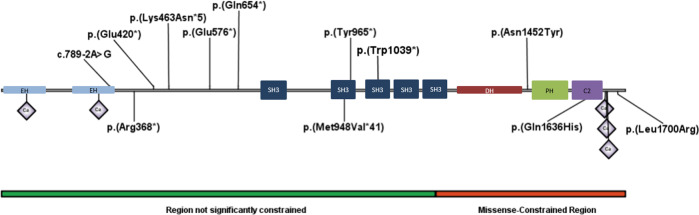
International datasharing allowed the identification of ten additional patients (Table 1) from 8 families with heterozygous truncating variants (7/8) or missense variant (1/8) affecting the two ITSN1 isoforms (Fig. 1). All have normal chromosomal micro-array.
Fig. 1. Representation of ITSN1 protein.
Identified variants in this study (up) and in the scientific literature (down): p.(Met948Val*41) reported by Yuen et al. [12]; p.(Leu1700Arg) reported by Rubeis et al. [13]; p.(Gln1636His) reported by DDD, [14] and p.(Arg368*) reported by Fromer et al. [15].
In total, we identified nine males, one female, and four previously reported patients (Table 1). The age ranged from 2 to 27 years (median of 7 years). Neurodevelopmental disorders were diagnosed in all patients and included ID or global developmental delay (10/10) and ASD (12/13). Seizures free (EEG not performed) was noted in 3/10 patients one year to onset of puberty and controlled by valproate/topiramate (patient 1) or carbamazepine (patient 5). All patients showed speech delay and/or language impairment (10/10). They said their first words at normal age but presented a very slow progression, most of them spoke in short sentence with poor vocabulary at the last examination. Five patients (1, 3, 4, 7, and 9) displayed a regression of speech, sometimes not definitive, sometimes leading to speech arrest, from 13-months to 18-year-old for patient 1, and from 15 month for patient 3 and from 18- to 24 months in patient 4. Eight patients presented severe behavioral troubles: attention-deficit hyperactivity disorder (ADHD) (5/10), aggressive outburst and self-mutilation (2/10), stereotypies (3/10), social interaction impairment (3/10), tantrums (1/10), and anxiety (2/10). More severe psychiatric disorders were found in two patients: one with psychosis, and one with schizophrenia (Table 1). These severe behavioral disorders required medication and sometimes day/full hospitalization. Treatment with Diazepam and Cyamemazine was instituted with Clonazepam when needed in patient 1 presenting anxiety, self-mutilation, aggressivity, and anorexia. At the age of 20, he was admitted to a psychiatric hospital all week and his state of anxiety remained high despite his treatment. He developed more and more rituals. Patient 4 showed ADHD, anxiety, and developed a psychosis, he is currently treated and well controlled with Aripiprazole and Sertraline. Patients 6 and 7 have ADHD treated by biphentin or CBD oil Charlotte’s WebTM. Brain MRI was performed in three patients and displayed no or minor abnormalities (Table 1). Two patients (1 and 7) presented hypotonia at birth and two others (patients 3 and 4) developed a hypotonia at 3 years old. All patients started walking at normal age (range to 10–18 months) and they now walk unsupported. Minor and inconstant dysmorphic features were observed. Gastro-intestinal problems including constipation, diarrhea, and gastroesophageal reflux was reported in 4/6 patients.
In ten families, parents were available for segregationally analysis. In seven of these patients, the reported variant was de novo but in two patients (patient 4 and 9) it was inherited from paucisymptomatic father. The father of patient 4 presented ADHD and the father of patient 8 reported having learning difficulties and autistic features. Two affected sibling of patient 8 (patients 9 and 10) presented the variant inherited from the father and one unaffected brother showed absent variant. Neither of the fathers have undergone a formal neuropsychological evaluation. None of the paternal grandparents are available for segregationally analysis.
Discussion
In addition to pathophysiological data showing that ITSN1 plays an important role in brain development, the ITSN1 gene could be considered as a candidate gene for neurodevelopmental disorders [12, 15]. However, the four previously published patients were described in the supplemental data of four meta-analysis studies [12, 15]. Very limited clinical data is available, but those patients presented with ASD with or without ID. In this study, we identified ten novel patients (Table 1), confirming the involvement of ITSN1 in a neurodevelopmental disorder spectrum, including ASD, ID, major behavioral difficulties and/or verbal impairment. We present detailed clinical and genetic data from a cohort of ITSN1 cases with a neurodevelopmental disorder. We explore the spectrum of symptoms and show that premature codon stop in the first half of the protein are reported in 9/12 families. The high intolerance to inactivation for ITSN1 is reported by GnomAD database (https://gnomad.broadinstitute.org/) with an associated pLI (probability of loss-of-function intolerance) score of 1, LOEUF (loss-of-function observed/expected upper bound fraction) score of 0.22 and a HI (HaploInsufficiency) index score of 24.17 by Decipher (https://decipher.sanger.ac.uk/gene/ITSN1#overview/clinical-info). All identified variants are absent in GnomAD database. In addition, we report 3/12 families with a heterozygous missense variant, clustering in C-terminal region of ITSN1 protein without splicing defect predicted by Human Splicing Finder software (http://www.umd.be/HSF/index.html). Missense variants were predicted probably damaging by PolyPhen-2 (scores for p.(Asn1452Tyr), p.(Leu1700Arg) and p.(Gln1636His) were respectively 0.952, 0.989 and 0.966) and deleterious by SIFT (Sorting Intolerant From Tolerant) (respectively 0, 0.04 and 0.04). The CADD Phred score (Combined Annotation Dependent Depletion) was between 25.3 and 29.2.”GnomAD database reported the intolerance of ITSN1 gene to support missense variants (misZ-score = 3.61). Furthermore, missense variants are located in important regional missense constraint (amino-acid 1199–1722), (cci = 0.49, p value = 1.80 × 10−12) that might explain by C-terminal region are provided in 3D structure of the protein [16]. We suggest that ITSN1 haploinsufficiency and the missense variants clustering in C-terminal region are implicated in a neurodevelopmental disorder, however future functional studies are needed to confirm and explore pathophysiological mechanisms.
Several studies have demonstrated the role of ITSN1 in brain development, in particular mice models showing that the expression of the a brain-specific pattern [9]. The knock-out mice displayed cerebral anomalies including defects of neuronal migration and synaptic plasticity of the hippocampus and the cortex, and anomalies of secretion and transport of synaptic vesicles. Jakob et al, demonstrated that ITSN1 interacts with Reelin receptors and is an essential component of Reelin pathway involved in neuronal migration causing similar brain malformations in mice. In human, recessive variants in REELIN (RELN) are associated with lissencephaly [#MIM 257320] and heterozygous variants are reported in autosomal dominant epilepsy [#MIM 616436] and ASD [17]. Interestingly, the hippocampus plays an important role in spatial memory and learning and anomalies of hippocampal structure or function are associated with autism [18, 19]. ITSN1 knock-out mice have previously revealed an alteration in spatial learning and memory [11]. The function of ITSN1 in hippocampus development supports the involvement of this gene in neurodevelopment disorders and in particular in ASD. Unfortunately, we cannot explore the potential brain malformations in ITSN1 patients reported in this study. Indeed, their behavioral difficulties posed an obstacle for performing brain MRI. Only a single patient (Patient 7) underwent brain MRI showing minor cerebral defects including prominent perivascular spaces. The marfanoid habitus described specifically in Patient 1 (Supplemental data) can be explain by an additional identified and presumed disease-causing variant in the RET gene. Pathogenic variants in the RET gene are associated with multiple endocrine neoplasia IIA (MIM #171400) that included medullary thyroid carcinoma and variable phenotypic spectrum including marfanoid habitus [20].
In conclusion, we report ten patients from eight unrelated families with heterozygous premature truncating or clustered missense variants highlighting ITSN1 gene as a new gene for neurodevelopmental disorders including ASD.
Supplementary information
Acknowledgements
The authors thank the family for participating and supporting this study. We thank the University of Burgundy Centre de Calcul (CCuB) for technical support and management of the informatics platform, and the Genematcher plateform for datasharing. This work was supported by grants from Dijon University Hospital, the ISITE-BFC (PIA ANR) and the European Union through the FEDER programs. Several authors of this publication are members of the European Reference Network for Developmental Anomalies and Intellectual Disability (ERN-ITHACA).
Funding
This work was funded by the French Ministry of Health (PHRC national 2008) and the Regional Council of Burgundy / Dijon University hospital (PARI 2011).
Data availability
The datasets generated during and/or analysed during the current study are available in the ClinVAR database (https://www.ncbi.nlm.nih.gov/clinvar/; submission SCV001806780- SCV001806787). The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
Consents obtained in the clinical setting. The IRB approval was granted by our institute.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00985-9.
References
- 1.Fernandez-Marmiesse A, Gouveia S, Couce ML. NGS Technologies as a Turning Point in Rare Disease Research, Diagnosis and Treatment. Curr Med Chem. 2018;25:404–32. doi: 10.2174/0929867324666170718101946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel DR, Apple R, Kanungo S, Akkal A. Intellectual disability: definitions, evaluation and principles of treatment, Pediatric Med. 2018;1. 10.21037/pm.2018.12.02.
- 3.Tărlungeanu DC, Novarino G. Genomics in neurodevelopmental disorders: an avenue to personalized medicine. Exp Mol Med. 2018;50:100.. doi: 10.1038/s12276-018-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvill GL, Mefford HC. Next-Generation Sequencing in Intellectual Disability. J Pediatr Genet. 2015;4:128–35. doi: 10.1055/s-0035-1564439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stojanovic JR, Miletic A, Peterlin B, Maver A, Mijovic M, Borlja N, et al. Diagnostic and clinical utility of clinical exome sequencing in children with moderate and severe global developmental delay/intellectual disability. J Child Neurol. 2020;35:116–31. doi: 10.1177/0883073819879835. [DOI] [PubMed] [Google Scholar]
- 6.Rossi M, El-Khechen D, Black MH, Farwell Hagman KD, Tang S, Powis Z. Outcomes of diagnostic exome sequencing in patients with diagnosed or suspected autism spectrum disorders. Pediatr Neurol. 2017;70:34–43.e2. doi: 10.1016/j.pediatrneurol.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 7.Gryaznova T, Gubar O, Burdyniuk M, Kropyvko S, Rynditch A. WIP/ITSN1 complex is involved in cellular vesicle trafficking and formation of filopodia-like protrusions. Gene. 2018;674:49–56. doi: 10.1016/j.gene.2018.06.078. [DOI] [PubMed] [Google Scholar]
- 8.Alvisi G, Paolini L, Contarini A, Zambarda C, Di Antonio V, Colosini A, et al. Intersectin goes nuclear: secret life of an endocytic protein. Biochem J. 2018;475:1455–72. doi: 10.1042/BCJ20170897. [DOI] [PubMed] [Google Scholar]
- 9.Jakob B, Kochlamazashvili G, Jäpel M, Gauhar A, Bock HH, Maritzen T, et al. Intersectin 1 is a component of the Reelin pathway to regulate neuronal migration and synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2017;114:5533–8. doi: 10.1073/pnas.1704447114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dergai M, Skrypkina I, Dergai O, Tsyba L, Novokhatska O, Filonenko V, et al. Identification and characterization of a novel mammalian isoform of the endocytic adaptor ITSN1. Gene. 2011;485:120–129. doi: 10.1016/j.gene.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Malakooti N, Pritchard MA, Chen F, Yu Y, Sgambelloni C, Adlard PA, et al. The Long Isoform of Intersectin-1 Has a Role in Learning and Memory. Front Behav Neurosci. 2020;14:24. doi: 10.3389/fnbeh.2020.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen RKC, Merico D, Bookman M, Howe JL, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–11. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samocha KE, Kosmicki JA, Karczewski KJ, O’Donnell-Luria AH, Pierce-Hoffman E, MacArthur DG, et al. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017;148353. 10.1101/148353.
- 17.Lammert DB, Howell BW. RELN Mutations in Autism Spectrum Disorder. Front Cell Neurosci. 2016;10:84.. doi: 10.3389/fncel.2016.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper RA, Richter FR, Bays PM, Plaisted-Grant KC, Baron-Cohen S, Simons JS. Reduced Hippocampal Functional Connectivity During Episodic Memory Retrieval in Autism. Cereb Cortex. 2017;27:888–902. doi: 10.1093/cercor/bhw417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ring M, Derwent CLT, Gaigg SB, Bowler DM. Structural learning difficulties implicate altered hippocampal functioning in adults with autism spectrum disorder. J Abnorm Psychol. 2017;126:793–804. doi: 10.1037/abn0000277. [DOI] [PubMed] [Google Scholar]
- 20.Fassbender WJ, Krohn-Grimberghe B, Görtz B, Litzlbauer D, Stracke H, Raue F, et al. Multiple endocrine neoplasia (MEN)–an overview and case report–patient with sporadic bilateral pheochromocytoma, hyperparathyroidism and marfanoid habitus. Anticancer Res. 2000;20:4877–87. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the ClinVAR database (https://www.ncbi.nlm.nih.gov/clinvar/; submission SCV001806780- SCV001806787). The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.