Abstract
SKP1-CUL1-F-box (SCF) ubiquitin ligases play fundamental roles in cellular functions. Typically, substrate phosphorylation is required for SCF recognition and subsequent degradation. However, phospho-dependent substrates remain largely unidentified. Here, using quantitative phoshoproteome approach, we performed a system-wide investigation of phospho-dependent SCF substrates. This strategy identified diverse phospho-dependent candidates. Biochemical verification revealed a mechanism by which SCFFBXO22 recognizes the motif XXPpSPXPXX as a conserved phosphodegron to target substrates for destruction. We further demonstrated BAG3, a HSP70 co-chaperone, is a bona fide substrate of SCFFBXO22. FBXO22 mediates BAG3 ubiquitination and degradation that requires ERK-dependent BAG3 phosphorylation at S377. FBXO22 depletion or expression of a stable BAG3 S377A mutant promotes tumor growth via defects in apoptosis and cell cycle progression in vitro and in vivo. In conclusion, our study identified broad phosphorylation-dependent SCF substrates and demonstrated a phosphodegron recognized by FBXO22 and a novel ERK-FBXO22-BAG3 axis involved in tumorigenesis.
Subject terms: Ubiquitylation, Proteomics
Introduction
The ubiquitin (Ub)-proteasome system (UPS) plays a prominent role in maintaining cellular homeostasis, which mediates 80–85% of total protein degradation events in eukaryotes [1, 2]. The selectivity of Ub-mediated proteolysis is primarily determined by the E3 ligases, which typically recognize the degradation signals of unneeded protein substrates to facilitate regulatory proteasomal degradation [3, 4]. Among them, the Cullin-RING ligases (CRLs) constitute the largest class E3 ligase family with more than 200 documented members [5]. The CRL1 ligases, better known as S phase kinase-associated protein 1 (SKP1)-cullin 1 (CUL1)-F box protein (SCF) complexes, are the best characterized among all CRLs [6]. The SCF complex consists of the constant cullin 1, a RING domain-containing component RBX1, an adapter SKP1, and one of ~70 various F-box proteins [7, 8]. Most F-box proteins, which are responsible for substrate recognition, can specifically bind to the phosphorylated motifs (phosphodegron) of their substrates, which are regulated by diverse kinases [9, 10]. For example, βTrCP binds the phosphodegron DSGXXS (both S residues are phosphorylated), and FBXW7 binds the phosphodegron TPPXS (the T and S residues are phosphorylated) [11–13]. However, given a large number of human F-box proteins, the identities of phosphodegrons for the majority of SCFs remain largely unknown.
In recent years, several high-throughput methodologies have been developed to systematically identify SCF substrates, including the global protein stability (GPS) platform [14], quantitative ubiquitination proteomics, [15, 16] and parallel SCF adaptor capture proteomics [17, 18], which has greatly advanced the understanding of the functional diversity of SCFs. However, these methods have not yet provided a system-wide view as to how phosphorylation events regulate cellular homeostasis via SCF-mediated phosphorylated substrate degradation. Therefore, a combination of simultaneous quantitative analysis of the SCF-perturbed proteome and phosphoproteome could enable a system-wide characterization of phosphorylation-dependent substrates and phosphodegrons, which would be important for better understanding the physiological and pathological functions of SCF E3 ligases.
BAG (Bcl-2-associated athanogene) is a family of proteins that are well recognized for their anti-apoptotic functions in protein quality control [19, 20]. Dysregulation of BAG3 is associated with many diseases, such as cancer, myopathy, and neurodegenerative disorders [21, 22]. Nevertheless, the current understanding of BAG3 is focused on its regulatory roles in the protein quality control of its substrate proteins. It remains largely unknown how the fate of BAG3 itself is regulated in cell physiology and pathology.
Here, by utilizing mass spectrometry-based proteomic and phosphoproteomic approaches, we carried out a system-wide quantitative analysis of potential phosphorylation-dependent SCF substrates. This study identified dozens of potential new substrates with broad cellular functions. Importantly, we identified BAG3 as a bona fide phosphorylated substrate of SCFFBXO22, which plays an important role in tumorigenesis.
Results
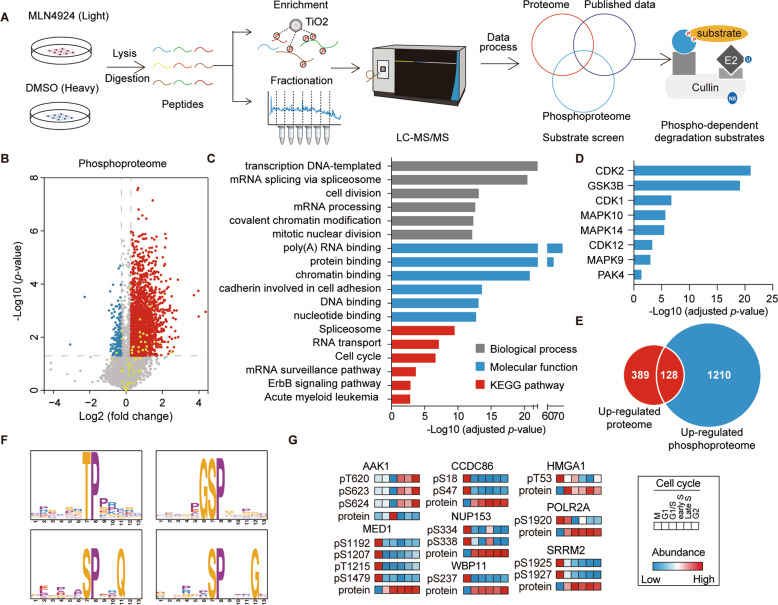
Systematic screening for phospho-dependent SCF substrates
We reasoned that, when the activities of CRLs were inhibited, phosphorylation levels of their phospho-dependent substrates would increase due to the impared degradation of these phosphorylated substrates. In agreement, the global phosphorylation levels of HEK 293T cell lysates were significantly increased after 4 h of MLN4924 treatment (1 μM), a widely used protein neddylation inhibitor for CRL inactivation [23] (Fig. S1A). The increased global phosphorylation levels was not due to altered cell cycle progress, as 1 µM MLN4924 for 4 h treatment did not affect the cell cycle progress (Fig. S1B). To identify phospho-dependent degradation substrate proteins of CRLs system-wide, we used a mass-spectrometry-based stable isotope labeling with amino acids in cell culture (SILAC) approach to quantify the altered proteome and phosphoproteome induced by MLN4924 (Fig. 1A). This workflow enabled quantification of 8,286 proteins, of which 517 proteins increased in abundance with p-value < 0.05 (by t-test) in two replicate analyses (Fig. S1C). In addition, we quantified 12,276 phosphorylation sites (FDR < 5%, localization probability > 0.75) on 4,322 phosphoproteins. Among them, 2,800 phosphorylation sites were up- or downregulated more than 1.2-fold. We found 33 phosphosites, annotated as protein degradation-associated modifications in the PhosphoSitePlus database, that were upregulated (Fig. 1B and Table S1). Bioinformatics analysis showed that the MLN4924 upregulated phosphoproteome (L/H > 1.2) was significantly enriched in diverse biological processes (Fig. 1C and Table S1). Kinase Enrichment Analysis (KEA) of the upregulated phosphoproteome resulted in the enrichment of CDKs, GSK3B, and MAPKs, suggesting these kinase-associated pathways were likely activated upon MLN4924 inhibition (Fig. 1D). Many SCF substrates play critical roles in cell-cycle regulation [24], and degradation of these substrates only accounts for a small percentage (10–20%) of the protein in the total cell lysate [25]. Therefore, we used the criteria of a 1.2-fold increase in both protein expression and phosphorylation in samples treated with MLN4924 for initial screening of potential SCF substrates. We found that 128 proteins and 369 phosphosites met our screening criteria (Fig. 1E and Table S1). Motif analysis identified several enriched motifs including one similar to the phosphodegron motif (TPPxE/S) for FBXW7 (Fig. 1F, Fig. S1D) [26]. To determine whether these phosphorylated proteins participated in cell cycle regulation, we compared our results to a published mitosis phosphoproteome dataset [27]. Subsequently, 8 substrate proteins and 85 phosphosites were identified from our dataset. For example, phosphorylation at Ser18 in CCDC86 and Ser1920 in POLR2A fluctuated dynamically and were negatively correlated with their protein levels during the cell cycle (Fig. 1G and Table S1).
Fig. 1. Systematic screening for phospho-dependent substrates of SCF complexes.
A The diagram shows the experimental workflow for systematic screening of phospho-dependent degradation substrates. B The volcano plot illustrates the altered phosphoproteome following 4 h of MLN4924 treatment in HEK 293T cells. Red and blue dots represent significantly up- and downregulated phosphosites, respectively. Yellow dots represent phosphosites functionally annotated as protein degradation-associated regulatory sites (“ON_FUNCTION” annotated as protein degradation, ubiquitination, and protein stabilization in PhosphoSitePlus database). C The histogram shows the enriched pathways (adjusted p-value < 0.05) of the significantly upregulated phosphoproteome using the DAVID bioinformatics database (v6.8). D Enriched kinases from KEA of the upregulated phosphoproteome upon MLN4924 inhibition. A Benjamini adjusted p-value of 0.05 was set as cutoff. E The overlap between the upregulated proteome and phoshoproteome. (L/H ratio > 1.2, p-value < 0.05 by t-test). F Representative motif enrichment analysis by MoMo software using Motif-X algorithms. G The diagram shows the candidates with an inverse relationship between protein expression and phosphorylation throughout the cell cycle based on a published mitosis phosphoproteome dataset [27]. Candidates should fit the following criteria: at least a fourfold change in protein abundance and a twofold change in phosphorylation throughout the cell cycle. Maximum protein abundance and minimum phosphorylation levels should simultaneously occur in one of the six-cell cycle phases (M, G1/S, Early S, Late S, and G2) (or vice versa).
Identification and biochemical validation of SCF candidate substrates
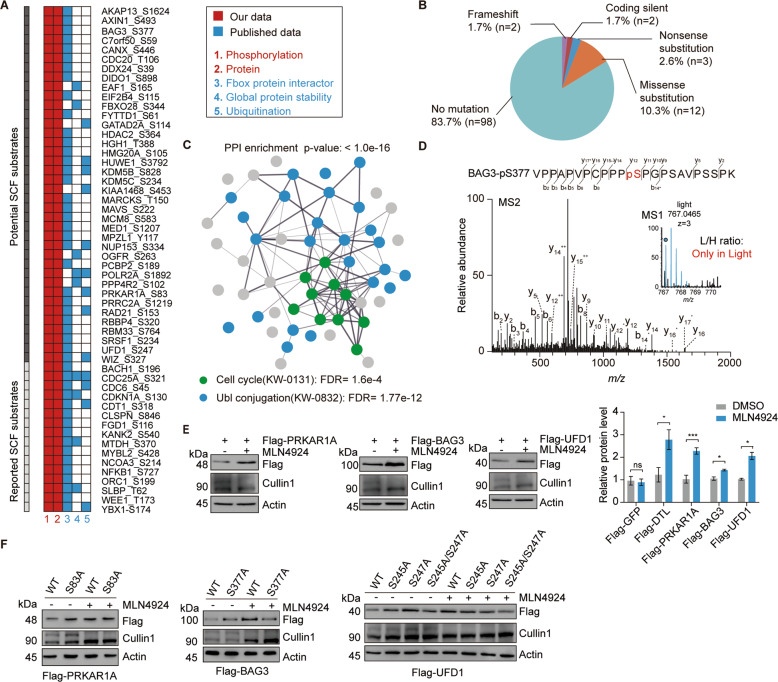
To identify new phospho-dependent SCF substrates, we considered a protein identified in our dataset as a potential substrate if it overlapped with either the SCF-associated interactome (BioGRID database), MLN4924-dependent ubiquitinome (ubiquitination downregulated by MLN4924 > 2-fold), or GPS dataset [14]. Using these criteria, we identified 16 known SCF substrates as well as 38 previously unreported potential new SCF substrates (Fig. 2A). Notably, among these potential substrates, 17% of their phosphosites (19 sites) were reported to be mutated in cancers according to the COSMIC (Catalogue of Somatic Mutations in Cancer) database (Fig. 2B, Fig. S2A and Table S2). Network analysis using the STRING database identified high-confidence interactions among 54 substrate proteins (Fig. 2C and Table S2). These substrates were highly enriched in the cell cycle (FDR = 1.60e−4) and ubiquitin-like protein (Ubl) conjugation (FDR = 1.77e−12).
Fig. 2. Identification and biochemical validation of SCF candidate substrates.
A The diagram displays the candidate substrates meeting the criteria as follows: (1) Phosphorylation upregulated upon MLN4924 treatment by more than 1.2-fold in our phosphoproteomic data, (2) protein upregulated upon MLN4924 treatment by more than 1.2-fold in our proteomic data, (3) interacting with either one of the F-box proteins from BioGRID, (4) ubiquitination downregulated by MLN4924 by more than twofold from a published dataset CRL substrate screening [14], or (5) protein stability governed by CUL1 from a published dataset screening for substrates of CRLs. The most significantly changed phosphosites of the protein candidates are shown. B The pie chart shows the percentages of the mutation types of upregulated phosphosites (n = 117) according to the COSMIC database. C The protein–protein interaction (PPI) was analyzed by the STRING database and a PPI enrichment was obtained with p-value < 1.0 × 10−16. D The annotated MS/MS spectrum of the phosphopeptide containing the S377 phosphorylated residue of BAG3 is shown. H: HEK-293T cells treated with DMSO, L: HEK-293T cells treated with MLN4924. E Immunoblot analysis of Flag-tagged candidate substrates to examine protein stabilization after MLN4924 treatment. Flag-GFP was considered as the negative control. DTL was considered as the positive control. Data are presented as mean ± SEM. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001 versus control groups (Student’s t-test), n = 3 replicates. F Immunoblot analysis of mutated phosphosites on protein stabilization.
We next chose three representative candidates that participated in different pathways for further biochemical validation, including BAG3, UFD1, and PRKAR1A (Fig. 2D and Fig. S2B). The reported SCF substrate DTL was used as a positive control. After ectopically expressed as Flag fusion proteins in HEK 293T cells, all these proteins significantly accumulated after 4 h of MLN4924 treatment (Fig. 2E and Fig. S2C). Consistently, when the dominant-negative cullin 1 (DN-CUL1) [28] was overexpressed to inactivate the SCF complexes, these proteins were significantly accumulated (Fig. S2D). In addition, the drastic accumulation of BAG3 protein was also observed in cullin1 knockout (KO) cells (Fig. S2E). Together, these results suggested that these proteins belonged to SCF substrates. We next mutated the identified phosphorylated serine sites of these proteins to alanine to evaluate their potential regulatory roles on protein degradation. The WT proteins were accumulated upon MLN4924 treatment. However, the phospho-mutant proteins did not respond to MLN4924 treatment, as their basal protein levels were similar to those of the MLN4924-treated WT counterparts (Fig. 2F). Consistently, the stability of these mutants was significantly increased compared with their WT counterparts (Fig. S2F). These results suggest that these phosphorylation sites are critical for SCF-dependent degradation of the corresponding proteins.
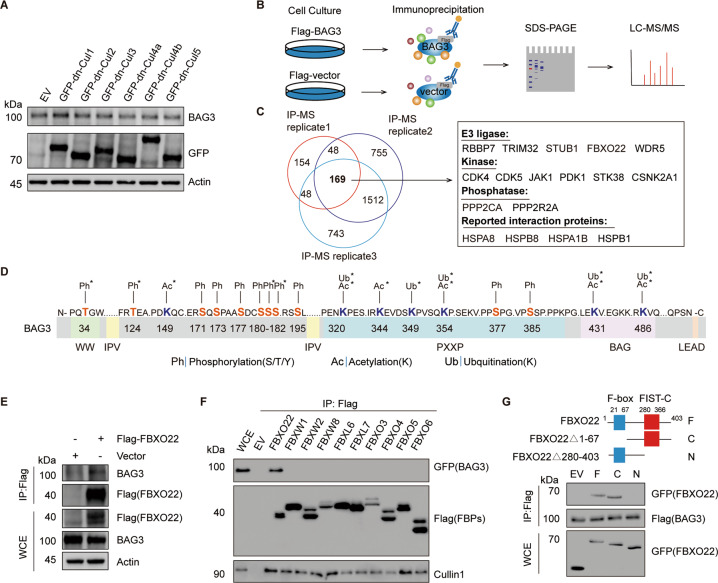
BAG3 is a bona fide substrate of SCFFBXO22
To further demonstrate the utility of our phosphoproteome data in SCF substrate identification, we selected BAG3 for further investigation. MLN4924 treatment resulted in the accumulation of BAG3 protein (Fig. S3A). As MLN4924 inactivates all cullins, we overexpressed six dominant-negative cullins (DN-CUL1, DN-CUL2, DN-CUL3, DN-CUL4A, and DN-CUL4B, DN-CUL5) in HEK 293T cells to investigate whether BAG3 was regulated by other CRLs. We found that BAG3 protein was only accumulated in DN-CUL1 transfected cells (Fig. 3A and Fig. S3B), indicating BAG3 is a putative substrate of the SCF E3 ligase. To identify which SCF complex is required for BAG3 destruction, the BAG3 protein complex was purified by immunoprecipitation (IP) in the presence of MLN4924 and subjected to MS analysis (Fig. 3B). Proteins identified from Flag-BAG3 IP group only or ≥5-fold (emPAI ratio) than the control group were considered. We identified 169 proteins that were associated with BAG3 from all the three biological replicates. Bioinformatics analysis showed protein folding associated pathways were significantly enriched in the BAG3 interactome (Fig. S3C–E and Table S3). Among these, several well-known BAG3 interacting proteins were identified [29]. Interestingly, we found that BAG3 tightly interacted with multiple different types of protein modification enzymes including E3 ligases, kinases, and phosphatases (Fig. 3C). Mass spectrometry analysis further identified five new ubiquitination, four new phosphorylation, and six new acetylation sites according to the PhosphoSitePlus database (Fig. 3D and Fig. S3F). These post-translational modifications were likely to be regulated by the protein-modifying enzymes identified in the BAG3 interactome.
Fig. 3. BAG3 is a bona fide substrate of SCFFBXO22.
A HEK 293 T cells were transfected with GFP empty vector (EV) or indicated GFP-DN-Culs for 36 h. Cell lysates were subjected to immunoblotting as indicated. B The diagram shows the experimental process for the MS-based interactome analysis of BAG3. C Overlap between three BAG3 interactome analyses. The E3 ligases, kinases, phosphatase, and reported interaction proteins identified in our dataset are shown. D The diagram shows all the identified phosphorylation, ubiquitination, mono-methylation, and acetylation sites on BAG3. Asterisk denotes the newly discovered post-translational modification sites. E Immunoblot analysis of the interaction between FBXO22 and BAG3. HEK 293T cells were transfected with Flag-FBXO22 for 36 h under MLN4924 treatment. Lysates were immunoprecipitated with anti-Flag M2 resin and analyzed by immunoblotting as indicated. WCE: whole-cell extracts. F Immunoblot analysis examining the specificity of the interaction between F-box proteins and BAG3. HEK 293T cells were co-transfected with EV or the indicated Flag-tagged F-box proteins (FBPs) plasmids and GFP-BAG3 for 36 h. WCE were immunoprecipitated by anti-Flag M2 resin and immunoblotted as indicated. G A schematic diagram showing the various constructs of FBXO22 used for binding assays with BAG3. In addition to full FBXO22 (F), two deletion constructs were also used: C: FBXO22 68–403 and N: FBXO22 1–279. HEK 293T cells were co-transfected with Flag-BAG3 and the indicated GFP-FBXO22 plasmids for 36 h. WCE were immunoprecipitated by anti-Flag M2 resin and immunoblotted as indicated.
Interestingly, FBXO22 was the only F-box protein associated with BAG3 among the E3 ligases identified in our dataset. Next, we confirmed the protein–protein interaction between BAG3 and FBXO22 by IP experiments via ectopic expression of either Flag-FBXO22 or Flag-BAG3 (Fig. 3E, and S3G–H). This interaction could be further increased by MLN4924 treatment (Fig. S3I). We next screened a library of F-box proteins and found that although all F-box proteins were associated with cullin1, FBXO22 was the only one that specifically bound to BAG3 (Fig. 3F). Through protein interaction domain mapping, we found that FBXO22 physically interacted with BAG3 through its C-terminal FIST-C domain (Fig. 3G).
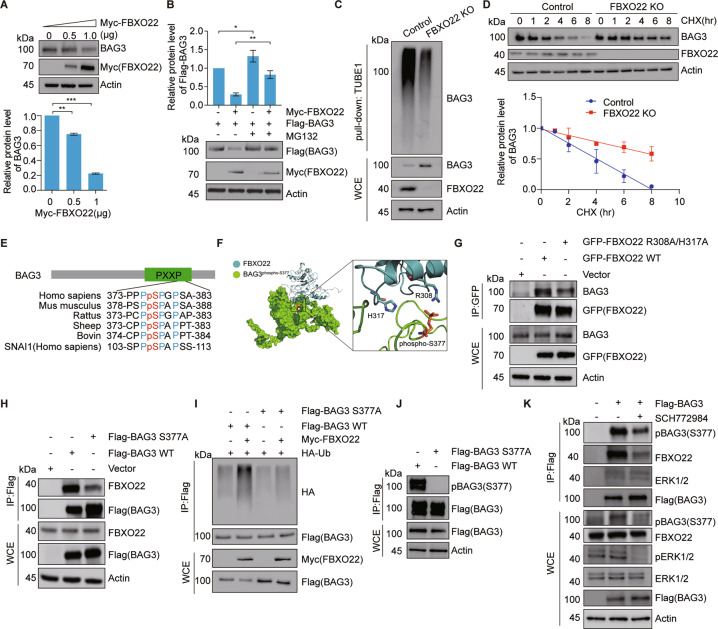
ERK-induced BAG3 S377 phosphorylation generates a phosphodegron recognized by SCFFBXO22 for ubiquitination and degradation
To gain a detailed mechanistic insight as to how FBXO22 regulates BAG3 degradation, we overexpressed Myc-FBXO22 in HEK 293T cells, which resulted in a marked reduction in both exogenous and endogenous BAG3 levels (Fig. 4A and Fig. S4A), but without affecting its mRNA level (Fig. S4B). Therefore, we proposed that the FBXO22-induced BAG3 reduction was due to the enhancement of BAG3 proteolysis, as shown by the rescue of BAG3 levels with a proteasome inhibitor MG132 and an increase in the ubiquitinated BAG3 form, as well as a decrease in the BAG3 half-life (Fig. 4B, S4C, and S4D). Depletion of FBXO22 by CRISPR/Cas9 system induced a decrease in ubiquitinated BAG3 and an increase of BAG3 stability (Figs. 4C and D). Our proteomics data showed the phosphorylation of BAG3 S377 was upregulated by MLN4924 treatment. To investigate whether S377 is a regulatory site for FBXO22-dependent BAG3 degradation, we performed a conserved sequence alignment analysis and found that this amino acid was highly conserved across different species. Interestingly, the amino acid residues around phosphorylated S377 containing PPPpSPGPSA were very similar to SNAI1 (SPPpSPAPSS), a reported substrate of FBXO22 (Fig. 4E) [30]. We therefore hypothesized that the XXPpSPXPXX motif might be a putative phosphodegron recognized by FBXO22. To test this hypothesis, we carried out structure-based computational analyses to investigate the potential recognition mechanism. Since currently there is no available crystal structure for these two proteins, the I-TASSER webserver was employed for structural modeling [31]. The modeling results were then subjected to protein docking prediction through the Haddock program, which retrieved that FBXO22 recognizes S377 phosphorylated BAG3 through R308 and H317 of its C-terminal FIST-C domain (Fig. 4F). Compared with FBXO22 WT, the ability of FBXO22 RH/AA mutant to recognize BAG3 protein is significantly reduced (Fig. 4G and Fig. S4E). When we used a BAG3 S377A mutant, the binding of the BAG3 S377A mutant to FBXO22 was similarly compromised (Fig. 4H and Fig. S4F). Moreover, the expression of FBXO22 failed to affect the ubiquitination and the stability of the BAG3 S377A mutant (Fig. 4I and Fig. S4G). Thus, our results suggested a critical role of phospho-S377 in mediating the binding of FBXO22 to BAG3 for subsequent ubiquitination. We next developed a phospho-specific antibody that exclusively detected BAG3 phospho-S377 (Fig. S4H). This antibody was able to detect BAG3 WT, but not a BAG3 S377A mutant (Fig. 4J). In agreement, the protein levels of BAG3 and BAG3 phospho-S377 were both reduced in BAG3 knockdown (KD) cells (Fig. S4I). These data suggest that the antibody specifically recognizes phosphorylated BAG3 at S377 both in vitro and in vivo. We hypothesized that if BAG3 phospho-S377 was required for BAG3 ubiquitination and degradation, both the phosphorylated and unphosphorylated protein forms of BAG3 would be accumulated in the absence of FBXO22. Indeed, both BAG3 phospho-S377 and BAG3 proteins were increased in FBXO22 KO cells compared with WT cells (Fig. S4J and S4K). Thus, the above results demonstrated that FBXO22-mediated degradation of BAG3 required phospho-S377 in its XXPpSPXPXX degron motif.
Fig. 4. FBXO22-mediated BAG3 ubiquitination and degradation requires BAG3 phosphorylation at S377.
A Immunoblot analysis of BAG3 levels after transfected with the indicated doses of Myc-FBXO22 plasmids. The quantification plot was based on scanning densitometry analysis and the relative protein levels were normalized to the control. Data are presented as mean ± SEM, n = 3 replicates. *p < 0.05, **p < 0.01, ***p < 0.001 versus control groups (Student’s t-test). B Proteasome inhibitor MG132 blocks FBXO22-induced down-regulation of BAG3. HEK 293T cells were co-transfected with Myc-FBXO22 and Flag-BAG3 for 36 h and treated with 20 μM MG132 for 5 h. Protein extracts were subjected to immunoblotting as indicated. The quantification plot was based on scanning densitometry analysis and presented as mean ± SEM, n = 3 replicates. *p < 0.05, **p < 0.01, ***p < 0.001 versus control groups (Student’s t-test). C Knockout of FBXO22 down-regulates ubiquitination of BAG3. FBXO22 KO HEK 293T cells were generated by CRISPR/Cas9 assays. Cells were treated with 20 μM MG132 for 5 h. WCE were pulled down by Tandem Ubiquitin Binding Entity 1 (TUBE1) resin [59] for ubiquitinated proteins enrichment and immunoblotted as indicated. D Knockout of FBXO22 increases the stability of BAG3. Control or FBXO22 KO cells were treated with cycloheximide for the indicated time. Protein extracts were immunoblotted for the indicated proteins. The quantification plot was based on scanning densitometry analysis and relative protein levels were normalized to the control. Data are presented as mean ± SEM, n = 3 replicates. E Alignment of amino acids corresponding to the PXXP sequence with BAG3 orthologs and another FBXO22 substrate SNAI1. F Structural model of the protein–protein interaction between FBXO22 (blue) and S377 phosphorylated BAG3 (green). Phospho-S377 was the critical phosphorylation site in the interaction. G Immunoblot analysis of the interaction between BAG3 and the FBXO22 mutant. HEK 293 T cells were transfected with GFP-FBXO22 or GFP-FBXO22-R308A/H317A mutated plasmids for 36 h. WCE were immunoprecipitated by anti-GFP beads and immunoblotted as indicated. H BAG3 S377 mutation attenuates the interaction between FBXO22 and BAG3. HEK 293 T cells were transfected with Flag-BAG3 WT or Flag-BAG3 S377A mutant for 36 h. WCE were immunoprecipitated by anti-Flag M2 resin and immunoblotted as indicated. I BAG3 S377 mutation decreases ubiquitination of BAG3. HEK 293T cells were co-transfected with Myc-FBXO22, HA-ubiquitin, and Flag-BAG3 WT or Flag-BAG3 S377A mutant for 36 h. WCE were immunoprecipitated by anti-Flag M2 resin and immunoblotted as indicated. J Antibody specificity detection of anti-phospho-BAG3 (S377) antibody. HEK 293T cells were transfected with Flag-BAG3 WT or Flag-BAG3 S377A mutant for 36 h. WCE were immunoprecipitated by anti-Flag M2 resin and immunoblotted as indicated. K ERK inhibition decreases phosphorylation of BAG3 S377, increases protein levels of BAG3, and attenuates the interaction of BAG3 and FBXO22. HEK 293T cells were transfected with EV or Flag-BAG3 for 24 h and treated with ERK1/2 inhibitor SCH772984. WCE were immunoprecipitated by anti-Flag M2 resin and immunoblotted as indicated.
Interestingly, the protein kinase ERK could phosphorylate BAG3 [32]. Consistently, ERK was also present in the BAG3 interactome in our IP experiment (Table S3). We found that ERK was associated with both BAG3 and pBAG3 (S377), the latter was decreased in response to the ERKi SCH772984 treatment. Pharmacological inhibition of ERK by SCH772984 [33] resulted in decreased phosphorylation of ERK and BAG3 phospho-S377, an increase in total BAG3 protein, and the reduced interaction between BAG3 and FBXO22 (Fig. 4K and Fig. S4L). These data suggest ERK is responsible for BAG3 phosphorylation at S377 and the subsequent degradation of BAG3.
XXPpSPXPXX is a conserved phosphodegron recognized by FBXO22
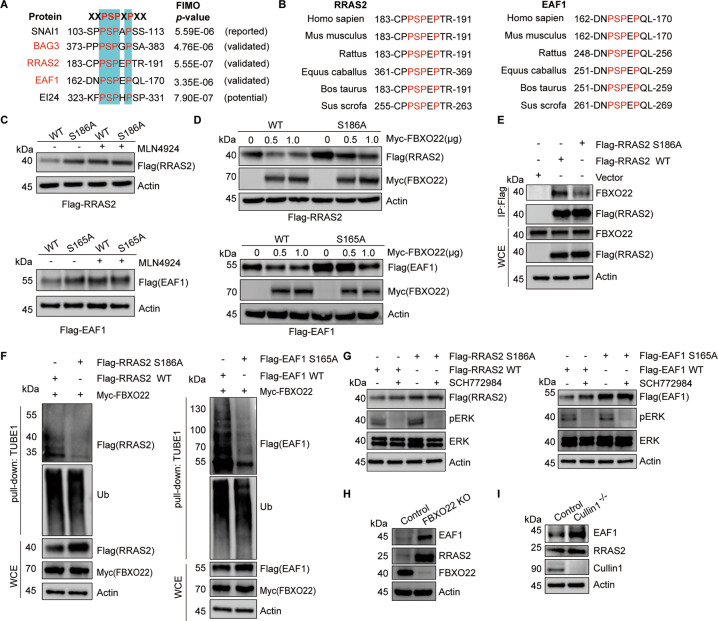
In order to examine whether XXPSPXPXX identified in BAG3 could be a conserved phosphodegron, we used FIMO software [34] to identify proteins bearing a similar motif in the human proteome. In this analysis, 1,345 short protein sequences on 1,026 proteins met the criterion of p-value ≤ 1.0e−05 according to the previous report [35]. We next compared these protein sequences with the upregulated phosphoproteome impacted by MLN4924 (Table S1). This analysis led to the identification of 31 sequences on 22 proteins matched to this putative phosphodegron, which could be potential FBXO22 substrates (Fig. 5A, Table S4). In addition to BAG3, many of these proteins were reported to play roles in cacogenesis, such as RRAS2 and EAF1, which were then chosen for further biochemical validation. Sequence alignment across different species revealed that this putative phosphodegron in these two proteins is highly conservative (Fig. 5B). Next, we mutated the corresponding phosphorylated serine sites (RRAS2 S186A, EAF1 S165A) to evaluate the impact on their degradation. The WT proteins, but not the mutants, were significantly accumulated after 4 h MLN4924 treatment, suggesting phosphorylation is required for sequential proteasomal degradation (Fig. 5C). Ectopic expression of FBXO22 resulted in a marked reduction of both exogenous WT RRAS2 and EAF1 proteins in a dose-dependent manner. In contrast, the protein levels of these mutants were unchanged in the presence of exogenous FBXO22 (Fig. 5D). In addition, the RRAS2 and EAF1 mutants could barely recognize FBXO22 (Fig. 5E and Fig. S5A). In line with it, FBXO22 can only affect the ubiquitination and stability of the WT RRAS2 and EAF1 proteins (Fig. 5F and Fig. S5B). Thus, these results suggest that the phosphorylation of this putative phosphodegron on both RRAS2 and EAF1 is required for FBXO22 recognition. As ERK has been reported to be able to phosphorylate RRAS2 at S186 [36], we then used ERKi to test whether suppressing the activity of ERK could prevent RRAS2 degradation. As expected, ERKi treatment caused the accumulation of RRAS2 WT protein, but not RRAS2 S186A. Similarly, the EAF1 WT protein, but not the EAF1 S165A, was accumulated in response to ERKi treatment (Fig. 5G). Like BAG3, the endogenous EAF1 and RRAS2 proteins, but not their mRNA levels, were elevated in FBXO22 KO cells (Fig. 5H and S5C). FBXO22 KO or cullin 1 KO also increased the stability of RRAS2 and EAF1 (Fig. S5D and Fig. 5I). Collectively, these results suggest that ERK-mediated phosphorylation of this putative phosphodegron is required for FBXO22 recognition and degradation.
Fig. 5. Validation of multiple substrates bearing XXPpSPXPXX as the phosphodegron for SCFFBXO22.
A Alignment of amino acids corresponding to the XXPSPXPXX sequence in BAG3, RRAS2, EAF1, MEN1, EI24 and SNAI1. B Alignment of amino acids corresponding to the XXPSPXPXX sequence across different species with RRAS2 and EAF1. C Immunoblot analysis of mutated phosphosites on protein stabilization. HEK 293T cells were transfected with the indicated Flag-tagged WT or the point mutated candidate substrates, and treated with 1 μM MLN4924 for 4 h before lysis. Lysates were subjected to immunoblotting as indicated. D Immunoblot analysis of indicated protein levels after co-transfected with the indicated Flag-tagged WT or the mutated plasmids and the indicated doses of Myc-FBXO22 plasmids. E HEK 293T cells were transfected with the indicated Flag-RRAS2 WT or the mutant plasmids. WCE were immunoprecipitated by anti-Flag M2 resin. Immunoblot analysis of WCE and IP as indicated. F FBXO22 regulates the ubiquitination of RRAS2 and EAF1. HEK 293T cells were transfected with Myc-FBXO22 and the indicated Flag-WT or the mutant plasmids for 36 h, and then treated with 20 μM MG132 for 5 h. WCE were pulled down by TUBE1 resin for ubiquitinated proteins enrichment and immunoblotted as indicated. G ERK inhibition increases protein levels of RRAS2 and EAF1. HEK 293T cells were transfected with indicated Flag-tagged WT or the mutate plasmids for 24 h and treated with SCH772984 for an additional 24 h. Protein extracts were subjected to immunoblotting as indicated. H Immunoblot analysis of RRAS2 and EAF1 protein levels in control and FBXO22-KO cells with indicated antibodies. I Immunoblot analysis of RRAS2 and EAF1 protein levels in control and cullin 1 KO cells with indicated antibodies.
Failure to degrade BAG3 promotes tumor growth
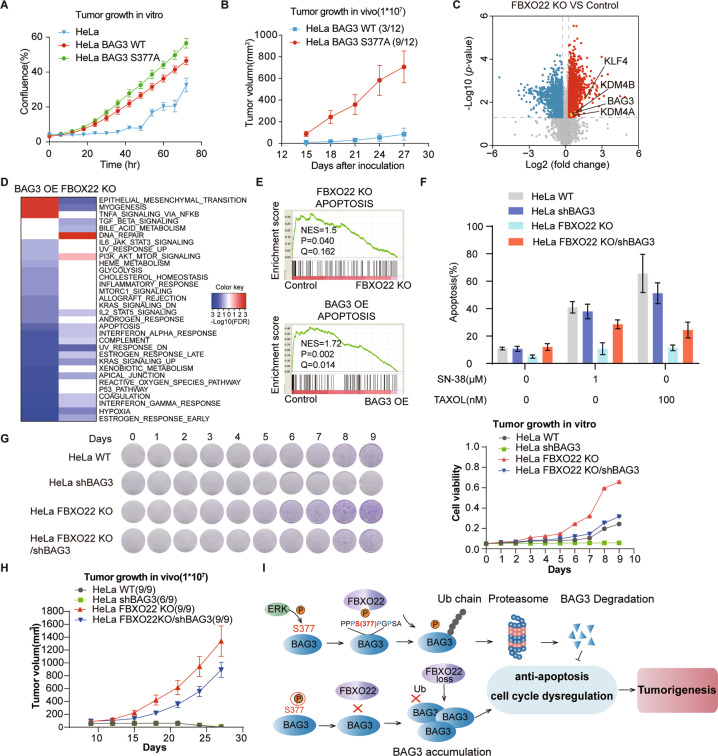
As aberrant expression of BAG3 is related to cancer development [37], we next investigated the possible role of ERK-FBXO22-BAG3 axis in tumorigenesis. HeLa cells stably expressing BAG3 WT exhibited an increased proliferative capacity relative to control cells, suggesting an oncogenic role for BAG3. Interestingly, cell proliferation was enhanced in cells expressing the BAG3 S377A mutant (Fig. 6A). Consistently, cells stably expressing BAG3 S377A possessed stronger cell migration ability (Fig. S6A). As many reported SCF substrates are regulated during cell cycle progression, we then asked whether this was also true for BAG3. We found that BAG3 protein levels fluctuated during the cell cycle, accumulated at G2/M, and declined after cells entered into G1 phase. However, protein levels of BAG3 S377A remained unchanged during the whole cell cycle (Fig. S6B). In cells expressing BAG3 S377A, there was a decreased percentage of G1 phase cells 4 h after nocodazole release and an increased percentage of S phase cells 12 h later (Fig. S6C). The function of BAG3 was further assessed in nude mice bearing subcutaneous human cervical cancer xenografts. We found that ectopic expression of the BAG3 S377A mutant significantly promoted tumor growth compared with the BAG3 WT control, as assessed by tumor size measurement (Fig. 6B, S6D).
Fig. 6. Failure to degrade BAG3 promotes tumor growth.
A BAG3 S377A mutation promotes tumor growth in vitro. HeLa cells stably expressing EV, BAG3 WT, or BAG3 S377A were cultured for 72 h, and the cell growth rate was measured by the IncuCyte Live-Cell Analysis System. Data are presented as mean ± SEM, n = 3 replicates. B BAG3 S377A mutation promotes tumor growth in vivo. HeLa cells stably expressing BAG3 WT or S377A were prone to tumorigenesis in nude mice. Each nude mouse was subcutaneously injected with 1 × 107 HeLa cells stably expressing BAG3 WT or BAG3 S377A for four weeks. Data are presented as mean ± SEM, n = 12 for each group. In the BAG3 WT group, the tumor formation rate was 3/12 (25%). In BAG3 S377A group, the tumor formation rate was 9/12 (75%). Tumor volume was measured every 3 days after mice were injected with cells. C The volcano plot illustrates the proteome changes between FBXO22 KO HeLa cells and control cells. Protein median Log2 fold-change (FBXO22 KO/ control) of two biological replicates was plotted against the -Log10 p-value (by t-test). Cutoffs of p = 0.05 and 1.2-fold change are marked by dotted lines. The reported substrates of FBXO22 quantified in our dataset (KLF4, KDM4B, and KDM4A) and BAG3 in this study are marked by yellow dots. D The heatmap shows the enriched hallmarks of the proteome changes between BAG3 OE and FBXO22 KO cells. The color is according to −Log10 transformed FDR q-value: red indicates pathways enriched in the treated (BAG3 OE or FBXO22 KO) group and blue indicates pathways enriched in the control group. The blank indicates not significantly enriched or not applicable (N/A). E Apoptosis enrichment plots in the BAG3 OE and FBXO22 depletion groups. F Statistical analysis of apoptosis assays. Data are presented as mean ± SEM, n = 3 replicates. G The colony formation assay for HeLa WT, HeLa shBAG3, HeLa FBXO22 KO, and HeLa FBXO22 KO/shBAG3 cells. Cells were grown in 6-well plates, cultured for the indicated days, and stained with crystal violet. Data are presented as mean ± SEM, n = 3 replicates. H BAG3 KD attenuates the effect of FBXO22 KO in promoting tumor growth in vivo. HeLa WT cells, HeLa shBAG3 cells, FBXO22 KO HeLa cells or FBXO22 KO/shBAG3 HeLa cells were prone to tumorigenesis in nude mice. Each nude mouse was subcutaneously injected with 1 × 107 cells for four weeks. Data are presented as mean ± SEM, n = 9 for each group. In the HeLa WT group, the tumor formation rate was 9/9. In the HeLa shBAG3 group, the tumor formation rate was 6/9. In the FBXO22 KO group, the tumor formation rate was 9/9. In the FBXO22 KO/shBAG3 group, the tumor formation rate was 9/9. Tumor volume was measured every 3 days after mice were injected with cells. I Proposed model for ERK-FBXO22-BAG3 axis-dependent physiological and pathological impacts.
We speculated that FBXO22 may have a reverse effect compared with BAG3. Indeed, in FBXO22 KO HeLa cells, we found that the proliferation and migration abilities were significantly enhanced, which was phenocopied in cells stably expressing BAG3 S377A (Fig. S6E-G). To further understand the FBXO22-BAG3 axis, we carried out quantitative proteomics to investigate the global impact of FBXO22 KO or BAG3 overexpression (OE) on the cellular proteome (Fig. S6H). Consistently, the protein level of BAG3 was increased in FBXO22 KO cells (Fig. 6C, Fig. S6I, and Table S5). Gene set enrichment analysis (GSEA) was conducted to compare significantly enriched hallmark pathways between the BAG3 OE proteome and FBXO22 KO proteome. Many pathways that were significantly enriched in the downregulated proteome of both BAG3 overexpressing cells and FBXO22 KO cells (Fig. 6D and Table S5). Notably, as BAG3 is well-recognized for its anti-apoptotic functions, the enrichment of the apoptosis signature in both the FBXO22 KO cells and BAG3 OE cells further suggested a pro-apoptotic function of FBXO22 via regulation of BAG3 stability (Fig. 6E and Table S5). Indeed, FBXO22 KO cells were significantly resistant to both spontaneous and drug-induced apoptosis, which could be largely restored by silencing BAG3 (Fig. 6F). BAG3 silencing in FBXO22 KO cells significantly decreased the cell proliferation and migration abilities relative to FBXO22 KO cells (Fig. 6G, Fig. S6J and Fig. S6K). In subcutaneous xenografts, knockout of FBXO22 significantly enhanced tumor growth, which was largely reversed by BAG3 depletion (Fig. 6H and Fig. S6L). It is worth noting that depletion of BAG3 in FBXO22 KO cells did not reduce the abundance of RRAS2 and EAF1 proteins, which might explain why BAG3 depletion cannot fully rescue the malignant phenotypes of FBXO22 KO cells (Fig. S6M).
In conclusion, we uncovered a critical role for the ERK-FBXO22-BAG3 axis in the regulation of tumorigenesis (Fig. 6I).
Discussion
To date, large-scale identification of phosphorylated substrates recognized by SCF has not been attempted. To address this question, we performed a simultaneous MLN4924-dependent quantitative proteome and phosphoproteome analysis, as MLN4924 treatment will result in global accumulation of phosphorylated SCF substrates that cannot be further ubiquitinated and degraded.
Using more stringent criteria, we further identified 54 potential SCF substrates, including 38 new ones. Interestingly, we found that 17% of these phosphorylated proteins were reported to be mutated in tumors, suggesting that these phosphorylation sites may result in the accumulation and activation of substrate proteins by inhibiting binding to the corresponding SCF, which eventually leads to tumorigenesis. In addition, phosphorylation of the substrates might also inhibit the binding to the F-box protein in some cases [38, 39].
As a proof of concept, we chose the newly validated SCF substrate BAG3 for further mechanistic investigation. We discovered that BAG3 is a substrate of SCFFBXO22. FBXO22 used its C-terminal to interact with BAG3, which required the prior phosphorylation of BAG3 by ERK at S377. Intriguingly, we found that expression of a stable S377A mutant BAG3, but not BAG3 WT, significantly promoted cancer cell growth and metastasis in vitro and tumorigenesis in vivo. Therefore, our study suggests that the upstream regulation of BAG3 by FBXO22 likely plays an essential role in tumorigenesis and metastasis repression. Indeed, loss of FBXO22-mediated degradation of Bach1 has been recently reported to stimulate lung cancer metastasis [40]. FBXO22 knockdown increased cell invasiveness via elevated levels of HDM2 [41], and lower expression of FBXO22 is associated with worse prognosis in ER-positive and HER2-negative breast cancer patients [42].
FBXO22 recognizes and degrades ERK-mediated phosphorylated proteins on their specific amino acid sequences might be a widely existed mechanism in mammalian cells. In addition to BAG3, sequence alignment and phosphoproteomic analysis revealed a variety of potential substrates bearing this new phosphodegron (Table S4). Among them, we further validated that RRAS2 and EAF1 proteins, which also contain the XXPpSPXPXX motif, can be phosphorylated by ERK and recognized by SCFFBXO22 for subsequent degradation. ERKi treatment caused the accumulation of these proteins, and a single amino acid mutation of this motif blocked ERK- and FBXO22-mediated proteasomal degradation.
Materials and methods
Cell lines
All cell lines used in this paper were from ATCC. HEK 293T, HEK 293T cells stably expressing BAG3, FBXO22 KO HEK 293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium containing 10% fetal bovine serum (FBS, 10091148, GIBCO) and 1% penicillin and streptomycin solution. HeLa cells, HeLa cells stably expressing BAG3, HeLa cells stably expressing BAG3 S377A, BAG3 KD HeLa cells, HeLa cells stably expressing FBXO22 and FBXO22 KO HeLa cells were cultured in DMEM medium containing 10% FBS and 1% penicillin and streptomycin solution. All cells were grown in 37 °C supplied with 5% CO2.
Xenograft models
All BALB/c nude mice were purchased from BEIJING HFK BIOSCIENCE Co. LTD and housed at specific facility. Cancer cells were implanted subcutaneously on the axilla of BALB/c nude mice (6–8-week old, female). The studies were conducted in compliance with the Institutional Animal Care and Use Committee of institutional ethical guidelines in Shanghai Institute of Materia Medica, Chinese Academy of Sciences. The mice were divided into groups randomly and 9 or 12 mice per group were required. The indicated cells were implanted into mice with DMEM medium. Tumor size was measured every three days. The size was calculated by the formula (length × width × width)/2.
Reagents
Where specified, the following reagents were added: SCH772984 (HY-50846, MedChem Express Company), MLN4924 (HY-70062, MedChem Express Company), SN-38 (Selleck S4908), Paclitaxel (TAXOL, Selleck S1150), Cycloheximide (HY-12320, MedChem Express Company), Lipofectamine 2000 Transfection Reagent (11668019, Thermo fisher Scientific), TiO2 (Titanium Dioxide, 5020–75000, GL Sciences), Modified sequencing grade trypsin (HLS TRY001N, HuaLishi Scientific), rLys-C (HLS LYS001C, HuaLishi Scientific).
Anti-BAG3 (Ab47124, Abcam), anti-phospho-BAG3 (S377, Shanghai Immune Biotech, Ltd., Shanghai, China, this paper), anti-Phospho-(Ser/Thr) Phe (Ab17464, Abcam), anti-HA(Sigma-Aldrich, H6908), anti-Flag M2 (Sigma-Aldrich, A2220), anti-FBXO22 (Proteintech, 13606-1-AP), anti-Cyclin B1 (Cell Signaling Technology, 4138S), anti-Myc (Cell Signaling Technology, 2276), anti-β-Actin (Cell Signaling Technology, 5125 s), anti-Phospho-p44/42 MAPK (Erk1/2) (Cell Signaling Technology, 4370), anti-p44/42 MAPK (Erk1/2) (Cell Signaling Technology, 4695S), anti-GFP, (Beyotime, AG281), anti-RRAS2 (Proteintech, 12530-1-AP), anti-EAF1 (Proteintech, 13787-1-AP), anti-Cullin 1 (Santa Cruz, sc-17775), Agarose-TUBE1(LifeSensors, UM401), anti-GFP beads (AlpaLife, KTSM1301).
SILAC based cell culture
For SILAC labeling, HEK 293T cells were labeled in DMEM medium with 12C6 lysine and 12C614N4 arginine (light group) and DMEM medium with 13C6 lysine and 13C615N4 arginine (heavy group), containing 2 mM L-glutamine, 1% penicillin and streptomycin and 10% dialyzed FBS (04-011-1A, Biological Industry). The labeling efficiency for the heavy group was determined by LC-MS/MS analysis before the proteomics study. When the labeling efficacy for heavy group was above 95%, the light group was treated with 1.0 μM MLN4924 for 4 h and the heavy group was treated with DMSO for 4 h. After treatment, cells were washed with chilled Dulbecco’s phosphate buffer saline (D-PBS). Both the light and heavy cells were harvested and lysed with lysis buffer (8 M urea in 50 mM NH4HCO3, supplemented with protease inhibitor cocktail and phosphatase inhibitor) on ice.
Sample preparation for mass spectrometry analysis
For protein profiling and phosphoproteomics analysis, the cell lysate was sonicated and centrifuged at 4 °C. The concentrations of supernatant were determined using BCA assay (P0009, Beyotime). Heavy and light labeled cell lysates were mixed at equal amount, and then the protein mixture was reduced by dithiothreitol at 56 °C for 30 min and alkylated by iodoacetamide in dark at room temperature for another 30 min. The alkylation reaction was quenched by cysteine at room temperature for another 30 min. rLys-C was added at enzyme-to-protein ratio of 1:100 (w/w) for digestion at 37 °C for 3 h. Then the sample solution was subjected to a fourfold dilution with 50 mM NH4HCO3 and sequencing grade trypsin was added at an enzyme-to-protein ratio of 1:50 (w/w) for digestion at 37 °C for 16 h. The peptide solution was desalted with SepPak C18 cartridges. Peptides for protein profile analysis were fractionated by high pH reverse phase HPLC using a Waters X-Bridge® BEH C18 column (130 Å, 3.5 μM particles, 4.6 × 100 mm, Waters). Then the separated peptide solutions were combined into 15 fractions and vacuum-dried for further analysis. Two biological replicate analyses were carried out.
Peptides for phosphoproteome analysis were enriched using TiO2 beads based on a reported protocol [43]. In brief, peptides were dissolved in loading buffer (6% TFA in 80% ACN, supplemented with 1 M lactic acid), and then incubated with TiO2 beads for 30 min at room temperature. After incubation, TiO2 beads were then loaded into a StageTip with C18 membrane plug, then washed with loading buffer and wash buffer (0.5% TFA in 50% ACN) for six times. The phosphopeptides were eluted from the beads with elution buffer (10% NH3H2O). Eluted phosphopeptides were fractionated into 6 fractions using an in-house made C18 StageTip and vacuum-dried for further analysis. Two biological replicate analyses were carried out.
For immunoprecipitation-mass spectrometry (IP-MS) analysis, samples were separated by SDS-PAGE and visualized by Coomassie Brilliant Blue (CBB) staining. The target gels lane were cut off, destained with 50% ethanol, and subjected to in-gel digestion as previously described [44]. In brief, gels were cut into slices and dehydrated with ACN. Dehydrated gel slices were reduced by dithiothreitol, alkylated by iodoacetamide, and washed by 100 mM NH4HCO3. Then gel slices were digested with sequencing grade trypsin at 37 °C overnight. After tryptic digestion, peptides were extracted from gel slices and vacuum-dried for further analysis.
Nano-HPLC−MS/MS analysis
Peptide separation was performed using an in-house made reverse phase C18 analytical column (75 μM ID × 25 cm length, 3 μM particle size) with a linear gradient of 5−31% solvent B (0.1% FA in 90% ACN) for 74 min and 31−80% solvent B for 16 min at a constant flow rate of 300 nL/min on an EASY-nLC 1000 system (Thermo Fisher Scientific, Waltham, MA). The eluted peptides were ionized and introduced to Obitrap Fusion (Thermo Fisher Scientific, Waltham, MA) using NSI source. Ions with m/z 300−1,400 were detected in the Orbitrap at a resolution of 120,000 at m/z 200. The automatic gain control (AGC) was set as 1 × 106. Data-dependent tandem MS analysis was performed using a top speed approach (cycle time of 3 s). The intensity threshold for fragmentation was set to 5,000. The precursor ions were isolated and sequentially fragmentized by higher collision dissociation (HCD) with a normalized collision energy of 32%, and then ion fragments were detected in the ion trap. The dynamic exclusion duration was set as 65 s, and the isolation window was 1 m/z. AGC with 5 × 103 was used to prevent overfilling of the ion trap.
Plasmids construction
The cDNAs encoding BAG3, FBXO22, DTL, PRKAR1A, UFD1, RRAS2, and EAF1 were PCR-amplified from a HEK 293T cell cDNA library. Full-length cDNAs were constructed by cloning the open reading frame of those genes into pBabe-Flag vector, pcDNA-Myc-BioID (Addgene, Cambridge, MA) or pEGFP-C1 vector by using standard procedures. The open reading frame of DN-CUL1, DN-CUL2, DN-CUL3, DN-CUL4a, DN-CUL4b, and DN-CUL5 were PCR-amplified from cDNA library either, then subcloned into pEGFP-C1 vector. Site-specific mutation of BAG3, UFD1, PRKAR1A, RRAS2, and EAF1 was introduced using PCR amplification with PrimeSTAR GXL DNA Polymerase. All constructs were confirmed by sequencing. The primer sequences were in Table S6.
Immunoblotting analysis
For Immunoblot analysis, cells were lysed with 2× loading buffer (100 mM Tris-Cl, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mM DTT) on ice for 30 min. The lysate was boiled at 99 °C for 5 min, and then cooled on ice for another 5 min. The lysate was separated in 12% polyacrylamide gel and then transferred to 0.45 μM nitrocellulose membrane. The membrane was blocked in 5% BSA in PBS buffer with 0.2% tween-20 (PBST) for 1 h at room temperature and immunoblotted with appropriate antibodies at 4 °C overnight. Washing the membrane for three times with PBST buffer and incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature. Washing the membrane for three times with PBST buffer, then the membranes were imaged by chemiluminescence (Clinx, chemiscope 6000). The relative band intensity was quantified by software Image J (Version 1.8.0).
Knock-out cell model construction
The knock-out cell model was obtained by CRISPR/Cas9 technology. In briefly, designed single-stranded DNA oligonucleotides targeted to the coding sites of the gene according to the CCTOP website (https://crispr.cos.uni-heidelberg.de/). Add the CACCG in the front of the forward primer if the base of the gDNA is not G, otherwise, add the CACC instead, and the reverse primer was the complement sequence (FBXO22: forward primer: CACCGGGTCACAATCCCAAGGACT, reverse primer: AAACAGTCCTTGGGATTGTGACCC. Cullin 1: forward primer: CACCGTGCCTACCTCAATAGACAT, reverse primer: AAACATGTCTATTGAGGTAGGCAC). Anneal the two-strand oligonucleotides to generate a double-stranded oligonucleotide (ds oligonucleotide), and then cloned into the vector PX459 (Addgene, Cambridge, MA). Cells were applied to transfected with 2 μg of sgRNA plasmid by Lipofectamine 2000 according to transfection protocol. After 48 h, cells were transferred into 96-well plates to obtain individual clones. The genomic fragments containing the target nucleotide sequence in the center were PCR-amplified and sequenced. The protein depletion was identified by Western blot.
Immunoprecipitation
For IP experiments, cells were lysed with IP buffer (100 mM NaCl, 20 mM Tris-cl pH 8.0, 0.5 mM EDTA, 0.5% (v/v) Nonidet P-40) with protease inhibitor cocktail and phosphorylate inhibitor for 30 min on ice. Sonicated the cell lysate with 30% power for 5 min, then the lysates were centrifuged at 21,300 × g for 5 min. The supernatant was transferred to a new tube. 50 μL samples were taken as the whole cell lysate. The residual supernatant was incubated with appropriate antibody beads for 16 h at 4 °C in a rotating wheel. Immunocomplexes were washed for seven times with IP buffer. Proteins were eluted by heating at 99 °C for 5 min in 2× loading buffer, and then run the SDS-PAGE for immunoblot with appropriate antibodies. For LC-MS analysis, immunocomplexes were washed for seven times with IP buffer, and then the flag peptides solution was separated with SDS-PAGE and stained with CBB. The target gels were cut off for the following LC-MS analysis.
Structure modeling and protein docking
Protein structure modeling was carried out with the I-TASSER webserver [31]. Full-length sequences of BAG3 and FBXO22 were uploaded onto the webserver and the modeling was run with default settings. Five structure clusters were obtained as results for each protein and the final models were selected according to the overall conformations and the C-scores calculated by the server. Phosphorylation of S377 in BAG3 was introduced through the Discovery Studio package. After that, the predicted BAG3 (phospho-S377) structure was docked towards FBXO22 using the Haddock2.2 webserver [45], with the phospho-S377 residue as an active residue directly involved in the protein–protein interaction. Haddock retrieved 171 complex models and clustered them into three structural ensembles. The best structure within the top cluster based on the Haddock calculations was chosen as our final result.
Tandem Mass Tag labeling based quantitative proteomics
Cells were harvested and lysed with lysis buffer (8 M urea in 50 mM NH4HCO3, supplemented with protease inhibitor cocktail). Cell lysates were reduced, alkylated and tryptic digested into peptides as previously described. Samples were desalted using SepPak C18 cartridges (Waters, Milford, MA). The desalted sample was subjected to Tandem Mass Tag (TMT) labeling following the manufacturer’s protocol. The labeling efficiency of labeled peptides determined by LC-MS/MS was >98%. Then peptides labeled with different tags were mixed at equal amounts. The peptide mixture was fractionated into 20 fractions by high pH reverse phase HPLC using a Waters X-Bridge® BEH C18 column (130 Å pore size, 3.5 μM particle size, 4.6 × 250 mm, Waters). The sample was analyzed by Nano-HPLC−MS/MS.
Cell proliferation assay
We used two methods for the cell proliferation assay. For IncuCyte system analysis, cells were seeded in 12-well plates at a density of 5,000 cells per well for overnight and then change fresh growth medium. Subsequently, the cell culture plates were placed into the live-cell analysis system IncuCyte (Essen BioScience Ltd., Hertfordshire, UK), and images were acquired every 6 h by automated real-time assessment for 72 h. Growth curves were plotted as the change in confluence percentage. For Cell Counting Kit-8 (CCK-8) assay, 3,000 Cells were grown in 96-well plates and incubated for 1–7 days. Cell viability was measured with CCK-8 (K04, DOJINDO) according to the manufacturer’s protocol. At each time, 10 μL CCK-8 was added into each well and incubated at 37 °C for 2 h. The optical density (OD) value was measured with Microplate Reader (Biotek) at 450 nm. The cell viability was calculated with three replicates.
Protein stability assay
Cells were grown in six-well plates (2 × 105 cells/well) in DMEM medium with 10% FBS, and then transfected with indicated plasmid and 5 µl lipofectamine 2000 for transient transfection following the manufacturer’s protocol. After 24 h, cells were treated with 25 μg/mL of cycloheximide (CHX) and harvested at the indicated times for immunoblot analysis with the appropriated antibodies.
Lentivirus infection for stable cell lines
Cells were seeded in six-well plates (2 × 105 cells/well) containing DMEM supplement with 10% FBS. Then cells were transfected with 1 µg target plasmid and lentivirus expression plasmid, packaging plasmid (including 0.5 µg gag/pol and 0.5 µg VSVG) using lipofectamine 2000. Forty-eight hours later, the culture supernatant was harvested, and added to the medium of cultured cells with a ratio of 1:1 (v/v) including polybrene (1 μg/ml) for 24 h infection. Single-cell clones were selected in the presence of 1.5 μg/ml puromycin for 2–4 weeks in 96-well plates. Protein overexpression was identified by Western blot.
Quantitative real-time PCR analysis
RNA was extracted from cells with RNAprep pure Cell/Bacteria Kit (DP430, Tiangen). cDNA was reverse transcribed with PrimeScript RT Master Mix (RR036A, TaKaRa). The DNA was amplificated with PCR instrument (A600). The BAG3 primers (Forward: 5′-ACCCTTTCCATGTCTATCCC-3′, Reverse: 5′-CGTTCTGCTCGTGTATCACC-3′). The RRAS2 primers (Forward: 5′-AGGAGAAGTACCGGCTCGT-3′, Reverse: 5′-CGCCAGTCCTCATATACTGTTC-3′). The EAF1 primers (Forward: 5′-AGGAGAGCTTCAAGTTGGCAA-3′, Reverse: 5′-TCTGGTAAGGCCGTTTGTTCC-3′). Quantitative real-time PCR (RT-qPCR) was performed according to the instruction of SYBR Premix ExTaq II (RR820Q, TaKaRa) with ABI7300 PCR system (Thermo Scientific). The relative quantification results were calculated using the ▵▵CT method. β-actin was used as the normalization gene.
Knock-down cell model construction
The BAG3 shRNA were synthesized (Forward primer: GATCGCAAAGAGGTGGATTCTAAACTCAAGAGGTTTAGAATCCACCTCTTTGCTTTTTG, Reverse primer: AATTCAAAAAGCAAAGAGGTGGATTCTAAACCTCTTGAGTTTAGAATCCACCTCTTTGC) and constructed into the Plvx-shRNA1 vector. Then cells were seeded into six-well plates and transfected with 2 μg of shRNA plasmid by Lipofectamine 2000 according to transfection protocol. After 48 h, cells were planted into 96-well plates and cultured with 1.5 μg/ml puromycin to obtain individual clones. The protein expression was identified by western blot using BAG3 antibody.
Cell apoptosis assay
Cells were planted into six-well plates and treated with the indicated concentrations of SN-38 and Paclitaxel for 48 h. Then the cells were harvested and washed in cold D-PBS for three times. Cells were resuspended in 100 μl of 1× binding buffer with 5 μl of Annexin V-FITC and 5 μl of propidium iodide (PI) staining solution (A211-02, Vazyme) and incubated for 10 min at room temperature. After incubation, the cell suspension was added with 400 μl of 1× binding buffer and analyzed in FACSCalibur (BD). The data was processed by the software FlowJo. The statistical analysis was analyzed with three replicates.
Colony formation assay
Cells were grown in 12-well plates at a density of 3,000 cells and incubated for 10 days. The culture medium was refreshed every three days. At each time point, cells were fixed with 4% paraformaldehyde for 30 min and washed with D-PBS, then stained with 0.1% crystal violet for 10 min. The cell colonies were photographed with microscope (Olympus). The experiment was repeated for three times. For the quantification of colonies, 30% sodium acetate was added to test the crystal violet OD value for each well. The statistical analysis was analyzed with three replicates.
Cell migration assay
For migration assay, cells were added to 24-well Millicell hanging inserts (Millipore, MCEP24H48) and cultured for 36 h or 48 h in DMEM medium not including FBS. The lower champers used the DMEM medium including FBS. At the right time, the insert champers were fixed with 4% paraformaldehyde and washed with D-PBS, then stained with 0.1% crystal violet. The cells in the inner wall were erased by cotton swab gently. The migrated cells were photographed with microscope.
Cell cycle assay
Cells were seeded into 6-well plates at a density of 5 × 105 cells/well. For G1/S synchronization, cells were blocked by 2 mM thymidine for 12 h, and then cells were washed twice with cold D-PBS. Cells were incubated with 100 ng/mL nocodazole for another 12 h, then washed twice with cold D-PBS and cultured in fresh DMEM medium and harvested at indicated times. The cells were harvested at the indicated time and fixed with ice-cold 70% ethanol. The fixed cells were washed with D-PBS and stained with 50 μg/ml propidium iodide (PI) for 10 min at 37 °C. The cell cycle proportions were measured through FACSCalibur flow cytometer (BD). The cell cycle distribution was analyzed by the software FlowJo.
Quantitative proteomic and phosphoproteomic data analysis
All the acquired MS/MS data files (.raw) were processed with MaxQuant (v1.5.3.8) based on Andromeda search engine [46]. Homo sapiens database from UniProt (update on 08/27/2018) with a reversed decoy database was used for data processing. For SILAC based proteomics analysis, Carbamidomethyl (C) was specified as the fixed modification and variable modifications were oxidation (M) and acetylation (Protein N-term). For SILAC based phosphoproteomics analysis, Carbamidomethyl (C) was specified as the fixed modification and variable modifications were oxidation (M), acetylation (Protein N-term), and phosphorylation (S/T/Y). Other MaxQuant search parameters were the default settings. SILAC ratios for all proteins and phosphorylation peptides were normalized using DJ-1 [47], β-Tubulin, and GAPDH protein ratios [48]. Statistical p-value was calculated in Excel using the paired student t-test. P-value < 0.05 and fold-change > 1.2 was considered as significant change. Besides, proteins and phosphopeptides that were only identified in SILAC light group or SILAC heavy group in two replicates were also considered as significantly changed proteins and phosphopeptides.
For TMT-based proteomics analysis, variable modifications were oxidation (M) and acetylation (Protein N-term). The TMT label on lysine residue and peptide N-termini (+229.163 Da) and Carbamidomethyl (C) were fixed modifications. Other MaxQuant search parameters were the default settings. To quantify the up- and downregulated proteins, the reporter ion intensity was normalized by median-centering, and the ratio (treat/control) was calculated by compare reporter ion intensity from different groups. Statistical p-value was calculated using the paired student t-test. P-value < 0.05 and fold-change > 1.2 was consider as significant change.
IP-MS data analysis
Acquired IP-MS raw data were processed with Mascot software (version 2.3.01, Matrix Science Ltd., London, UK). Homo sapiens database from UniProt (update on 08/27/2018) was used for data processing. Search parameters were specified as follows: mass error was ±10 ppm for parent ions and ±0.5 Da for fragment ions. The enzyme was specified as trypsin with a maximum of two missing cleavages. Carbamidomethyl (C) was specified as the fixed modification and oxidation (M) was specified as variable modification. Peptide ion score and protein score cutoff were 20. For BAG3 protein modification analysis, acetylation (Protein N-term), acetylation (K), mono-methylation (K/R), GlyGly (K), and phosphorylation (S/T/Y) were additionally specified as variable modifications.
Bioinformatics analysis and data integration
DAVID bioinformatics functional annotation tool was used to identify enriched GO terms and KEGG pathway [49]. The significance of fold enrichment was calculated using a Benjamini adjust p-value < 0.05. KEA2 (http://www.maayanlab.net/KEA2) was used to predict kinase activity [50]. MLN4924 upregulated phosphosites were used as input. Enrichment analysis was performed using Literature-Based Kinase-Substrate Library with Phosphosites option. A p-value of 0.05 is used as the cutoff for significance. Motif analysis was carried out using MoMo software tool [51] and visualized using WebLogo [52]. Protein interaction network was analyzed by STRING (v11.0) database [53] and visualized by Cytoscape (v 3.7.2) [54]. Gene set enrichment analysis (GSEA, v4.0.0) [55] was performed using gene set collections “H” (hallmark gene sets) from MSigDB as bioinformatics functional tool. FDR q-value of 0.25 and p-value of 0.05 was used as enrichment cutoff. FIMO (v 5.1.1) was used to find protein sequences with motifs described as position-specific scoring matrices [34]. FIMO is part of the MEME Suite software toolkit.
Protein–protein interaction data were obtained from BioGRID database (release 3.4.163) [56]. Physical interactome data from both low-throughput and high-throughput experiment was downloaded. “Regulatory sites” from PhosphoSitePlus (updated 14/12/2017) were downloaded for annotation of the known molecular function of phosphosites [57]. Mutation data from COSMIC (release v87) was used to discover mutated phosphosites [58].
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 5.0.0). All data were presented as mean ± SEM. Unpaired two-tailed Student’s t-test with equal variance was performed. Statistical details of each experiment can be found in the figure legends. Significances are indicated with asterisks: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, and not significant (ns) p > 0.05.
Supplementary information
Author contributions
Conceptualization, MT and BL; Methodology, PL, XC, SL, and XJ; Formal Analysis, XC, LZ, DN, and ZL; Investigation, PL, XC, SL, XJ, WD, LZ, XW, YC, JJ, WL, and LP; Resources, MT, BL, MH, and JZ; Data Curation, PL, XC, SL, and XJ; Writing, MT, BL, PL, and XC; Visualization, PL, XC, SL, WD, and XJ; Supervision, MT and BL; Funding Acquisition, MT, BL, WD, and JZ; All authors reviewed and approved the manuscript.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplementary materials. Mass spectrometry data have been deposited to the iProX (84), a full member of the ProteomeXchange consortium (http://www.iprox.cn), accession ID: IPX0001809000.
Ethics statement
Our studies did not include human participants, human data, or human tissue. The animal studies were conducted in compliance with the Institutional Animal Care and Use Committee of institutional ethical guidelines in Shanghai Institute of Materia Medica, Chinese Academy of Sciences (2019-03-GMY-10).
Funding statement
This work was supported by the Natural Science Foundation of China (Nos. 32071432, 81872888, 81821005, 91753203, 81773018, 71473074), National Key R&D Program of China (No. 2020YFE020220), the National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (No. 2018ZX09711002-004), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA12050406), the Shanghai Science and Technology Committee (No. 19JC1416300), Innovative Research Team of High-Level Local Universities in Shanghai (SSMU-ZDCX20181202) and the K. C. Wong Education Foundation.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by A. Degterev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ping Liu, Xiaoji Cong, Shengjie Liao, Xinglong Jia.
Contributor Information
Bin Liu, Email: liubin@jou.edu.cn.
Minjia Tan, Email: mjtan@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00827-7.
References
- 1.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 2.Jang HH. Regulation of protein degradation by proteasomes in cancer. J Cancer Prev. 2018;23:153–61. doi: 10.15430/JCP.2018.23.4.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–5. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 4.Lucas X, Ciulli A. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr Opin Struct Biol. 2017;44:101–10. doi: 10.1016/j.sbi.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–51. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 7.Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–30. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 8.Reitsma JM, Liu X, Reichermeier KM, Moradian A, Sweredoski MJ, Hess S, et al. Composition and regulation of the cellular repertoire of SCF ubiquitin ligases. Cell. 2017;171:1326–39.e1314. doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–19. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 10.Ang XL, Wade Harper J. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–70. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- 11.Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, et al. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–74. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye X, Nalepa G, Welcker M, Kessler BM, Spooner E, Qin J, et al. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem. 2004;279:50110–9. doi: 10.1074/jbc.M409226200. [DOI] [PubMed] [Google Scholar]
- 13.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–56. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 14.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–74. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Jiang S, Li M, Xiong X, Zhu M, Li D, et al. Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN. Nat Commun. 2018;9:4770. doi: 10.1038/s41467-018-07185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan MK, Lim HJ, Bennett EJ, Shi Y, Harper JW. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol Cell. 2013;52:9–24. doi: 10.1016/j.molcel.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TY, Siesser PF, Rossman KL, Goldfarb D, Mackinnon K, Yan F, et al. Substrate trapping proteomics reveals targets of the betaTrCP2/FBXW11 ubiquitin ligase. Mol Cell Biol. 2015;35:167–81. doi: 10.1128/MCB.00857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 20.Behl C, Breaking BAG. The co-chaperone BAG3 in health and disease. Trends Pharm Sci. 2016;37:672–88. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 21.De Marco M, Basile A, Iorio V, Festa M, Falco A, Ranieri B, et al. Role of BAG3 in cancer progression: a therapeutic opportunity. Semin Cell Dev Biol. 2018;78:85–92. doi: 10.1016/j.semcdb.2017.08.049. [DOI] [PubMed] [Google Scholar]
- 22.Meister-Broekema M, Freilich R, Jagadeesan C, Rauch JN, Bengoechea R, Motley WW, et al. Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks. Nat Commun. 2018;9:5342. doi: 10.1038/s41467-018-07718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–81. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 25.Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell. 2016;166:1198–214.e1124. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yalla K, Elliott C, Day JP, Findlay J, Barratt S, Hughes ZA, et al. FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system. Mol Psychiatry. 2018;23:1278–86. doi: 10.1038/mp.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 28.Biswas M, Phan D, Watanabe M, Chan JY. The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J Biol Chem. 2011;286:39282–9. doi: 10.1074/jbc.M111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hishiya A, Kitazawa T, Takayama S. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ Res. 2010;107:1220–31. doi: 10.1161/CIRCRESAHA.110.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R, Xie HY, Qian JX, Huang YN, Yang F, Zhang FL, et al. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018;78:5274–86. doi: 10.1158/0008-5472.CAN-17-3647. [DOI] [PubMed] [Google Scholar]
- 31.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue L, Wang P, Cao P, Zhu JK, Tao WA. Identification of extracellular signal-regulated kinase 1 (ERK1) direct substrates using stable isotope labeled kinase assay-linked phosphoproteomics. Mol Cell Proteom. 2014;13:3199–210. doi: 10.1074/mcp.O114.038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–35. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Low TY, Peng M, Magliozzi R, Mohammed S, Guardavaccaro D, Heck AJ. A systems-wide screen identifies substrates of the SCFbetaTrCP ubiquitin ligase. Sci Signal. 2014;7:rs8. doi: 10.1126/scisignal.2005882. [DOI] [PubMed] [Google Scholar]
- 36.Fremin C, Guegan JP, Plutoni C, Mahaffey J, Philips MR, Emery G, et al. ERK1/2-induced phosphorylation of R-Ras GTPases stimulates their oncogenic potential. Oncogene. 2016;35:5692–8. doi: 10.1038/onc.2016.122. [DOI] [PubMed] [Google Scholar]
- 37.Huang X, Yan J, Zhang M, Wang YF, Chen Y, Fu XH, et al. Targeting epigenetic crosstalk as a therapeutic strategy for EZH2-Aberrant solid tumors. Cell. 2018;175:186−+. doi: 10.1016/j.cell.2018.08.058. [DOI] [PubMed] [Google Scholar]
- 38.Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, et al. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472–80. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong YT, Simoneschi D, Keegan S, Melville D, Adler NS, Saraf A, et al. The ULK1-FBXW5-SEC23B nexus controls autophagy. Elife. 2018;7:e42253. [DOI] [PMC free article] [PubMed]
- 40.Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of bach1. Cell. 2019;178:316–29.e318. doi: 10.1016/j.cell.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai J, Wu K, Cao MH, Yang Y, Pan Y, Liu H, et al. SCF(FBXO22) targets HDM2 for degradation and modulates breast cancer cell invasion and metastasis. Proc Natl Acad Sci USA. 2019;116:11754–63. doi: 10.1073/pnas.1820990116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johmura Y, Maeda I, Suzuki N, Wu W, Goda A, Morita M, et al. Fbxo22-mediated KDM4B degradation determines selective estrogen receptor modulator activity in breast cancer. J Clin Invest. 2018;128:5603–19. doi: 10.1172/JCI121679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–35. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 44.Nie L, Shuai L, Zhu M, Liu P, Xie ZF, Jiang S, et al. The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Mol Cell Proteom. 2017;16:1324–34. doi: 10.1074/mcp.M117.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Zundert GCP, Rodrigues J, Trellet M, Schmitz C, Kastritis PL, Karaca E, et al. The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428:720–5. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Wisniewski JR, Mann M. A proteomics approach to the protein normalization problem: selection of unvarying proteins for MS-based proteomics and western blotting. J Proteome Res. 2016;15:2321–6. doi: 10.1021/acs.jproteome.6b00403. [DOI] [PubMed] [Google Scholar]
- 48.Larance M, Ahmad Y, Kirkwood KJ, Ly T, Lamond AI. Global subcellular characterization of protein degradation using quantitative proteomics. Mol Cell Proteom. 2013;12:638–50. doi: 10.1074/mcp.M112.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Lachmann A, Ma’ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–6. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng A, Grant CE, Noble WS, Bailey TL. MoMo: discovery of statistically significant post-translational modification motifs. Bioinformatics. 2019;35:2774–82. doi: 10.1093/bioinformatics/bty1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70. [DOI] [PMC free article] [PubMed]
- 58.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–8. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplementary materials. Mass spectrometry data have been deposited to the iProX (84), a full member of the ProteomeXchange consortium (http://www.iprox.cn), accession ID: IPX0001809000.