Abstract
Colorectal cancer (CRC) is the third most diagnosed cancer and the second leading cause of cancer-related deaths. However, there are few effective therapeutic targets for CRC patients. Here, we found that CDK15 was highly expressed in human CRC and negatively correlated with patient prognosis and overall survival in tissue microarray. Knockdown of CDK15 suppressed cell proliferation and anchorage-independent growth of CRC cells and inhibited tumor growth in cell line-derived xenograft (CDX) model. Importantly, knockout of CDK15 in mice retarded AOM/DSS-induced tumorigenesis and CDK15 silencing by lentivirus significantly suppressed tumor progression in patient-derived xenograft (PDX) model. Mechanistically, CDK15 could bind PAK4 and phosphorylate PAK4 at S291 site. Phosphorylation of PAK4 at the S291 residue promoted cell proliferation and anchorage-independent growth through β-catenin/c-Myc, MEK/ERK signaling pathway in CRC. Moreover, inhibition of PAK4 reversed the tumorigenic function of CDK15 in CRC cells and pharmacological targeting PAK4 suppressed tumor growth in PDX models. Thus, our data reveal the pivotal role of CDK15 in CRC progression and demonstrate CDK15 promotes CRC tumorigenesis by phosphorylating PAK4. Hence, the CDK15-PAK4 axis may serve as a novel therapeutic target for CRC.
Subject terms: Cancer models, Oncogenes
Introduction
Colorectal cancer (CRC) is a leading cause of cancer-related deaths worldwide [1]. CRC incidence and death rate have continued to rise in adults under 50 in recent years [2]. However, only few available therapeutic targets are effective in a subset of patients due to molecular heterogeneity and acquired resistance [3]. There remains an unmet need to exploit oncogenic divers and identify novel effective therapeutic targets for CRC.
Cell-cycle regulation resides at the core of tumor initiation and progression. CDKs (cyclin-dependent kinases) are critical regulators of the cell cycle and closely associated with cancer. CDKs, comprising CDK 1–20, involved in numerous critical cellular processes including cell cycle (CDK 1–4, 6), transcription (CDK 7–10), translation (CDK 1, 11), and mRNA processing (CDK 10–13) as well as differentiation of neurons (CDK5) [4]. Dysregulation of CDKs promotes the occurrence and development of tumors. Several CDK inhibitors have been developed and achieved good clinical efficacy [5, 6]. Meanwhile, the newly proposed family members PFTAIRE (CDK14–CDK15) and PCTAIRE (CDK16–CDK18) have attracted increasing attention as they have been shown to promote progression of various cancers including colon cancer [7–9]. CDK15 is a highly conserved homologous gene present in PFTAIRE and PCTAIRE family [10] and closely associated with cancer [11, 12]. However, the study of CDK15 in CRC cancer is few and unclear, thus, the role of CDK15 and potential of CDK15 as therapeutic target in CRC is worthy of further investigation.
Herein, we report that CDK15 is a critical oncogenic factor capable of predicting adverse CRC prognosis. CDK15 promotes proliferation and colony formation of cells in vitro; CDK15 knockout suppresses AOM/DSS-induced tumorigenesis in mice and CDK15 silencing by lentivirus inhibits tumor growth in patient-derived xenograft (PDX) models. Mechanistically, CDK15 exerts its oncogenic function by binding PAK4 and phosphorylating PAK4 at the S291 site, which subsequently is demonstrated to play an important role in CRC. Taken together, this study systematically demonstrates that CDK15 promotes CRC tumorigenesis through phosphorylating PAK4 at the S291 residue, and provides a novel therapeutic target for CRC.
Methods
Reagents and antibodies
PF-3758309 (purity ≥ 98% from HPLC and NMR analysis) was purchased from Shanghai Rechem Science Co., Ltd. (Shanghai, China). The following antibodies were used in our study: anti-CDK15 (Cat#PA5-28595, Invitrogen), anti-CDK15 (Cat#TA811934, ORIGENE), anti-PAK4 (Cat#sc-390507, Santa Cruz), anti- phospho-β-catenin (Ser675) (Cat#4176, Cell Signaling Technology), anti-β-catenin (Cat # 8480, Cell Signaling Technology), anti-phospho-ERK1/2 (Thr202/Tyr204)(Cat# 4370, Cell Signaling Technology), Anti-ERK1/2 (Cat#4695, Cell Signaling Technology), anti-phospho-MEK1/2 (Ser217/221)(Cat# 9154, Cell Signaling Technology), anti-MEK1/2 (Cat# 4694, Cell Signaling Technology), anti-c-Myc antibody (Cat#ab32072, Abcam), anti-Flag (Cat #F1804, Sigma), anti-HA (Cat #3724, Cell Signaling Technology), anti-His (Cat #ab137839, Abcam), anti-GAPDH (#HRP-60004, Proteintech), anti-β-actin (#HRP-60008, Proteintech).
Plasmid construction
Eukaryotic expression constructs, including pcDNA3.1-CDK15 and pcDNA3.1-PAK4 were obtained from Youbao Biotechnology Company (Changsha, China). Packaging vectors (PLKO.1, pMD2.0 G, and psPAX2) were from Addgene Inc (Cambridge, MA, USA). To construct various expression vectors, Flag-CDK15, HA-CDK15, His-CDK15, mutant Flag-PAK4, Flag-PAK4, and His-PAK4 were amplified by PCR with designed specific primers (provided in Supplementary Table 1).
Cell culture and transfection
The human normal colon epithelial cell line CCD18-co and CRC cell lines (SW480, HCT116, SW620, HCT-15, and HT-29) were purchased from the American Type Culture Collection (ATCC). CCD18-co was cultured in minimum essential medium, SW480 and SW620 cells were cultured in L-15, HCT116 cells were cultured in McCoy’s 5 A and HCT-15 in RPIM-1640. HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium. All cell lines utilized in this study were mycoplasma-free and authenticated by cellular morphology and STR analysis. All media contained penicillin (100 units/mL), streptomycin (100 μg/mL), and 10% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit-Haemek, Israel). Cells were maintained at 5% CO2 and 37 °C in a humidified atmosphere. Transfection was performed using Lipofectamine 2000 (11668-019, Invitrogen Life Technologies, Carlsbad, CA, USA) for CRC cell lines and Simple-Fect Reagent (Signaling Dawn Biotech, Wuhan, Hubei, China) for HEK293T cells, according to the manufacturer’s instructions.
Cell proliferation and anchorage-independent growth assays
Cell proliferation was determined using the MTT assay. Viable cells were seeded into 96-well plates (1.0–3.5 × 103 cells per well) and absorbance was measured at 0, 24, 48, and 72 h after seeding the cells. The colony formation capacity of CRC cell lines was examined by an anchorage-independent growth assay. A total of 8000 viable cancer cells were suspended in triplicate in 1 mL maintenance medium mixed with 2 mM glutamine, 5 μg/mL gentamycin, and 0.3% soft agar in the top layer. The base layer was replaced with 2 mL 0.5% soft agar in six-well plates. After 7–21 days, colonies were counted and photographed under a microscope. The number of colony was determined using Image-Pro Plus software (v.6.0) program (Media Cybernetics, Rockville, MD).
Lentivirus construction and infection
For protein knockdown, shRNA-PLKO.1 was co-transfected with psPAX2 and pMD2.G at a 1:1:1 mass ratio into HEK293T cells using Simple-Fect Reagent (Signaling Dawn Biotech, Wuhan, Hubei, China). The shRNA sequences were designed as described in Supplementary Table 1. Two days after transfection, the virus particles were collected and filtered through a 0.45 µm filter. Cells were infected with 8 mg/mL polybrene (#107689, Sigma–Aldrich) and selected with puromycin following the manufacturer’s instructions.
Western blot analysis
Cell pellets were resuspended in RIPA lysis buffer and incubated on ice for 30 min (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.25% deoxycholic acid disodium salt, 1% NP40, 150 mM NaCl, 0.1% SDS). After centrifugation at 12,000 × g for 10 min at 4 °C, the supernatant fractions were harvested. Protein concentration was detected using the BCA Quantification kit (Solarbio, Beijing, China, Cat#PC0020). Next, 20–50 μg of protein extract was resolved by SDS/PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked for 1 h at room temperature (RT) followed by an appropriate primary antibody overnight at 4 °C and subsequently incubated with a secondary antibody conjugated with horseradish peroxidase for 2 h at RT. The specific protein bands in the membrane were visualized using an enhanced chemiluminescence (ECL) reagent and Amersham Imager 600 (GE Healthcare Life Science, Pittsburgh, PA, SA).
Immunohistochemistry
Human CRC tissue microarray was purchased from Shanghai Outdo Biotech Company (Shanghai, China). Clinical CRC tissue specimens were obtained from the Affiliated Cancer Hospital of Zhengzhou University. All specimens were embedded in paraffin, deparaffinized, and rehydrated in advance. Antigen retrieval was subsequently performed in boiling sodium citrate buffer (10 mmol/L, pH 6.0) for 90 s. After incubation with 3% H2O2 for 10 min, the slides were blocked with 10% goat serum albumin for 30 min at RT. Slides were then treated with specific primary antibodies at 4 °C in a humidified chamber overnight, followed by incubation with the secondary antibody for 30 min at RT. Finally, slides were stained using 3,3’-diaminobenzidine and counterstained with hematoxylin. Immunohistochemistry (IHC) staining was quantified by calculating the percentage of positive cells using the Image-Pro Plus software (v.6.0) program (Media Cybernetics, Rockville, MD, USA).
Immunoprecipitation
Cell pellets were incubated with lysis buffer (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% NP40, 150 mM NaCl) containing protease inhibitors and PMSF for 30 min at 4 °C. After quantification, appropriate cell lysates were incubated with specific antibodies, rotating overnight at 4 °C, 30 μL of protein A/G agarose beads (#sc-2003, Santa Cruz) was then added to each sample and rotated for 2 h at 4 °C. The beads were washed four times with lysis buffer, and the immune complexes were eluted at 95 °C for 5 min with 6× loading buffer. The immunoprecipitated complexes were then separated by SDS/PAGE and subjected to western blot analysis.
Purification of recombinant proteins
For protein purification, the pet28a-his expression system was established and transfected into Escherichia coli strain BL-21. Protein expressed in BL-21 was induced at 16 °C overnight with 0.5 mM IPTG. BL-21 cells were resuspended and disrupted with lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 1 mM phenylmethanesulfonylfluoride, 1% Triton X-100) through sonication. The supernatants of the bacterial lysates were incubated with NI-NTA agarose for 4 h at 4 °C (QIAGEN, Germantown, MD, USA). The beads were washed four times with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 60 mM imidazole), and purified proteins were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 300 mM imidazole). Protein purity was determined using SDS-PAGE followed by Coomassie brilliant blue (CBB) staining.
Pull down assay and mass spectrometry
CDK15-NI-NTA agarose complex purified from BL-21 cells was incubated with CRC lysates (CDK15-NI-NTA agarose only and CRC lysates only as control) for 4 h at 4 °C. The CDK15-NI-NTA agarose complex was then washed four times in buffer (50 mM NaH2PO4, 300 mM NaCl, 60 mM imidazole) and subjected to SDS/PAGE. Using CBB staining, discrepant gel lanes were cut down and prepared for mass spectrometry.
In vitro kinase assay
For in vitro kinase assays, pcDNA3.1-CDK15-Flag was transfected into HEK293T cells. After 36 h, cells were collected, and active CDK15 was purified from HEK293T cells. Active CDK15 recombinant protein (50 ng) and 200 ng of wild-type (WT) PAK4 or PAK4 mutant recombinant protein were incubated with 250 μM ATP. The reactions were run in kinase buffer (20 mmol/L HEPES, pH 7.4, 10 mmol/L MgCl2, 5 mmol/L EGTA, 150 mmol/L NaCl, 20 mmol/L β-glycerol phosphate) for 30 min at 30 °C and stopped using 6× SDS sample buffer. Proteins were resolved by SDS-PAGE and assessed by western blot analysis.
Cell-derived xenograft mouse model
This study was approved by the Ethics Committee of Zhengzhou University (Zhengzhou, Henan, China). Six- to eight-week-old NU/NU mice (Vital River Labs, Beijing, China) were randomly divided into three groups as follows: shNT (n = 10); shCDK15-3 (n = 10); shCDK15-7 (n = 10). Cells infected with the indicated lentivirus (SW480: 1 × 107 cells; HCT116: 2 × 106 cells) were injected subcutaneously into the right flank of mice. Tumor volumes were measured using a Vernier caliper and calculated as V = (length) × (width) × (height) × 0.52 [13]. Mice were euthanized and tumors were extracted when tumor volume reached 1000 mm3 (cubic millimeter).
Patient-derived xenograft mouse model
This study was approved by the Ethics Committee of Zhengzhou University (Zhengzhou, Henan, China). For all the cancer tissues utilized in the study, written informed consent was obtained from the patients. Six- to eight-week-old severe combined immunodeficient (SCID) female mice (Vital River Labs, Beijing, China) were selected for animal experiments. Human tumor specimens were collected at the time of surgery from patients in the Affiliated Cancer Hospital of Zhengzhou University. Cancer tissues were cut into the same weight pieces and implanted into the backs of mice. When tumors reached an average volume of approximately 150 mm3, mice were randomly divided into different groups. For lentivirus treatment groups (shNT (n = 8), shCDK15-3 (n = 8), shCDK15-7 (n = 8)), the lentiviruses were produced in HEK293T cells as previously described. Then, the indicated viruses were concentrated by centrifugation at 30,000 rpm for 4 h at 4 °C. Mice in different groups received the corresponding viruses via intratumoral injection at a dose of 1 × 108 pfu/100 μL per mouse every three days for a total of four times. For the PF-3758309 treatment (vehicle (n = 9–10), 5 mg/kg (n = 9–10), 20 mg/kg (n = 9–10)), PF-3758309 or 0.9% NaCl was intraperitoneally injected once per day for 20 days. Tumor volumes were measured as described for cell line-derived xenograft (CDX). Mice were euthanized and tumors were extracted for further IHC analysis when tumor volume reached 1000 mm3.
Azoxymethane/ Dextran Sodium Sulfate (AOM/DSS) induced tumor in mice
All studies were approved by the Ethics Committee of Zhengzhou University (Zhengzhou, Henan, China). BALB/c mice (CDK15 wild-type (WT) and knockout (KO)) were purchased from the Jackson Laboratory (Cat#023307). Mice were genotyped by PCR analysis with specific primers (Supplementary Table 1) according to the protocol of Jackson Laboratory. Mice (6–8 weeks old) were randomly divided into four groups: (1) WT vehicle (n = 6), (2) KO-vehicle (n = 6), (3) WT-AOM/DSS (n = 14), and (4) KO-AOM/DSS (n = 14). The AOM/DSS-treated mice were injected once intraperitoneally with 10 mg/kg AOM (Cat # A5486, Sigma Chemical). After 2 weeks, 2% DSS (Cat#160110; MP Biomedicals, LLC) was added to the drinking water for 1 week, followed by regular water for two weeks, which was administered for two rounds. The body weight and stool were continuously monitored until the mice were euthanized at the 12th weeks. Tumors macroscopically visible on the colorectal mucous were counted, and the weight and length of the colorectum were measured. Colorectums were photographed and fixed in 4% formalin for histological analysis.
Statistical analysis
All statistical results were expressed as mean values ± SD, and P < 0.05 was considered statistically significant. Significant differences were determined by Student’s t-test or one-way ANOVA using GraphPad Prism 8.0 or SPSS 22.0 software.
Results
CDK15 is overexpressed in CRC and elevated CDK15 predicts an adverse prognosis in CRC patients
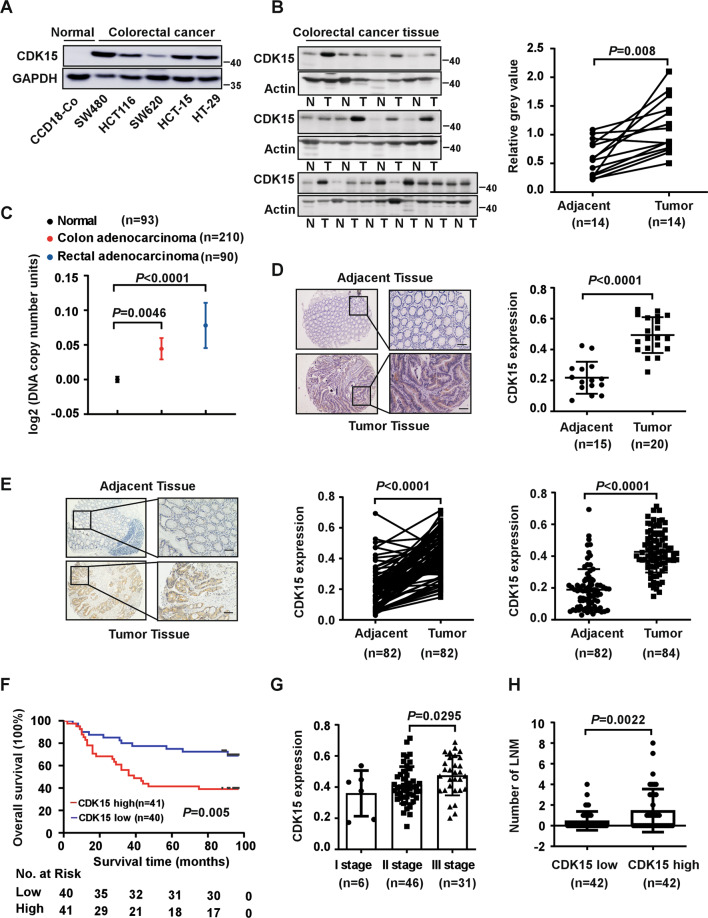
To explore the function of CDK15 in CRC, the expression of CDK15 was determined by western blot in different CRC cell lines, as well as in 14 paired CRC and adjacent tissues, which were collected from patients during surgery at the Affiliated Cancer Hospital of Zhengzhou University. CDK15 is significantly upregulated in CRC cells compared to the normal colon epithelial cell line CCD18-Co (Fig. 1A). Moreover, CRC tissues showed significantly elevated CDK15 expression compared to adjacent tissues (Fig. 1B). In addition, data retrieved from the Oncomine database demonstrates that CDK15 DNA copy number is higher in colon adenocarcinoma and rectal adenocarcinoma compared to normal tissue (Fig. 1C). Furthermore, IHC staining in unpaired clinical CRC tissues donated from the Affiliated Cancer Hospital of Zhengzhou University showed that CDK15 was localized in both the cytoplasm and nucleus, and stained more strongly in CRC compared to adjacent tissues (Fig. 1D). To further confirm the role of CDK15 in CRC, we used a commercial CRC tissue microarray containing 84 paired CRC tissues and adjacent tissues for IHC. Clinicopathological characteristics of CRC cases were shown in Supplementary Table 2. The distribution and staining intensity of CDK15 in tissue microarray (Fig. 1E) agreed with that observed in Fig.1D. Furthermore, clinical correlation analysis revealed that CDK15 expression was significantly associated with clinical stage and lymph node metastases (LNM) of CRC (Figs. 1G, 1H and Table 1). Moreover, Kaplan–Meier analysis showed that patients with high levels of CDK15 manifested a lower survival probability and had an adverse prognosis (Fig. 1F). These data suggest that CDK15 acts as an oncoprotein and plays an essential role in CRC tumorigenesis.
Fig. 1. CDK15 is upregulated in human colorectal cancer and is associated with patient survival probability.
A Expression of CDK15 in human colon epithelial cell line CCD18-Co and colorectal cancer (CRC) cell lines. B Expression of CDK15 in 14 paired human clinical samples with CRC and adjacent tissues. Right panel: CDK15 protein expression levels normalized according to β-actin levels. C DNA level of CDK15 in normal and CRC tissues retrieved from the Oncomine database. D, E Representative immunohistochemical staining images of human CRC tissues elucidated by CDK15 antibody (40× and 100×; Scale bar: 50 μm). Statistical analysis performed for immunohistochemical staining, and CDK15 expression denoted as a positive percentage (Tissue specimens in D from the Affiliated Cancer Hospital of Zhengzhou University include 15 adjacent tissues and 20 tumor tissues; Commercial tissue microarray in E from Shanghai Outdo Biotech Company contains 84 paired tumor tissues with clinical information). F Relationship between CDK15 expression level and overall survival for tissue microarray. G Expression of CDK15 in patients with different clinical stages for tissue microarray. H Number of lymph node metastases (LNM) stratified by CDK15 expression level. Data statistical analysis performed using Student’s paired t-test in B, E; Student’s unpaired t-test in C, D, E, G, H; Kaplan–Meier analysis in F. Error bars represent mean ± SD.
Table 1.
Correlation between CDK15 expression and clinicopathological characteristics of CRC.
| Characteristics | CDK15 expression | ||
|---|---|---|---|
| Low (n = 42) | High (n = 42) | P | |
| Gender | |||
|
Male Female |
21 (50.00%) 21 (50.00%) |
22 (52.38%) 20 (47.62%) |
>0.9999 |
| Age | |||
|
≤60 >60 |
8 (19.05%) 34 (80.95%) |
9 (21.43%) 33 (78.57%) |
>0.9999 |
| Tumor size | |||
|
≤5 cm >5 cm |
29 (69.05%) 13 (30.95%) |
25 (59.52%) 17 (40.48%) |
0.4949 |
| Histological grade | |||
|
Well/moderately Poorly |
38 (90.48%) 4 (9.52%) |
33 (78.57%) 9 (21.43%) |
0.2268 |
| pT status | |||
|
T1 + T2 T3 T4 |
3 (7.32%) 31 (75.61%) 7 (17.07%) |
3 (7.14%) 33 (78.57%) 6 (14.29%) |
0.9383 |
| pN status | |||
|
N0 N1 N2 |
31 (73.81%) 11 (26.19%) 0 (0%) |
21 (50.0%) 13 (30.95%) 8 (19.05%) |
0.0064 |
| Clinical stage | |||
|
I II III |
3 (7.32%) 28 (68.29%) 10 (24.39%) |
3 (7.14%) 18 (42.86%) 21 (50%) |
0.0482 |
All data are the number of patients (%).
Numbers do not equal to the total number due to missing data.
CDK15 promotes colorectal cancer cell growth in vitro and in vivo
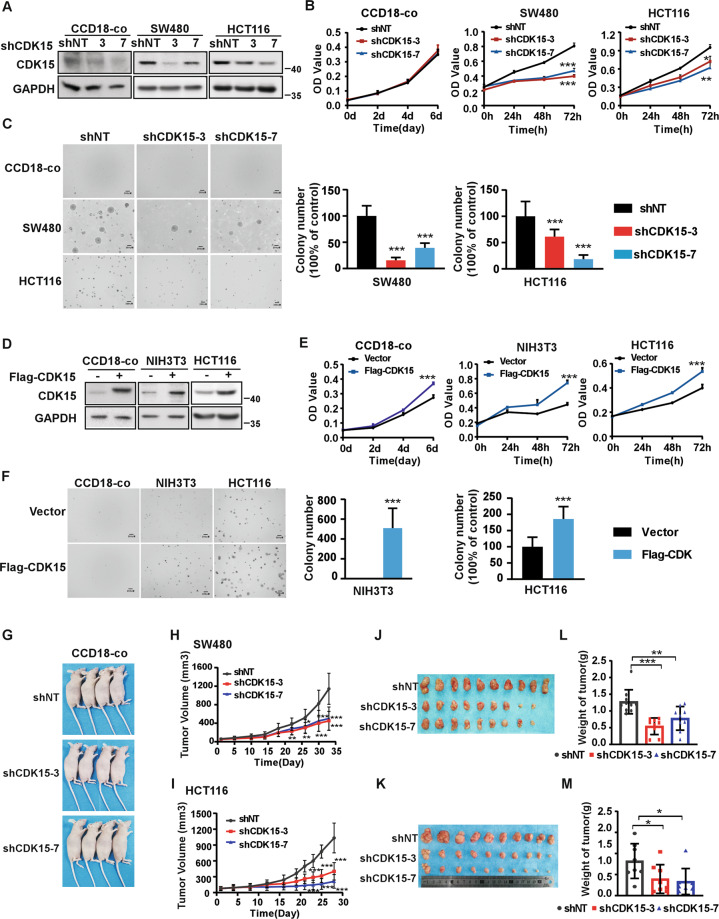
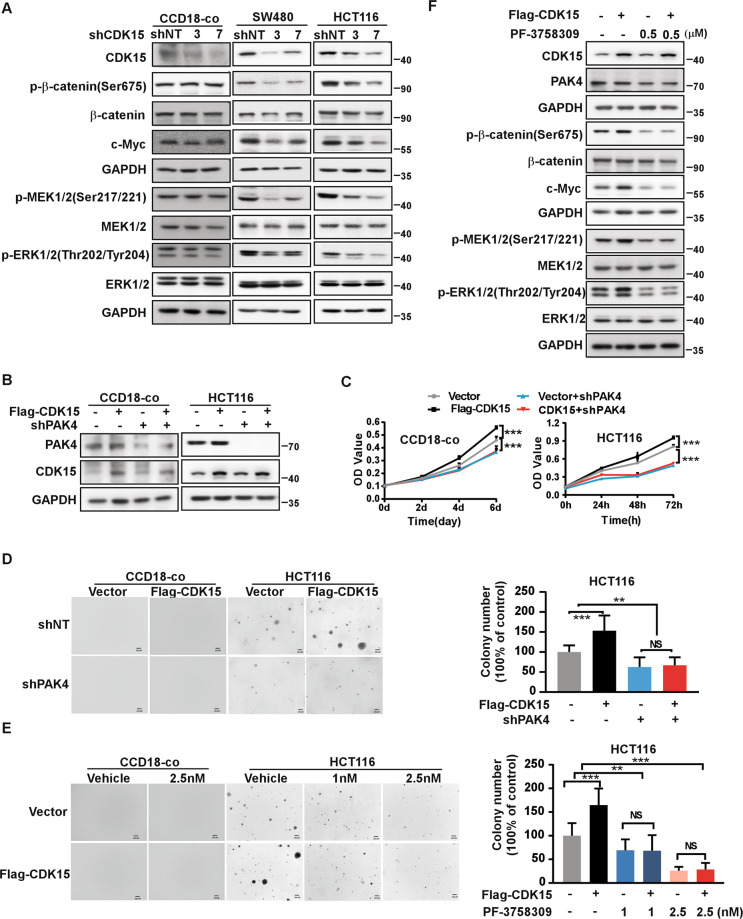
To investigate the role of CDK15 in CRC cells, we utilized two different shCDK15 sequences to generate CDK15 silencing in three CRC cells and in human colon epithelial cell line CCD18-co as a control (Fig. 2A, Fig. S1A). The results showed that CDK15 silencing dramatically reduced cell proliferation and suppressed the colony formation properties of CRC cells, while, cell proliferation of CCD18-co did not change significantly after CDK15 knockdown and CCD18-co did not form colony in soft agar. (Fig. 2B, C and Fig. S1B, C). Moreover, forced expression of CDK15 in CCD18-Co cells with very low CDK15 expression promoted cell proliferation (Fig. 2D, E). Overexpressed CDK15 in the mouse embryonic fibroblasts cell (NIH3T3) and CRC cell (HCT116 with CDK15 moderate expression) increased cell growth and colony formation (Fig. 2D, E, F). Those results further confirmed the enhancing effect elicited by elevated CDK15 on the proliferation and colony formation of CRC cells. To further explore the tumorigenic capacity of CDK15 in CRC cells, we used cell lines derived xenograft (CDX) model in nude mice subcutaneously injected with CDK15 silencing cells. The result showed that CCD18-co cell did not form transplanted tumors in CDX model (Fig. 2G), while, CDK15 knockdown remarkably suppressed tumor growth in CRC CDX models. The volume and growth rate of tumors in shCDK15 inoculated mice were significantly decreased compared to shNT inoculated mice (Fig. 2H, I), and the average body weight of mice did not differ significantly between different groups (Fig. S2A, C). Tumor size in the shCDK15 groups was much smaller than those in the shNT group (Fig. 2J, K). The final average tumor weight in shCDK15 groups was reduced compared to that in shNT group (Fig. 2L, M). Tumor growth inhibition normalized according to shNT was presented in Fig. S2B. Western blot analysis of tumor tissues confirmed the lower expression of CDK15 in tumor tissues with a lower growth rate (Fig. S2D). These results illuminate that CDK15 promotes aberrant cell proliferation and plays an important role in tumorigenesis of colorectal cancer cells in vitro and in vivo.
Fig. 2. CDK15 promotes colorectal cancer cell growth in vitro and in vivo.
A, D Cells with CDK15 knockdown and overexpression were established and the expression of CDK15 was determined by western blot. B, E Cell proliferation was measured by MTT assay. C, F Anchorage-independent growth from different cells with CDK15 knockdown or overexpression. Colonies were counted using Image J-Plus (Scale bar: 200 μm). Data represent statistical analysis of colony number in right panel. G CCD18-co cells stably infected with lentivirus (shNT, shCDK15-3, shCDK15-7) subcutaneously were injected into the right flank of nude mice (n = 4 mice per group). H, I SW480 and HCT116 cells stably infected with lentivirus (shNT, shCDK15-3, shCDK15-7) were subcutaneously injected into the right flank of nude mice (n = 9–10 mice per group). Tumor volume was monitored every 2–5 days. J, K Tumor photographs. L, M Tumor weight. Data from in vitro experiments were presented as means ± SD from triplicate experiments. All data statistical differences were evaluated using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
CDK15 knockout inhibits AOM/DSS-induced tumorigenesis in vivo
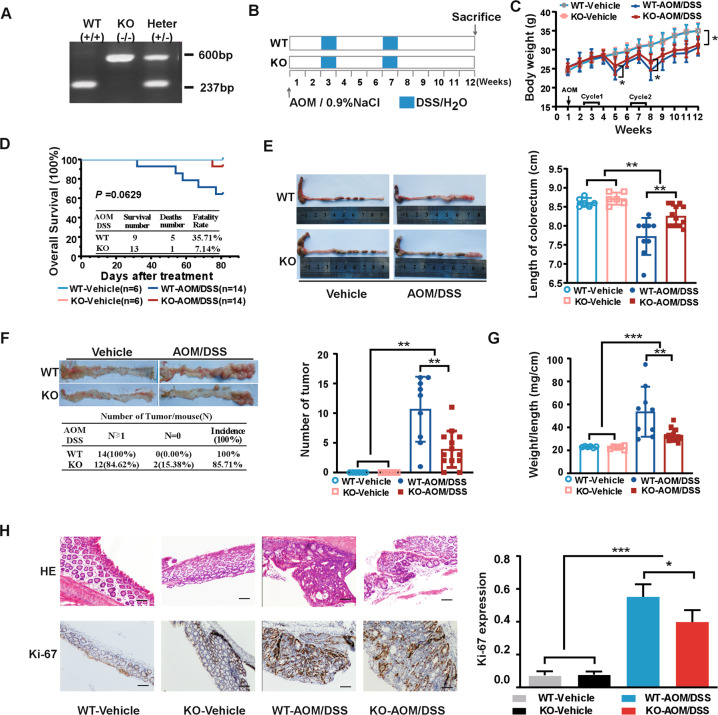
The results thus far prompted us to explore the functional significance of CDK15 in colon tumorigenesis using AOM/DSS-induced tumor model. First, PCR genotyping of CDK15 wild-type (WT), heterozygous (HT), and knockout (KO) mice was performed (Fig. 3A), and western blot was conducted to determine CDK15 expression in mice with different genotypes (Fig. S3A). Mice were grouped and treated according to the schematic diagram shown in Fig.3B. The results showed that significant body weight loss was observed in AOM/DSS-treated mice compared to control mice. Notably, WT mice treated with AOM/DSS suffered much severer body weight loss at the fifth and eighth weeks (Fig. 3C). In addition, following AOM/DSS treatment, WT mice exhibited earlier and more severe hemafecia on the 29th day compared to the 32nd day in KO mice, which appeared to be alleviated by CDK15 knockout. Moreover, the overall survival time of AOM/DSS-induced WT mice was lower than that of KO mice based on Kaplan–Meier survival curves with fatality rates of 35.71% (WT) and 7.14% (KO), respectively; however, these results were not significant (P = 0.0629; Fig. 3D).
Fig. 3. CDK15 knockout suppresses tumorigenesis in AOM/DSS-induced mice model.
A Example of PCR genotyping for CDK15 wild-type (WT), knockout (KO), and heterozygous (HT) mice. B Schematic diagram for AOM/DSS administration: six mice per group in WT and KO control groups, and 14 mice per group in AOM/DSS treatment groups. Mice were injected intraperitoneally with vehicle (0.9%NaCl) in control groups or 10 mg/kg AOM in experimental groups once at the first week, after which, control mice received regular water and experimental groups received 2% DSS exposure for two round. C Body weight in mice. Mice were weighed twice per week for total 12 weeks. D Effect of CDK15 on mice survival. E Representative macroscopic views of colorectum from mice. Right panel: length statistics of the colorectum in different groups (WT vehicle: n = 6; KO-vehicle: n = 6; WT-AOM/DSS: n = 9; KO-AOM/DSS: n = 13). F Tumor formation in different groups. Left panel: representative photos and incidence rate of tumors in the colorectum; Right panel: average number of tumors in different groups. G Effect of CDK15 on colorectal weight to length ratio. H Colorectum samples were harvested and stained with hematoxylin and eosin (H&E) and Ki-67 (100×; Scale bar: 50 μm). Right panel: expression level of Ki-67. All data were presented as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001.
In addition, anatomical results showed that AOM/DSS treatment mice presented with reduced colorectum length compared to control mice. Notably, much reduced colorectum length was observed in WT compared to KO mice following AOM/DSS treatment (7.72 ± 0.49 vs. 8.25 ± 0.27 cm, P = 0.0037; Fig.3E). Furthermore, more and larger tumors were also observed in WT mice compared to KO mice (tumor number, 10.7 ± 5.5 vs. 3.9 ± 3.1; P = 0.0014), while control groups have no tumor formed (Fig. 3F). Additionally, AOM/DSS-induced tumors in mice were primarily located in the distal portion of the colorectum. Colorectum weight in WT mice was also significantly increased compared with that in KO mice following AOM/DSS treatment (Fig. S3B). The ratio between weight and length of the colorectum was remarkably higher in WT mice than that in KO mice (53.6 ± 21.8 vs. 33.2 ± 5.5 mg/cm; P = 0.0038; Fig. 3G). Moreover, most colorectal neoplasms were histologically identified as adenomas in AOM/DSS-treated mice by hematoxylin and eosin (HE) staining (Fig. 3H). More severe dysplasia and loss of glandular structures in the mucosa was observed in colorectal sections of WT mice compared to KO mice after AOM/DSS treatment. Consistent with the colorectal tumor burden, Ki-67 expression was also significantly increased in WT mice compared to that in KO mice. These data demonstrate that CDK15 exerts a strong accelerative effect on colorectal tumorigenesis and greatly promotes the development of tumor induced by AOM/DSS.
CDK15 can bind PAK4 and phosphorylate its S291 residue
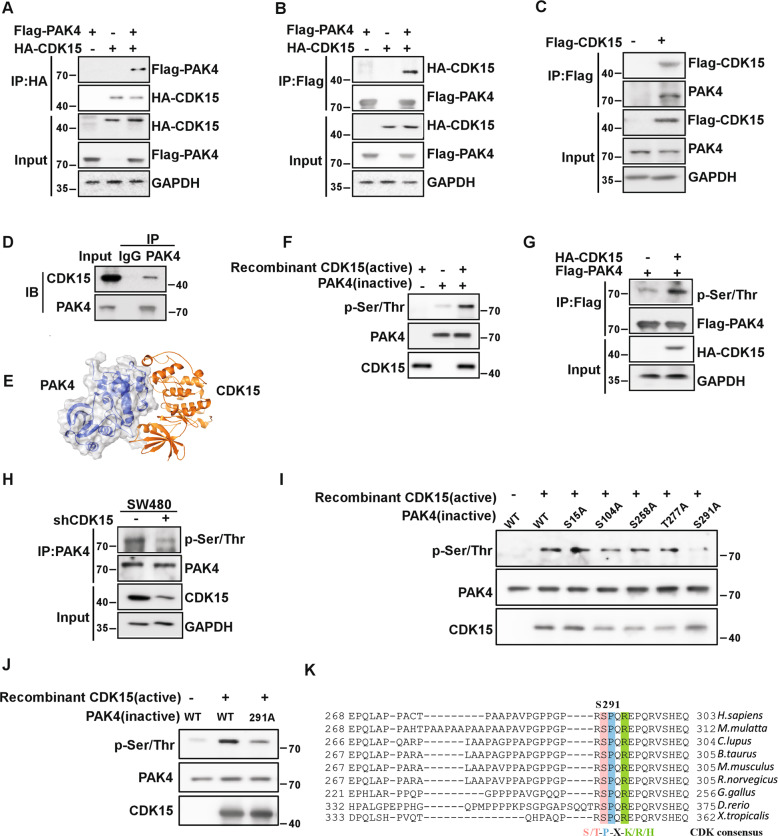
The data in this study suggest a strong oncogenic potential for CDK15 in colorectal cancer. However, the mechanism responsible for these effects remains unknown. Therefore, we extended our study to identify the potential substrates of CDK15 in colorectal cancer. Using pull down assays, 95 proteins were obtained, which were identified via mass spectrometry (MS). The top protein was found to be PAK4 according to the MS score. To confirm the interaction between CDK15 and PAK4, we performed co-immunoprecipitation assays with exogenously expressed CDK15 and PAK4 in HEK293T cells. HA-CDK15 was found to precipitate Flag-PAK4 from HEK293T cell lysates, and vice versa. (Fig. 4A, B). Furthermore, the endogenous interaction between CDK15 and PAK4 was validated in the colon cancer cell lines, HCT116 and SW480 (Fig. 4C, D). Computer modeling further confirm the interaction between CDK15 and PAK4 (Fig. 4E). Moreover, our results showed that CDK15 phosphorylated PAK4 in the in vitro kinase assay (Fig. 4F). Immunoblotting results showed that overexpressed CDK15 enhanced the phosphorylation level of PAK4 in HEK293T cells (Fig. 4G), while, CDK15 knockdown decreased the phosphorylation of PAK4 in SW480 cells (Fig. 4H). To further determine the potential residues of PAK4 that become phosphorylated by CDK15, we predicted five possible PAK4 phosphorylation sites in the Netphos2.0 server (Fig. S4). The results demonstrated that the S291 residue in PAK4 was likely the most relevant phosphorylation site for CDK15 in the in vitro kinase assay (Fig. 4I, J). More notably, we found that the motif containing S291 in PAK4 matched the consensus phosphorylation motif by CDK (S/T-P-X-K/R/H) [14] and the amino acid sequence around this site was highly evolutionarily conserved among multiple species suggesting that S291 was a functional phosphorylation site (Fig. 4K). Those findings suggest that PAK4 is a substrate of CDK15 and CDK15 can phosphorylates PAK4 at S291 residue.
Fig. 4. CDK15 binds and phosphorylates PAK4 in vitro.
A, B CDK15 and PAK4 bind with each other exogenously. Flag-PAK4, HA-CDK15, and mock plasmids were co-transfected into HEK293T cells. At 48 h after transfection, proteins were extracted. The FLAG- or HA-tagged proteins were immunoprecipitated with anti-HA or anti-Flag, and western blot was performed. C, D CDK15 and PAK4 bind with each other endogenously. HA-CDK15 and mock plasmids were co-transfected into HCT116 cells as indicated in C. At 48 h after transfection, proteins were extracted and immunoprecipitated with anti-HA. Endogenous PAK4 was detected by western blot in C. Proteins extracted from SW480 were immunoprecipitated by anti-PAK4, and immunoprecipitated complexes were detected using anti-CDK15, in D. E Modeling of CDK15 binding with PAK4. CDK15 is colored yellow, and PAK4 is colored blue with 70% transparency. F Active CDK15 was incubated with recombinant PAK4 protein in kinase reaction buffer for 30 min at 30 °C and stopped by SDS sample buffer. Phosphorylation signals were detected by western blot. Active CDK15 was purified from HEK293T cells, while recombinant PAK4 protein was purified from BL-21. G HEK293T cells were co-transfected with the indicated constructs. Cell lysates were immunoprecipitated with anti-Flag and immunoblotted with anti-p-Ser/Thr. H SW480 cells with stable knockdown of CDK15 were immunoprecipitated with anti-PAK4, and the immunoprecipitated complex was detected by anti-p-Ser/Thr in western blot. I Active CDK15 was incubated with the indicated PAK4 mutated protein in kinase reaction buffer. Phosphorylation signals were detected by western blot. J Active CDK15 incubated with PAK4 protein harboring S291A mutation in kinase reaction buffer and then were subjected to western blot. K Sequence alignment of CDK15 phosphorylation consensus within PAK4 orthologs among different species. Phosphorylated serine residue is highlighted in red, proline at the n + 1 position is highlighted in blue, and arginine at the n + 3 position is highlighted in green.
PAK4 mediates the oncogenic effect of CDK15 in colorectal cancer
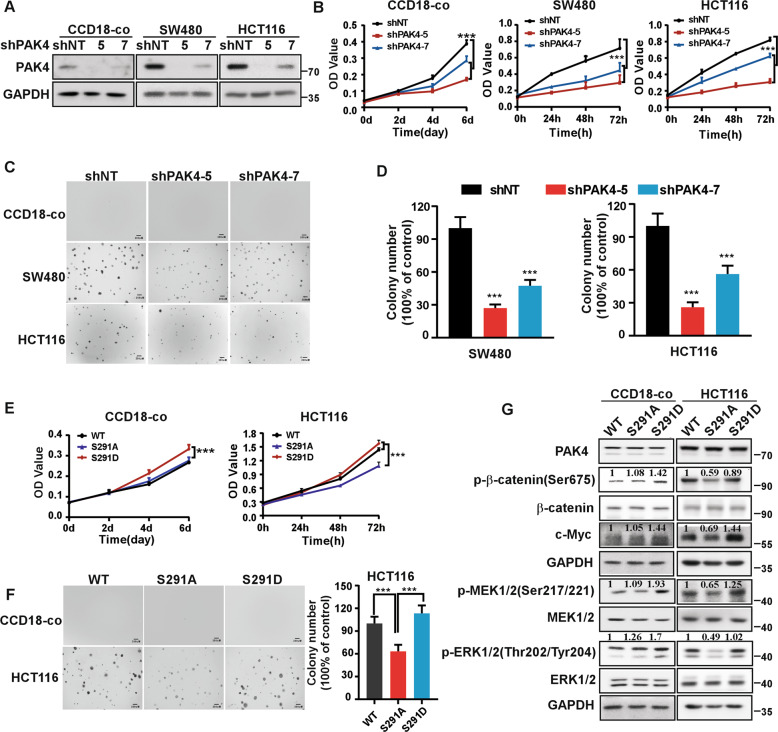
Studies have reported that PAK4 plays an important role in several cancers and regulates MEK/ERK and β-catenin signal pathways in cancer [15, 16]. In our results, levels of p-β-catenin(Ser675), c-Myc, p-MEK1/2(Ser217/221), and p-ERK1/2(Thr202/Tyr204) were decreased in CRC cells with CDK15 knockdown, while those signals had no change in CCD18-co cell after CDK15 knockdown (Fig. 5A). These results indicated that CDK15 silencing attenuated PAK4 downstream (β-catenin and MEK/ERK signaling pathway) in CRC cells with elevated CDK15, while had no effect in CCD18-co, which was consistent with its phenotype in Fig. 2B-C. Moreover, CDK15 overexpression increased cell proliferation, which was reversed by PAK4 knockdown in CCD18-co cells (Fig. 5B-D). Similarly, in HCT116 cells with PAK4 silencing and CDK15 overexpression, CDK15 overexpression increased cell proliferation and anchorage-independent cell growth compared with the control, while, the CDK15-induced cellular proliferation and colony growth were reversed following PAK4 knockdown (Fig. 5B-D). Furthermore, when CDK15 overexpressed cell treated with PF-3758309, the cell colony formation capacity was also reversed among different PF-3758309 concentration due to PAK4 inhibition (Fig. 5E and Fig. S5A). Correspondingly, western blot results showed that β-catenin and MEK/ERK signaling pathways, enhanced by CDK15 overexpression, were also reversed by PAK4 inhibition (Fig. 5F and Fig. S5B). Taken together, these results demonstrate that PAK4 is a substrate for CDK15 and mediates oncogenic function of CDK15 in CRC cells.
Fig. 5. PAK4 mediates the oncogenic effect of CDK15 in colorectal cancer.
A p-β-catenin(Ser675), c-Myc, p-MEK1/2 (Ser217/221), and p-ERK1/2 (Thr202/Tyr204) were detected by western blot after CDK15 knockdown in CCD18-co, SW480 and HCT116 cells. B Cells with PAK4 silencing and CDK15 overexpression were established. PAK4 and CDK15 expression was determined by western blot. C PAK4 knockdown reverses cell proliferation induced by CDK15 in CCD18-co and HCT116 cells. MTT assay was used to detect cell proliferation. D Anchorage-independent growth in CCD18-co and HCT116 cells with PAK4 silencing and CDK15 overexpression. Left panels: representative images (Scale bar: 200 μm). Right panels: Colonies were counted using Image J-Plus (Scale bar: 200 μm) and data represented statistical analysis of colony number ratio. E Anchorage-independent growth in CCD18-co and HCT116 cells treated with PAK4 inhibitor (PF-3758309). Left panel: representative images of colonies (Scale bar: 200 μm). Right panel: statistical analysis of the colony ratio. F Western blot to validate β-catenin and MEK/ERK signaling pathway in HCT116 cells with indicated treatment. Data were presented as mean values ± SD from triplicate experiments. Statistical differences were evaluated using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Phosphorylation of PAK4 at S291 site by CDK15 promotes cell proliferation
Given that the S291 residue of PAK4 can be phosphorylated by CDK15, and that PAK4 mediates the oncogenic effect of CDK15 in CRC, we next sought to determine the function of S291 residue in CRC. First, we demonstrated that PAK4 silencing reduced cell proliferation, as well as the number of colonies in SW480 and HCT116 cells. Moreover, cell proliferation of CCD18-co cell was also decreased significantly after PAK4 knockdown (Fig. 6A-D). The results suggest that PAK4 not only is necessary to maintain the proliferative activity of CCD18-co, but also plays a critical role in the proliferation of CRC cells. Next, we transfected PAK4-WT, PAK4-S291A, and PAK4-S291D into HCT116 and CCD18-co cells. Results showed that in HCT116 cells, PAK4-S291A transfection displayed decreased proliferation as well as fewer and smaller colonies compared to PAK4-WT transfection, while, PAK4-S291D was able to enhance the proliferation and colony formation capacity of cells (Fig. 6E, F). In addition, western blot results indicated that in PAK4-S291A transfected cells, β-catenin and MEK/ERK signals were decreased compared to PAK4-WT cells, while, PAK4-S291D effectively enhanced those signaling pathways (Fig. 6G and Fig. S5C). However, in CCD18-co cells, PAK4-S291D transfection displayed increased cell proliferation compared to PAK4-WT and PAK4-S291A. The proliferation between PAK4-WT and PAK4-S291A transfected cells did not show significant difference, which may due to that intracellular CDK15 protein level was too low to promote the phosphorylation of S291 in PAK4. Western blot data from CCD18-co cells showed consistent results (Fig. 6E-G). These findings suggest that abnormal phosphorylation of S291 site in PAK4 not only promotes cell proliferation of CCD18-Co cell, but also involves in the oncogenic function of PAK4 and plays an important role in CRC cells.
Fig. 6. CDK15-induced phosphorylation of PAK4 at S291 promotes cell proliferation.
A CCD18-co, SW480, and HCT116 cells with PAK4 knockdown were established. PAK4 expression was determined by western blot. B MTT assays demonstrate that PAK4 knockdown reduces cell proliferation in CCD18-co, SW480, and HCT116 cells. C Anchorage-independent growth in different PAK4 knockdown cells. D Number of colonies determined by Image J-Plus (Scale bar: 200 μm). Data represents statistical analysis of the colony ratio. E The proliferation ability in cells transfected with different S291 mutations of PAK4 was determined by MTT assay. F Anchorage-independent growth in cells transfected with different PAK4 S291 mutations (Scale bar: 200 μm). Data represent statistical analysis of the colony ratio in right panel. G β-catenin and MEK/ERK signals determined by western blot in cells transfected with different PAK4 S291 mutations. Densitometric analysis represents the ratio of phosphorylated protein/total protein. Data were presented as mean values ± SD from triplicate experiments. Statistical differences were evaluated using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
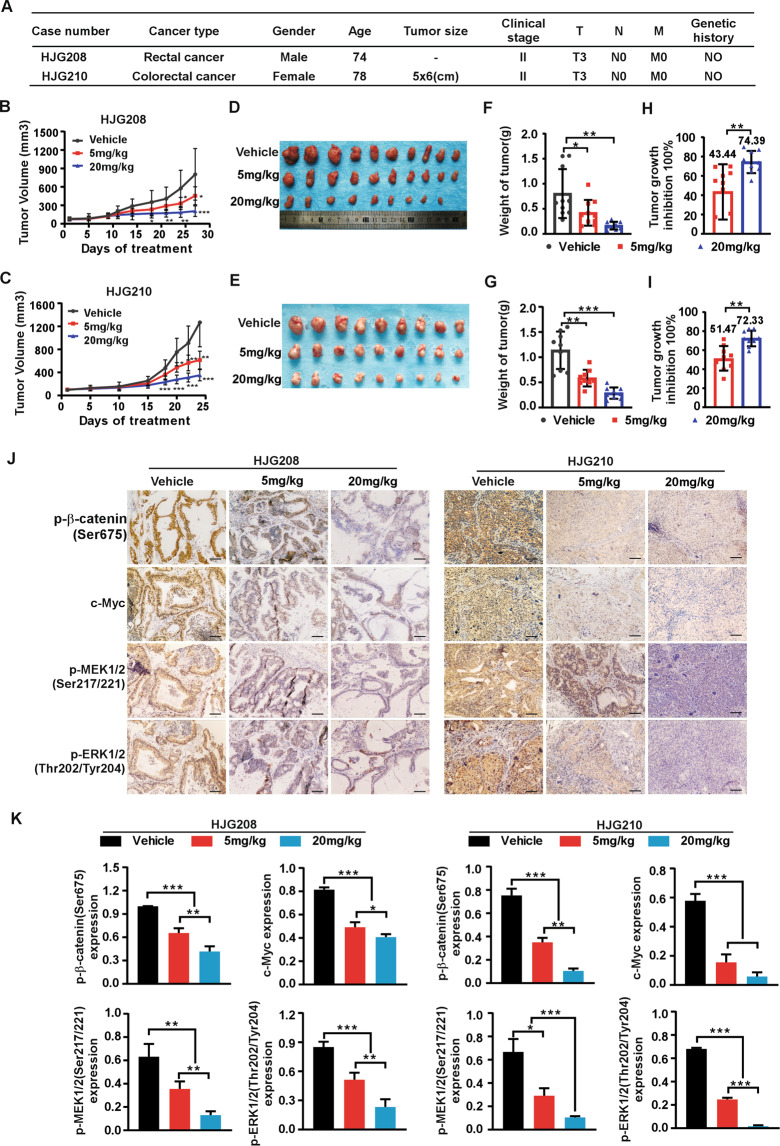
Targeting PAK4 delays colorectal tumor growth in patient-derived xenografts model
To further confirm the function of PAK4 in human CRC progression in vivo, we used patient-derived xenografts model (PDX) in SCID mice treated with PAK4 inhibitor, PF-3758309. We measured PAK4 expression among multiple clinical CRC tissues and chose two cases, HJG208 and HJG210, with high PAK4 protein level to develop the PDX models (Fig. S6). The clinical information for HJG208 and HJG210 was shown in Fig. 7A. The results revealed that targeting PAK4 remarkably suppressed tumor growth in PDX. Tumor volume and growth rate were significantly decreased in the treatment groups compared to the vehicle groups (0.9%NaCl), while, the average body weight had no significant difference between different groups (Fig. 7B, C and Fig. S7A). Tumor size in the treatment groups was much smaller than that in the vehicle groups (Fig. 7D, E). The final average tumor weight in the treatment groups was reduced compared to that in the vehicle groups (Fig. 7F, G). The tumor growth inhibition rate in the 20 mg/kg groups was significantly higher than that in 5 mg/kg groups (Fig. 7H, I). Moreover, IHC analysis showed that the levels of p-β-catenin(Ser675), c-Myc, p-MEK1/2(Ser217/221), and p-ERK1/2(Thr202/Tyr204) were decreased in treatment groups compared to vehicle group, which were consistent with the tumor burdens in different groups (Fig. 7J, K). These results confirm that PAK4 plays an important role in CRC and pharmacological targeting PAK4 suppresses CRC tumor growth in PDX in vivo.
Fig. 7. Targeting PAK4 delays tumor growth in patient-derived xenografts.
A Clinical information for HJG208 and HJG210 from patient’s cancer tissues. B, C 0.9% NaCl as vehicle, 5 mg/kg or 20 mg/kg PF-3758309 were intraperitoneally injected once per day for 20 days, and tumor volume was monitored every 2–5 days (n = 9–10 mice per group). D, E Tumor photographs. F, G Tumor weight and tumor growth inhibition (H, I) normalized to control group. J Levels of p-β-catenin (Ser675), c-Myc, p-MEK1/2 (Ser217/221), and p-ERK1/2 (Thr202/Tyr204) in harvested tumor tissues were assessed by immunohistochemistry. Representative photographs for each antibody in different groups are shown (100×; Scale bar: 50 μm). K Statistical analysis for immunohistochemistry staining. Statistical differences were evaluated using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent mean ± SD.
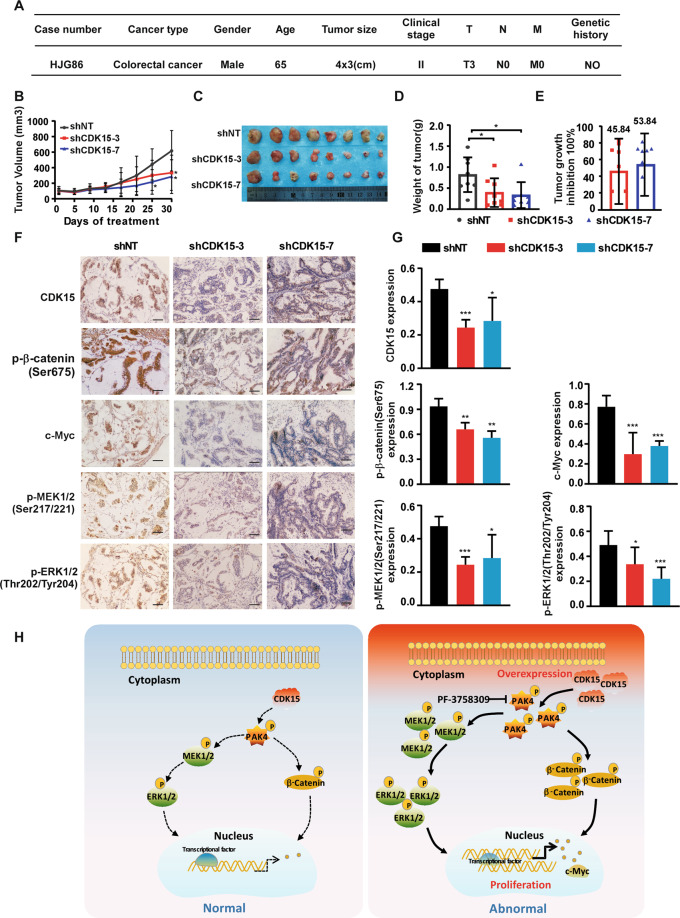
Lentivirus-mediated CDK15 silencing inhibits colorectal tumor growth in patient-derived xenografts model
To further verify the role of CDK15 in human CRC progression in vivo, the patient-derived xenografts (PDX) were intratumorally injected with lentivirus to knockdown CDK15 expression in the tumor [17]. CDK15 expression level was determined by western blot and case HJG86, with elevated CDK15, was chosen to establish the PDX model (Fig. S6). The clinical information of HJG86 was shown in Fig. 8A. The results showed that CDK15 silencing remarkably retarded tumor growth in PDX models. The volume and growth rate of tumors in shCDK15 groups were significantly decreased compared to shNT groups, while, the average body weight had no significant difference between the groups (Fig. 8B and Fig. S7B). Furthermore, tumors in the shCDK15 groups were much smaller than that in the shNT group (Fig. 8C). The final average tumor weight in shCDK15 group was also reduced compared to that in the shNT group (Fig. 8D). Tumor growth inhibition in shCDK15 groups was shown in Fig. 8E. Considering that PAK4 is a substrate of CDK15, and that CDK15 oncogenicity in CRC has been shown to be closely associated with the PAK4 signaling pathway, we examined PAK4 downstream signals on tumor extracts from each group through IHC (Fig. 8F, G). The results showed that after CDK15 knockdown in tumors by lentivirus infection, the levels of p-β-catenin(Ser675), c-Myc, p-MEK1/2(Ser217/221), and p-ERK1/2(Thr202/Tyr204) were also decreased, which were consistent with the changes of tumor burden in mice. Overall, these data provide strong evidences that CDK15 acts as an oncogene in CRC tumorigenesis and its silencing by lentivirus suppresses CRC tumor growth by regulating PAK4 signaling pathway in vivo.
Fig. 8. Lentivirus-mediated CDK15 silencing inhibits colorectal tumor growth in patient-derived xenografts.
A Clinical information for HJG86 from patient’s cancer tissues. B Mice received the lentiviruses (shNT, shCDK15-3, shCDK15-7) via intratumoral injection every 3 days for a total of four times. Tumor volume was monitored every 2–5 days for four continuous weeks (n = 8 mice per group). C Tumors photographs. D Tumor weight measured at the end of the study. E Tumor growth inhibition normalized to control group. F Levels of CDK15, p-β-catenin(Ser675), c-Myc, p-MEK1/2(Ser217/221), and p-ERK1/2 (Thr202/Tyr204) in harvested tissues were assessed by immunohistochemistry. Representative photographs in different groups are shown (100×; Scale bar: 50 μm). G Statistical analysis for immunohistochemistry staining. H Schematic model for the findings of this work: Aberrant CDK15 in CRC binds PAK4 and phosphorylates PAK4 at the S291 site. Accumulated phosphorylation of S291 upregulates the β-catenin/c-Myc and MEK/ERK signals, which in turn contribute to CRC tumor growth. Statistical differences were evaluated using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent mean ± SD.
Discussion
The newly proposed CDKs family members (CDK14-CDK16) have been reported to be closely associated with cancer progression. CDK14 acts as an oncogene in breast, esophageal gastric, and pancreatic cancer [18–20] and involves in Wnt signaling by regulating LRP6 [21]; CDK16 promotes progression of hepatocellular carcinoma, lung cancer, and melanomas by regulating cell cycle and apoptosis [8, 22–24]. CDK15, which is highly homologous to CDK14 and CDK16, has been reported to protect cancer cell from tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis [11], while, another paper shows CDK15 acts as a tumor suppressor in breast cancer cell invasion and metastasis [12]. In the present study, we illuminate that CDK15 is highly expressed in human CRC and high abundance of CDK15 manifests a lower survival probability in CRC patients. Moreover, CDK15 knockout mice and lentivirus-infected PDX model provide strong evidences that CDK15 plays an important role in the progression of CRC.
The AOM/DSS model is a powerful, reproducible initiation-promotion CRC model [25, 26]. Specifically, tumor development in this model mimics that observed in humans, with the tumors induced in mice accurately recapitulating the pathogenesis observed in humans [27, 28]. This model demonstrates CDK15 exerts a strong accelerative effect on colorectal tumorigenesis and clearly defined an oncogenic function for CDK15 in CRC, which help us better understand the role of CDK15 in cell behavior and tumor biology observed in humans. The PDX model has high stability and resemblance to human tumors and is the preferred tool for clinical translation research [29, 30]. In our study, CDK15 silencing significantly suppresses colorectal tumor growth in PDX model, which provides strong support for CDK15 as a novel therapeutic target for CRC therapy.
PAK4, a serine/threonine p21-activating kinase, plays an important role in a variety of cellular functions [31]. Dysregulation of PAK4 expression and activity is closely associated with cancer progression [32]. Our results show that PAK4 silencing decreases cell proliferation in CRC, which agrees with other studies that PAK4 is highly expressed in CRC tissues compared to normal tissues, and its silencing decreases cell proliferation, while increases apoptosis in CRC [33]. A myriad of PAK4 effectors, including MEK/ERK, Wnt/β-catenin, LIMK1/Cofilin, and PI3K/AKT signaling pathways, have been described in cancers [34–36]. However, the precise mechanisms associated with PAK4 regulation are not yet fully understood. Herein, our studies demonstrate that CDK15 can bind and phosphorylate PAK4 at S291 residue, subsequently enhances MEK/ERK and β-catenin signals, which are reported to be frequently aberrantly expressed in CRC and promote CRC progression [37, 38]. Moreover, the phosphorylation of S291 site by CDK15 promotes CRC cells proliferation and plays a critical role in PAK4 oncogenic function in CRC. Other studies found that phosphorylation of Ser474 in PAK4 enhances the transcriptional activity of CREB in prostate cancer [39] and PAK4(S445N) induces a transformed morphology in fibroblasts [40]. Another study argued that PAK4 activity and function are regulated by conformational changes rather than phosphorylation of residues residing within the activation loop of the protein [41]. Hence, PAK4 function appears to be regulated by multiple residue sites as well as conformational changes depend on different cancers and microenvironments.
PF-3758309, a PAK4 inhibitor, suppresses the proliferation of multiple human tumor cells in vitro and in vivo [42]. There have reports that pharmacological inhibition of PAK4 improved tumor response to anti-PD-1 immunotherapy in murine melanoma CDX model [43]. In addition, combined inhibitor treatment for LIMK1 and PAK4 exhibited good antitumor efficacy in breast cancer [44]. In our study, PAK4 inhibition by PF-3758309 significantly suppresses colorectal tumor growth in PDX models and exhibits potent preclinical antitumor efficacy in CRC suggesting that targeting CDK15-PAK4 axis might provide a potential preventative or therapeutic strategy for CRC patients.
All results in our study demonstrate that CDK15 is a critical oncogenic factor capable of predicting adverse prognoses and provide strong support for CDK15 as a novel therapeutic target for CRC patients. Furthermore, CDK15 exerts its oncogenic function by phosphorylating PAK4 at S291 site, which subsequently enhances the β-catenin, MEK/ERK signaling pathway to promote CRC progression. Hence, therapeutic approaches designed to inhibit CDK15-PAK4 axis represent an attractive strategy for CRC patients and are worthy of more investigation.
Supplementary information
Acknowledgements
We thank Dr. Fangfang Liu for advice on AOM/DSS-induced mice model. We are grateful to Wenna Nie for analysis of mass spectrometry and Ran Yang for preparation of immunohistochemical section.
Author contributions
CTH wrote the manuscript. CTH and RJD participated in designing the research and in analyzing data. CTH, XCJ, and QW performed in vivo study and participated in mouse experiment; NY, LY, LTZ, XJL, and MXX performed mouse experiments and helped with in vitro studies. YQ conducted the computer docking model. PX performed the pathological studies. YW performed the immunohistochemical studies. XL and ZGD conceived the study. All authors critically revised manuscript and approved the final version.
Funding
This work was supported by National Natural Science Foundation of China (No. 82073075, 81802795, 31301144); Key scientific research project plan of colleges and universities in Henan Province (No. 18A310034); Science and Technology Project of Henan Province (No. 182102310324, 202102310206); Training plan for young backbone teachers of Zhengzhou University (No. 2018ZDGGJS037); Training plan for young backbone teachers of Henan Province (No. 2020GGJS010); Basic research and Cultivation Fund for young teachers of Zhengzhou University (No. JC202035023); Science and technology innovation talents support plan of Henan Province (No.21HASTIT048).
Data availability
The data are available to academic researchers upon request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Written informed consent was obtained from all subjects. All animal experiments were conducted in agreement with the Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of Zhengzhou University (Zhengzhou, Henan, China).
Footnotes
Edited by H. Ichijo
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chuntian Huang, Ruijuan Du.
Contributor Information
Zigang Dong, Email: dongzg@zzu.edu.cn.
Xiang Li, Email: lixiang@zzu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00828-6.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 4.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, et al. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11:1275–6. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roskoski R., Jr Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol Res. 2019;139:471–88. doi: 10.1016/j.phrs.2018.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker SR, Mallinger A, Workman P, Clarke PA. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther. 2017;173:83–105. doi: 10.1016/j.pharmthera.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, Liu C, Liu F, Wang Y, Zhu M. Knockdown of PFTAIRE protein kinase 1 (PFTK1) inhibits proliferation, invasion, and EMT in colon cancer cells. Oncol Res. 2016;24:137–44. doi: 10.3727/096504016X14611963142218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie J, Li Y, Jiang K, Hu K, Zhang S, Dong X, et al. CDK16 phosphorylates and degrades p53 to promote radioresistance and predicts prognosis in lung cancer. Theranostics. 2018;8:650–62. doi: 10.7150/thno.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ning JF, Stanciu M, Humphrey MR, Gorham J, Wakimoto H, Nishihara R, et al. Myc targeted CDK18 promotes ATR and homologous recombination to mediate PARP inhibitor resistance in glioblastoma. Nat Commun. 2019;10:2910. doi: 10.1038/s41467-019-10993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park MH, Kim SY, Kim YJ, Chung YH. ALS2CR7 (CDK15) attenuates TRAIL induced apoptosis by inducing phosphorylation of survivin Thr34. Biochem Biophys Res Commun. 2014;450:129–34. doi: 10.1016/j.bbrc.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Dai X, Gong K, Song K, Tai F, Shi J. PA28alpha/beta promote breast cancer cell invasion and metastasis via down-regulation of CDK15. Front Oncol. 2019;9:1283. doi: 10.3389/fonc.2019.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–54. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 14.Dixon-Clarke SE, Shehata SN, Krojer T, Sharpe TD, von Delft F, Sakamoto K, et al. Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16. Biochem J. 2017;474:699–713. doi: 10.1042/BCJ20160941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Zhang X, Yang Z, Du H, Wu Z, Gong J, et al. MiR-145 regulates PAK4 via the MAPK pathway and exhibits an antitumor effect in human colon cells. Biochem Biophys Res Commun. 2012;427:444–9. doi: 10.1016/j.bbrc.2012.06.123. [DOI] [PubMed] [Google Scholar]
- 16.Rane CK, Minden A. P21 activated kinase signaling in cancer. Semin Cancer Biol. 2019;54:40–9. doi: 10.1016/j.semcancer.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Du R, Huang C, Chen H, Liu K, Xiang P, Yao N, et al. SDCBP/MDA-9/syntenin phosphorylation by AURKA promotes esophageal squamous cell carcinoma progression through the EGFR-PI3K-Akt signaling pathway. Oncogene. 2020;39:5405–19. doi: 10.1038/s41388-020-1369-2. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Wang Y, Jiang W, Ni R, Wang Y, Ni S. CDK14 involvement in proliferation migration and invasion of esophageal cancer. Ann Transl Med. 2019;7:681. doi: 10.21037/atm.2019.11.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Zou A, Ma L, Chen X, Wang L, Zeng X, et al. miR-455 inhibits breast cancer cell proliferation through targeting CDK14. Eur J Pharmacol. 2017;807:138–43. doi: 10.1016/j.ejphar.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Zhu H, Jin Y, Song Y. MiR-431 inhibits cell proliferation and induces cell apoptosis by targeting CDK14 in pancreatic cancer. Eur Rev Med Pharmacol Sci. 2018;22:4493–9. doi: 10.26355/eurrev_201807_15503. [DOI] [PubMed] [Google Scholar]
- 21.Davidson G, Shen J, Huang YL, Su Y, Karaulanov E, Bartscherer K, et al. Cell cycle control of wnt receptor activation. Dev Cell. 2009;17:788–99. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Qin X, Guo T, Liu P, Wu P, Liu Z. Up-regulation of CDK16 by multiple mechanisms in hepatocellular carcinoma promotes tumor progression. J Exp Clin Cancer Res. 2017;36:97. doi: 10.1186/s13046-017-0569-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Yanagi T, Krajewska M, Matsuzawa S, Reed JC. PCTAIRE1 phosphorylates p27 and regulates mitosis in cancer cells. Cancer Res. 2014;74:5795–807. doi: 10.1158/0008-5472.CAN-14-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanagi T, Reed JC, Matsuzawa S. PCTAIRE1 regulates p27 stability, apoptosis and tumor growth in malignant melanoma. Oncoscience. 2014;1:624–33. doi: 10.18632/oncoscience.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parang B, Barrett CW, Williams CS. AOM/DSS model of colitis-associated cancer. Methods Mol Biol. 2016;1422:297–307. doi: 10.1007/978-1-4939-3603-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SM, Kim N, Son HJ, Park JH, Nam RH, Ham MH, et al. The effect of sex on the azoxymethane/dextran sulfate sodium-treated mice model of colon cancer. J Cancer Prev. 2016;21:271–8. doi: 10.15430/JCP.2016.21.4.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, et al. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crncec I, Pathria P, Svinka J, Eferl R. Induction of colorectal cancer in mice and histomorphometric evaluation of tumors. Methods Mol Biol. 2015;1267:145–64. doi: 10.1007/978-1-4939-2297-0_7. [DOI] [PubMed] [Google Scholar]
- 29.Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–9. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borodovsky A, McQuiston TJ, Stetson D, Ahmed A, Whitston D, Zhang J, et al. Generation of stable PDX derived cell lines using conditional reprogramming. Mol Cancer. 2017;16:177. doi: 10.1186/s12943-017-0745-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar R, Sanawar R, Li X, Li F. Structure, biochemistry, and biology of PAK kinases. Gene. 2017;605:20–31. doi: 10.1016/j.gene.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King H, Nicholas NS, Wells CM. Role of p-21-activated kinases in cancer progression. Int Rev Cell Mol Biol. 2014;309:347–87. doi: 10.1016/B978-0-12-800255-1.00007-7. [DOI] [PubMed] [Google Scholar]
- 33.Wang M, Gao Q, Chen Y, Li Z, Yue L, Cao Y. PAK4, a target of miR-9-5p, promotes cell proliferation and inhibits apoptosis in colorectal cancer. Cell Mol Biol Lett. 2019;24:58. doi: 10.1186/s11658-019-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Tabusa H, Brooks T, Massey AJ. Knockdown of PAK4 or PAK1 inhibits the proliferation of mutant KRAS colon cancer cells independently of RAF/MEK/ERK and PI3K/AKT signaling. Mol Cancer Res. 2013;11:109–21. doi: 10.1158/1541-7786.MCR-12-0466. [DOI] [PubMed] [Google Scholar]
- 35.Vershinin Z, Feldman M, Chen A, Levy D. PAK4 methylation by SETD6 promotes the activation of the Wnt/beta-catenin pathway. J Biol Chem. 2016;291:6786–95. doi: 10.1074/jbc.M115.697292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whale AD, Dart A, Holt M, Jones GE, Wells CM. PAK4 kinase activity and somatic mutation promote carcinoma cell motility and influence inhibitor sensitivity. Oncogene. 2013;32:2114–20. doi: 10.1038/onc.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaj C, Schmidt EM, Lamprecht S, Hermeking H, Jung A, Kirchner T, et al. Oncogenic effects of high MAPK activity in colorectal cancer mark progenitor cells and persist irrespective of RAS mutations. Cancer Res. 2017;77:1763–74. doi: 10.1158/0008-5472.CAN-16-2821. [DOI] [PubMed] [Google Scholar]
- 38.Tenbaum SP, Ordonez-Moran P, Puig I, Chicote I, Arques O, Landolfi S, et al. beta-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med. 2012;18:892–901. doi: 10.1038/nm.2772. [DOI] [PubMed] [Google Scholar]
- 39.Park MH, Lee HS, Lee CS, You ST, Kim DJ, Park BH, et al. p21-Activated kinase 4 promotes prostate cancer progression through CREB. Oncogene. 2013;32:2475–82. doi: 10.1038/onc.2012.255. [DOI] [PubMed] [Google Scholar]
- 40.Qu J, Cammarano MS, Shi Q, Ha KC, de Lanerolle P, Minden A. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol Cell Biol. 2001;21:3523–33. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baskaran Y, Ng YW, Selamat W, Ling FT, Manser E. Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 2012;13:653–9. doi: 10.1038/embor.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray BW, Guo C, Piraino J, Westwick JK, Zhang C, Lamerdin J, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci USA. 2010;107:9446–51. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abril-Rodriguez G, Torrejon DY, Liu W, Zaretsky JM, Nowicki TS, Tsoi J, et al. PAK4 inhibition improves PD-1 blockade immunotherapy. Nat Cancer. 2020;1:46–58. doi: 10.1038/s43018-019-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao CC, Zhan MN, Liu WT, Jiao Y, Zhang YY, Lei Y, et al. Combined LIM kinase 1 and p21-Activated kinase 4 inhibitor treatment exhibits potent preclinical antitumor efficacy in breast cancer. Cancer Lett. 2020;493:120–7. doi: 10.1016/j.canlet.2020.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available to academic researchers upon request.