Highlights
-
•
Integrative analysis identified 130 core racial disparity associated genes.
-
•
The racial disparity associated genes were enriched for prostate cancer risk genes.
-
•
The racial disparity genes were more likely to be prognostic.
-
•
Kinases show differential phosphorylation levels between different races.
-
•
Transcription factors show differential regulatory activities between different races.
Keywords: Prostate cancer, African American, Network
Abbreviations: CA, Caucasian; AA, African American; DAG, racial disparity associated gene; CNV, copy number variation; PRAD, prostate adenocarcinoma; GEO, gene expression omnibus; RPPA, reverse phase protein array; FDR, false discovery rate; GWAS, genome-wide association study; CRPC, castration-resistant prostate cancer; GO, Gene Ontology
Abstract
Compared to Caucasians (CAs), African Americans (AAs) have a higher rate of incidence and mortality in prostate cancer and are prone to be diagnosed at later stages. To understand this racial disparity, molecular features of different types, including gene expression, DNA methylation and other genomic alterations, have been compared between tumor samples from the two races, but led to different disparity associated genes (DAGs). In this study, we applied a network-based algorithm to integrate a comprehensive set of genomic datasets and identified 130 core DAGs. Out of these genes, 78 were not identified by any individual dataset but prioritized and selected through network propagation. We found DAGs were highly enriched in several critical prostate cancer-related signaling transduction and cell cycle pathways and were more likely to be associated with patient prognosis in prostate cancer. Furthermore, DAGs were over-represented in prostate cancer risk genes identified from previous genome wide association studies. We also found DAGs were enriched in kinase and transcription factor encoding genes. Interestingly, for many of these prioritized kinases their association with racial disparity did not manifest from the original genomic/transcriptomic data but was reflected by their differential phosphorylation levels between AA and CA prostate tumor samples. Similarly, the disparity relevance of some transcription factors was not reflected at the mRNA or protein expression level, but at the activity level as demonstrated by their differential ability in regulating target gene expression. Our integrative analysis provided new candidate targets for improving prostate cancer treatment and addressing the racial disparity problem.
Introduction
Prostate cancer is the second leading cause of cancer-related death among men in the US [1]. The incidence and mortality of prostate cancer vary greatly among races. Specifically, African Americans (AAs) have about 75% higher morbidity and over twice the mortality rate of prostate cancer than Caucasians (CAs) [2]. Additionally, AAs with prostate cancer are usually diagnosed at a late stage and their disease progresses quickly [3,4]. Although external factors such as socioeconomic status, health insurance coverage, and household income have been found to contribute to racial disparity in prostate cancer [5], [6], [7], more research has focused on the impact of internal factors, the molecular features such as somatic mutation and gene expression between AA and CA prostate cancers [8], [9], [10]. Previous studies have shown distinct genetic profiles among AA prostate tumors, including PTEN loss, MYC amplification, and SPINK1 overexpression [10], [11], [12].
Although previous studies have extensively compared molecular features between AAs and CAs [[8], [9], [10],[13], [14], [15], [16]], their results are not always consistent and may even be conflicting. For example, some studies found AAs had higher frequencies of SPOP mutations [10,15], while others found low SPOP mutation frequencies in AAs compared with CAs [8]. Furthermore, there are still debates regarding the clinical implications of these differential molecular features between AA and CA, so it is inconclusive whether these molecular differences are causally related to racial disparity in prostate cancer. As an example, SPINK1 was found to be overexpressed in AA across many studies [8,10,12,15], but its role in the development of prostate cancer remains unclear. Several studies reported no association between SPINK1 expression and poorer clinical outcomes [17], [18], [19], while others found it could predict disease-specific and recurrence-free survival in prostate cancer [20,21].
During tumorigenesis, genetic and epigenetic aberrations of genes will integrate at the pathway level to determine the ultimate phenotypes. Considering race as a phenotype variation, the seemingly inconsistent observations at the gene level between different studies might be explained at the pathway or regulatory network level. Using the SPOP mutation as an example, it is possible that other genes in related pathways also contribute to racial disparity, e.g., the onco-protein AR and SRC in the AR signaling pathway that are regulated by SPOP-mediated degradation [22]. Indeed, it has been reported that differentially expressed genes between AA and CA prostate cancer are enriched in AR-related pathways [23]. Interestingly, differential expression of AR itself between the two races are still under debate [13,24]. Thus, in this study we propose to integrate different types of data (genomic, epigenomic, and transcriptomic) to better understand the inconsistency between them and to identify a core set of genes associated with racial disparity in prostate cancer.
Network-based methods have been widely used to integrate different data types. For example, hotnet2 [25] integrates CNV and somatic mutation data through gene interaction networks and uses a network diffusion approach to identify dysregulated pathways in certain cancer types. Nevertheless, hotnet2 only considers somatic aberration data and ignores other molecular aberrations such as DNA methylation and RNA expression. Therefore, NetICS [26] integrates more data types such as DNA methylation and RNA expression to identify mediator genes within networks, which are affected by upstream genetic aberrations and lead to altered downstream gene expressions. This approach has been proved to accurately predict cancer genes based on TCGA data and might be applied to other scenarios as well.
In this study, we assume that there is a core set of genes that receive the effects of upstream genetic alterations and trigger downstream abnormal gene expression through these networks, thereby driving the racial disparity in prostate cancer. Based on this assumption, we performed a series of comparative analyses between AAs and CAs using multiple genomic data types including copy number variation, somatic mutation, DNA methylation and RNA expression to identify differential altered genes between the two races. Detected differential alterations were further mapped into a protein-protein interaction network on gene level. By applying a random walk strategy to propagate influence of these alterations through the network, we prioritized genes associated with racial disparity and identified a set of 130 core disparity associated genes (DAGs). These DAGs had important clinical implications in prostate cancer, including prognosis, progression and development. Furthermore, we found that these DAGs dysregulated several pathways in prostate cancer, including MAPK signaling and AR signaling pathways. Potential targets for precision therapy may lie in these pathways and DAGs, providing new insights into handling the racial disparity in prostate cancer.
Materials and methods
Datasets
Somatic mutation, copy number variation (CNV), DNA methylation, RNA expression and clinical information for TCGA prostate adenocarcinoma (PRAD) were downloaded through Firehose portal (https://gdac.broadinstitute.org/). Specifically, mutation annotation files were downloaded, which contained mutation status of 24,058 genes for 332 samples. The original CNV data provided segmental chromosome CNVs. We calculated relative CNV for each segment by log2(copy-number/2). A gene's CNV was then determined by averaging the segment CNVs with which it overlaps, weighted by the ratio of overlapping length to the gene's length. Using such method, we obtained relative copy numbers of 23,311 genes for 492 samples. DNA methylation level in genes was represented by the averaged β values of their promoters, covering 21,855 genes in 498 samples. RNA-seq data included 20,502 genes for 497 samples. From the clinical data, we extracted 58 AAs and 415 CAs. Samples other than AA and CAs and those of unknown race were excluded. Protein expression data for prostate cancer were downloaded from TCGA from reverse phase protein array (RPPA) experiment, which included the expression level of 195 proteins in 352 prostate cancer samples (32 AA and 301 CA). For some protein, the dataset provides their abundance in both phosphorylated and unphosphorylated states.
Four processed microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database with accession IDs: GSE6956 [27], GSE17356 [28], GSE41969 [29], GSE91037 [30]. Specifically, the four datasets contained 87 (40 AA and 47 CA), 27 (14 AA and 13 CA), 802 (350 AA and 452 CA) and 26 samples (13 AA and 13 CA) respectively. Gene expression level was represented by expression level of corresponding probesets. For genes with multiple probesets, we selected the probe with maximum average intensity across all samples to represent the gene. Except for GSE91037, the other three provide Gleason scores of patients. Besides, GSE41969 provides age while GSE6956 and GSE 17356 provide tumor stage information. A summary of these datasets can be found in Table S1.
Identification of differential molecular alterations between AA and CA samples
To identify gene mutations with significant different distribution between AA and CA, we used Fisher's exact test to compare their gene mutation frequencies. A P value under 0.05 was considered as significant. Since the majority of genes have relative low mutation frequency and the number of samples are small, the statistical power for detecting variant mutation is low. Thus, we used unadjusted p-value (p<0.05) directly without considering confounding variables to ensure high sensitivity. To identify differential CNV, DNA methylation and mRNA expression between AA and CAs, we used R package “limma” [31]. Specifically, relative copy number, log2 transformed expression level and promoter methylation of genes were taken as inputs. To avoid potential impacts of factors other than race, we also adjusted clinical variables including age and Gleason score with “limma”. Adjusted p values under 0.05 were considered as significant. Since there are more than one mRNA expression dataset, meta-analysis was used to integrate results across datasets and to calculate a meta-p value for each gene based on Fisher's method. In total, we identified 39, 466, 2876 and 1340 genes with differential mutation, CNV, DNA methylation and RNA expression between AA and CAs.
Network propagation
To integrate pre-identified differential altered genes between AA and CAs, we mapped these identified genes into a protein-protein interaction network and applied a network-based approach to prioritize genes associated with racial disparity in prostate cancer. A network with 13,096 genes (nodes) and 137,686 interactions (edges) was built on human interactome data from a previous study [32]. R package “igraph” was used to construct the network [33]. We pruned duplicated edges between nodes and only kept the largest connected components within the network, resulting in 12,972 nodes and 134,800 edges remaining. Pre-identified differentially altered genes between AA and CAs were then mapped to the network as seeds with label “1”, while the rest were labelled “0”. To prioritize genes associated with racial disparity, we simplified the network propagation method developed by Dimitrakopoulos et al. [26], which was based on an insulated heat diffusion process [25]. Specifically, we normalized the adjacency matrix of the network by multiplying the inverse diagonal matrix of node degrees. A diffusion matrix was then defined as:
where W denotes the normalized adjacency matrix and β denotes the restart probability. F represents the diffusion matrix, in which Fij describes the influence of node i applying to node j. To find optimized β, we selected several nodes with different centrality to study the effect of β value on the network. In practice, we selected 5 nodes according to their centrality (minimum, 25% quantile, median, 75% quantile, and maximum) in the network after excluding nodes with zero centrality. As the centrality metric, we used betweenness centrality since it provides better insight into the connectivity of nodes, which plays a major role in propagation scores. We calculated the influence of the 5 nodes applying to their neighbors, and selected β values that maximize the sum of influence of the 5 nodes. Based on our calculations, the influence was maximized when β equals 0.45 (Fig. S1). The final propagation score was calculated as:
where S denotes the vector that contains the initial labels assigned to the nodes.
To determine the statistical significance of each gene's contribution to racial disparity rather than simply using its propagation score, we constructed 100 random networks to determine a false discovery rate (FDR) for each gene. Specifically, each random network was constructed by 100 random edge swaps of the original network, so that node degrees can be preserved. For each gene, its FDR was calculated by taking the proportion of propagation scores from the 100 random networks that were higher than its propagation score in the original network. Using a cut-off as FDR < 0.01, we identified 130 genes as core DAGs. To visualize the interactions among the 130 genes, we created a subnetwork of them by extracting the 130 nodes and related edges between them in the original network using Cytoscape [34].
Disease associated gene sets
Disease associated gene sets were downloaded from DisGeNET [35]. This database not only curated gene-disease associations from UNIPROT, CGI, ClinGen, Genomics England, CTD (human subset), PsyGeNET, and Orphanet, but also text-mined several gene-disease associations from MEDLINE abstracts. In total, 11,181 curated and 17,993 text-mined disease associated gene sets were retrieved. Fisher's exact tests were used to evaluate the enrichment differences of these gene sets between DAGs and non-DAGs. To correct p values from multiple tests, Benjamini-Hochberg method was used.
Gene set enrichment analysis
To investigate associated pathways of DAGs, we performed enrichment analysis of DAGs using KEGG pathways and GO terms. Specifically, 186 KEGG pathways and 10,185 GO terms were downloaded from C2 and C5 collection from MsigDB database [36]. Fisher's exact test was used to calculate statistical significance of enrichment of KEGG pathways and GO terms in DAGs. Benjamini-Hochberg method was used to correct p values from multiple tests.
Gene essentiality analysis
To examine the importance of the 130 DAGs in prostate cell line growth, we investigated gene essentialities of them. Gene essentiality data of prostate cancer cell lines was downloaded from DepMap database [37]. CERES score and dependency probability of 18,119 genes for VCAP cell line was retrieved [38]. The CERES score describes cell line growth rates after knock-out of given genes. Therefore, a low CERES score or a high dependency probability indicates a high essentiality of a gene. Gene essentialities between DAGs and non-DAGs were compared by Wilcoxon rank-sum test.
SNPs associated with prostate cancer
We downloaded a total of 616 risk SNPs for prostate cancer identified by genome-wide association study (GWAS) from NHGRI-EBI GWAS Catalog [39] on 02/14/2020. Associated studies are summarized in Table S2.
The minor allele frequencies of these SNPs in different populations from the 1000 Genomes Project [40] were obtained through The Ensembl Variant Effect Predictor (http://www.ensembl.org/vep) [41]. There are five super populations in the 1000 Genomes Project dataset: African, Ad Mixed American, East Asian, European, South Asian. As African and European populations are genetically closest to AA and CA, we considered minor allele frequencies in the two populations as those in AA and CA. Fisher's exact test was used to compare minor allele frequencies of SNPs between the two races. In total, we compared minor allele frequencies of 6 SNPs (corresponding to TP53, SMAD2, ADAMTSL4, SAMD9, AR, MAP3K1), whose associated genes were within the 130 DAGs.
Survival analysis
To identify a gene list associated with prognosis, we downloaded a dataset (Accession ID: GSE16560) [42] from the GEO database, which contains expression profiles of 6100 genes for 281 samples, and clinical variables including disease-specific survival. We used univariate Cox regression models to assess the prognostic value of each gene based on its continuous expression levels. Because the statistical power is relatively low, with only 13 genes having p values below 0.05 after Benjamini-Hochberg adjustments, we considered unadjusted p values without multi-test corrections below 0.05 as significant to identify more prognostic genes. Specifically, 878 prognostic genes were identified. To examine whether the 130 DAGs are enriched for prognostic genes compared to non-DAGs, Fisher's exact tests were performed. We selected two DAGs (YWHAZ and JUN) as examples to visualize their prognostic value by Kaplan-Meier plots. Specifically, the whole cohort (Accession ID: GSE16560) [42] was dichotomized according to the mean expression value of each gene, and log-rank tests were used to compare survival differences between high and low expression groups.
We used R package “survival” to perform survival analysis. Cox regression models were built using the function "coxph". Comparisons between two survival curves were made by using the "survdiff" function.
Kinase-substrate interactions
To investigate which kinase are enriched for DAGs in its substrates, we downloaded Kinase-substrate dataset PhosphoSitePlus [43]. 12,228 kinase-substrate interactions from humans were retrieved. We examined which kinase substrates were enriched with the identified DAGs using Fisher's exact tests. Kinases with fewer than 20 substrates were excluded. Benjamini-Hochberg adjusted p values below 0.05 were considered significant.
Inference of transcription factor regulatory activity
Transcription factor binding profiles were downloaded from the ChEA dataset [44]. A previously developed rank-based algorithm, BASE, was used to calculate regulatory activities for transcription factors based on the expression profile of their target genes [45]. The Wilcoxon rank-sum test was used to compare transcription factor regulatory activities between AA and CA.
Results
Overview of the study
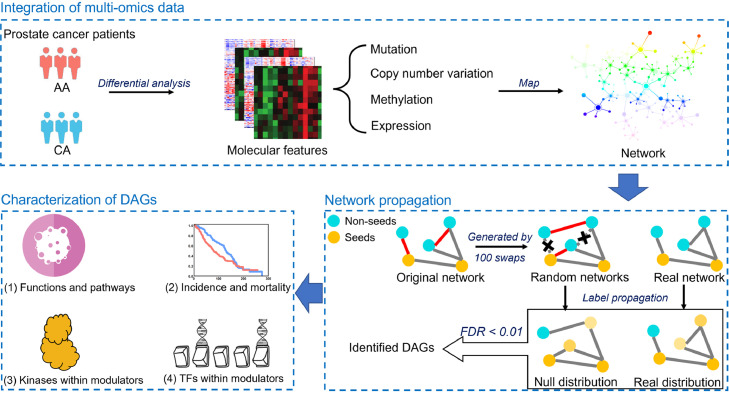
In this study, we collected a comprehensive list of genomic data that were generated to identify differential molecular features between AA and CAs in prostate cancer, including copy number variation (CNV), somatic mutation, DNA methylation, and gene expression. To better understand racial disparity, we employed a network-based method to integrate these data to identify a set of core DAGs using the pipeline shown in Fig. 1. First, for each type of data we performed comparative analyses between AA and CA prostate cancer patients to identify significantly differential molecular features on gene level. For gene expression analysis, multiple datasets were available and meta-analysis was performed to obtain combined results. Next, we integrated these results by projecting them into a curated protein-protein interaction network [32] and then adopted network propagation to prioritize genes. Statistical significance was determined by referring a null distribution obtained from topology-preserved network permutations. We identified a total of 130 DAGs that were significantly associated with racial disparity at the significance level of false discovery rate (FDR) <0.01. Finally, we performed a series of analyses to investigate the 130 DAGs in the context of their associated pathways, their contribution to genetic risk of prostate cancer and their prognostic value. In particular, we found kinases and transcription factors were highly enriched in these DAGs, and thus we investigated their potential functions in prostate cancer.
Fig. 1.
The diagram of this study. We performed comparative analysis to identify four types of differential molecular alterations (somatic mutation, CNV, DNA methylation, mRNA expression) between AA and CA. The detected alterations were then mapped to an interaction network on gene level. We next applied network propagation to prioritize genes associated with racial disparity in prostate cancer. Based on FDR < 0.01, we identified 130 DAGs underlying racial disparity and characterized them in terms of function and pathways, prognostic value as well as kinases and transcription factors within them.
Identification of molecular alterations of different types between AA and CA
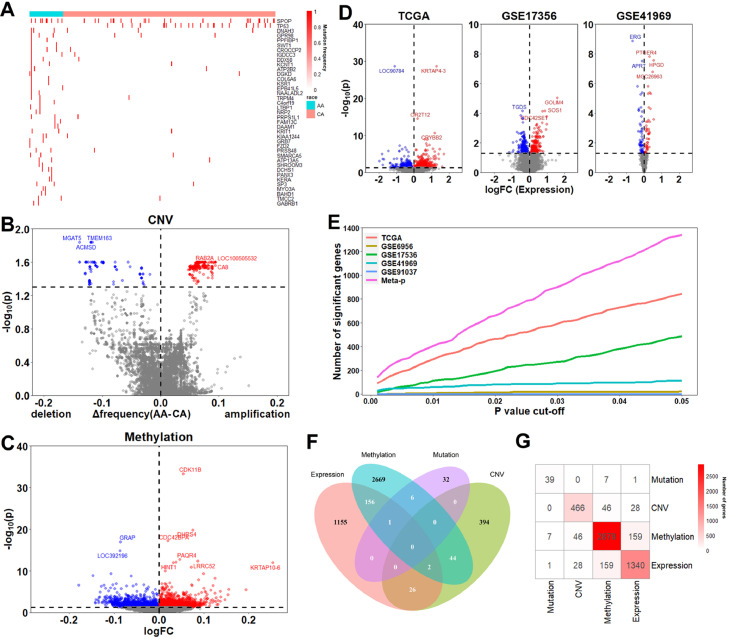
We compared molecular difference between AA and CA prostate cancer in four types of molecular features: somatic mutation, CNV, DNA methylation and RNA expression. We first examined differential somatic mutation events between AA and CAs in the TCGA prostate adenocarcinoma dataset (TCGA-PRAD). We compared number of point mutations of each gene between the two races and identified 39 genes with significantly differential mutation frequency at the significance level of p < 0.05 using fisher's exact test. Out of them, 38 were enriched in AA and 1 was enriched in CA (Fig. 2A). For example, SPOP mutation was found to be enriched in AA (p = 0.008), while TP53 mutation was found to be enriched in CA (p = 0.04). For CNV, we identified 466 differential CNV events between AA and CA at FDR < 0.05 from TCGA-PRAD dataset, of which 139 gene deletions and 327 gene amplifications were enriched in AA after adjusting for age and Gleason score using “limma” [31] (Fig. 2B).
Fig. 2.
Differential molecular alterations between AA and CA. (A–C): Identified differential mutated (A), copy number altered (B) and methylated (C) genes between AA and CA. (D): Differential expressed genes identified from TCGA, GSE17356, GSE41969 dataset. (E): Meta-analysis was used to combine p values from different mRNA expression datasets. (F): Total numbers of the differential altered genes between AA and CA from four data types. (G): Overlap of identified differential altered genes across four data types.
To identify differentially methylated genes between AA and CA, for each gene we calculated the average methylation level (β values) of measured CpG sites that were located in its promoter. Using the TCGA-PRAD data, we identified 2876 differentially methylated genes (1786 hyper- and 1110 hypomethylated in AA) at FDR < 0.05 after adjusting for clinical variables (Fig. 2C).
Using the TCGA-PRAD dataset, we identified 845 differentially expressed genes, with 542 up-regulated and 303 down-regulated in AA (Fig. 2D). In addition to the TCGA dataset, we examined four datasets with expression profiles of AA and CAs [27], [28], [29], [30]. We identified 487, 113, 21 differential expressed genes in GSE17356, GSE41969 and GSE6956 at FDR < 0.05 (Figs. 2D and S2). We didn't find any differential expressed genes in GSE91037, potentially due to its small sample size. Only a few differentially expressed genes were shared across datasets. Specifically, there were only 17 differentially expressed genes shared between TCGA-PRAD and GSE17356, the two datasets with the highest number of identified differentially expressed genes (Fig. S2). These results indicated high inconsistency across these datasets. Because of this, we performed a meta-analysis to combine the results from these datasets. As shown in Fig. 2E, we identified more differential expressed genes with meta-analysis. At the same threshold, 1340 differential expressed genes were identified, with 748 up-regulated and 592 down-regulated in AA across the five datasets.
Taking together, we identified 39, 466, 2878 and 1340 genes associated with differential mutation, CNV, methylation and expression between AA and CA. Specifically, 159 genes showed both differential methylation and expression, 46 genes showed both differential methylation and CNV, and 28 genes showed both differential CNV and expression between AA and CA (Fig. 2F and G, Table. S3). There were only seven genes showing differential methylation and one gene with differential expression within differentially mutated genes because of their small number.
Network-based data integration leads to the identification of 130 core genes associated with racial disparity
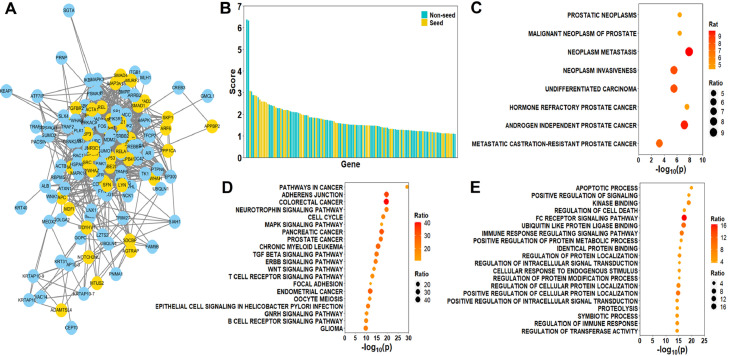
To identify core DAGs, we mapped previously identified genes as seeds into a human protein-protein network and applied a label propagation method to prioritize all genes within the network. Using FDR < 0.01, we identified 130 DAGs (Table S4). These 130 DAGs formed a subnetwork (Fig. 3A). Within them, 52 were seeds and 78 were non-seeds. Seeds and non-seeds had similar distributions of propagation scores, non-seeds did not necessarily have lower scores than seeds (Fig. 3B). Specifically, the AR gene, which drives tumorigenesis and progression of prostate cancer [46], was within the 130 DAGs, even though it's not a seed. Indeed, AR was previously reported to differ in related signaling pathways [23] and to correlate differently with carcinogen [24] between AA and CA, despite no differences in expression levels. These results suggested that network- based methods identified latent genes associated with racial disparity, which were not readily apparent from comparative analyses alone.
Fig. 3.
Functions and pathways of DAGs. (A): Subnetwork of the 130 DAGs. (B): Distribution of scores for the 130 DAGs. (C): DAGs were enriched with cancer-related disease gene sets. (Fisher's exact test) (D and E): DAGs were enriched with cancer-related and signaling transduction pathways from KEGG (D) and GO (E) term enrichment analysis. (Fisher's exact test).
The functions and pathways of DAGs
We then examined the function of these DAGs in known human diseases. To this end, we retrieved disease associated gene sets from DisGeNet [35]. As shown, gene sets associated with prostate cancer such as “Prostatic neoplasms” and “Malignant neoplasms of prostate cancer” were highly enriched in DAGs, four times compared to non-DAGs (Fig. 3C). Furthermore, gene sets associated with cancer severity such as “Neoplasm metastasis”, “Neoplasm invasiveness” and “Undifferentiated carcinoma” were also enriched in DAGs, 8–15 times more than non-DAGs (Fig. 3C). Specifically, gene sets related to castration-resistant prostate cancer (CRPC), the most lethal subtype of prostate cancer, such as “Hormone refractory prostate cancer”, “Androgen independent prostate cancer” and “Metastatic castration-resistant prostate cancer”, were all significantly enriched in DAGs (Fig. 3C). As previous studies reported AA and CA patients responded differently to CRPC treatment [47], DAGs might explain such racial disparity in CRPC treatments.
Next, we investigated which pathways the DAGs were involved in. As expected, we observed the DAGs were enriched for some cancer pathways including prostate cancer from KEGG pathways (Fig. 3D). Besides, several kinases-involved signaling pathways such as MAPK, TGF-β, ERBB and WNT pathways were found to be enriched in DAGs, 10–25 times compared to non-DAGs. In addition, cell cycle related pathways were also enriched in DAGs. We found similar results using enrichment analysis of Gene Ontology (GO) terms. Signaling-related GO terms (positive regulation of signaling and kinase binding) and cell cycle related GO terms (apoptotic process and regulation of cell death) were enriched in DAGs 3 to 6 times more than non-DAGs (Fig. 3E).
Association of DAGs with incidence and mortality in prostate cancer
To elucidate potential mechanisms by which these DAGs contribute to racial disparity in prostate cancer, we characterized them in several clinical implications.
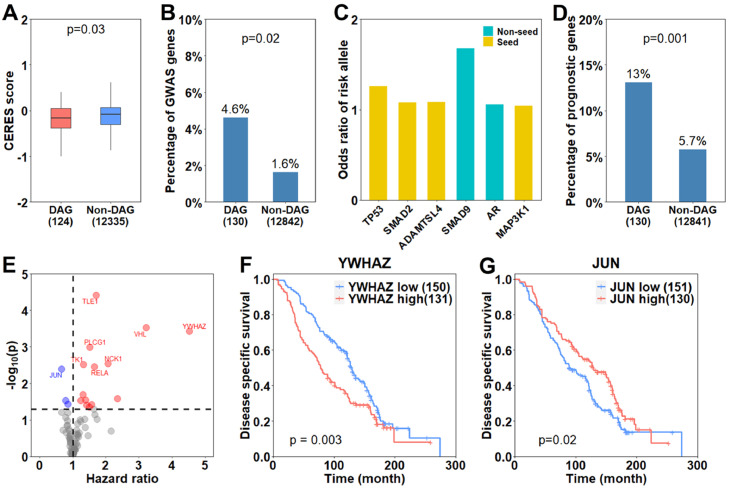
First, we examined gene essentiality of these DAGs in prostate cancer cell lines. The gene essentialities were downloaded from DepMap database [37], represented by CERES score and dependency probability. Low CERES scores or high dependency probabilities imply high essentiality of a gene for prostate cancer cell line growth. We observed that the DAGs tend to have lower CERES scores and higher dependency probabilities compared to non-DAGs (Figs. 4A and S3). Our results indicated that DAGs were more likely to be essential in prostate cancer compared to non-DAGs.
Fig. 4.
Association of DAGs with incidence risk and mortality of prostate cancer. (A): DAGs have higher essentialities compared to non-DAGs, represented by lower CERES score (Wilcoxon rank-sum test). (B): DAGs were enriched for GWAS genes compared to non-DAGs (Fisher's exact test). GWAS gene: genes corresponding to prostate cancer risk SNPs in GWAS studies. (C): Genetic risk of the six GWAS genes within a DAG as measured by corresponding risk allele's odds ratio. (D): The DAGs were enriched for prognostic genes compared to non-DAGs (Fisher's exact test). (E): Hazard ratio and corresponding p values for the prognostic DAGs. (F and G): Survival difference between patients with high and low expression of YWHAZ (F) and JUN (G) with mean as cut-off (log-rank test).
Second, we investigated association of the DAGs with prostate cancer incidence. To this end, we retrieved 559 genes (GWAS genes) associated with risk SNPs from genome-wide association studies (GWAS) [39]. We found that the DAGs were enriched with GWAS genes nearly three times to non-DAGs (p = 0.02, Fig. 4B), indicating potential roles of DAGs in prostate cancer incidence. Totally, there were six GWAS genes in DAGs, containing both seed and non-seed genes. The incidence risk of corresponding risk alleles of these six genes were represented by odds ratio as shown in Fig. 4C. Given AA and CA have different incidence rates of prostate cancers [2], we next examined if those DAGs, which were also GWAS genes, exhibited different allele frequencies between the two races. As shown, the six genes being both DAGs and GWAS genes significantly differed in their minor allele frequencies between AA and CA (Fig. S4A–F).
Third, we examined prognostic values of these DAGs. Specifically, we retrieved 878 genes prognostic for disease specific survival in prostate cancer from the GSE16560 dataset [42] at p < 0.05. As shown in Fig. 4D, the DAGs were enriched for more than twice as many prognostic genes as non-DAGs (p = 0.001). Out of the 17 prognostic DAGs, 3 were protective and 14 were hazardous (Fig. 4E). Among them, there are both seeds and non-seeds. For example, a seed gene, YWHAZ, which is associated with prostate cancer progression and cell proliferation [48], [49], [50], was in the 14 hazardous DAGs (Fig. 4F). Furthermore, non-seed genes such as JUN, an onco-gene regulates apoptosis [51], were also within the prognostic DAGs (Fig. 4G).
Taken together, these results suggested that the DAGs can affect incidence and mortality, which in turn might contribute to the racial disparity in prostate cancer.
Differential activity of disparity associated kinases between AA and CAs
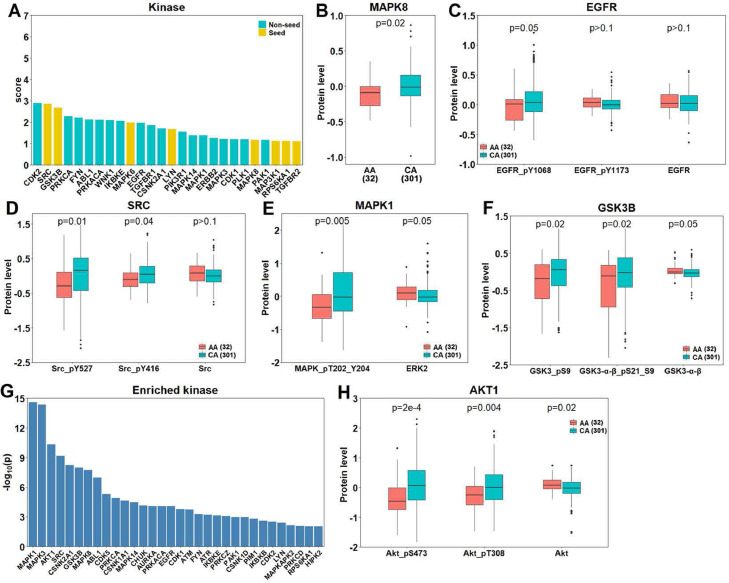
Kinases play important roles in signaling transduction and regulate a variety of cellular processes. Several dysregulated kinases and associated pathways have been observed in prostate cancer, including overexpression of SRC and elevated PI3K-AKT-mTOR pathway activity [52,53]. Therefore, we examined potential roles of kinases within the DAGs in racial disparity. There were 26 kinases as DAGs, with 8 seeds and 18 non-seeds (Fig. 5A). Using TCGA reverse phase protein array (RPPA) data, we compared protein levels of these kinases between AAs and CAs. As shown, AA has significantly higher levels of phosphorylated MAPK8 than CAs (Fig. 5B). In addition to phosphorylated kinases, TCGA RPPA includes some unphosphorylated kinases, allowing comparison of kinase protein levels both in the phosphorylated (activated) and unphosphorylated (inactivated) states between AA and CA. We found some kinases with comparable levels of inactivated states between AA and CA, but significant differences of activated states, such as EGFR and SRC (Fig. 5C and D). Furthermore, there were also some kinases that differed in AA and CA in opposite directions between inactivated and activated states, such as MAPK1 and GSK3B (Fig. 5E and F). To investigate whether the differences of kinase levels between AA and CA would remain significant after adjusting for clinical variables, we constructed a generalized linear model using kinase levels, age and Gleason score. Our results indicated the differences in protein levels of aforementioned kinases between AA and CA remain significant, even after taking into account other clinical variables (Table S5).
Fig. 5.
Kinases within DAGs exhibited differential activity states between AA and CA. (A): Propagation score of 26 kinases within DAGs. (B–F): Protein levels of MAPK8 (B), EGFR (C), SRC (D), MAPK1 (E) and GSK3B (F) between AA and CA (Wilcoxon rank-sum test). The position of the phosphorylated amino acid in a protein is followed by "_p". (G): 32 enriched kinases with the DAGs (Fisher's exact test). (H): Protein levels of AKT1 between AA and CA (Wilcoxon rank-sum test).
Since a kinase can phosphorylate many substrates including other kinases as well as itself [54], [55], [56], we next examined which kinases were enriched for DAGs in their substrate using a kinase-substrate dataset from PhosphoSitePlus [43]. At a threshold of adjusted p values below 0.01, we identified 32 kinases significantly enriched for the DAGs in their substrates, including 18 DAGs (Fig. 5G). Although not a DAG, AKT1 exhibited a very significant p value, ranking third, right after the two DAGs: MAPK1 and MAPK3. Indeed, AKT1 showed significantly differential activities between AA and CA, with a more elevated activity in the latter (Fig. 5H). This result remained significant after adjusting for the same clinical variables (Table S5). Taken together, our results suggested that the AA and CA differed significantly in activation states of kinases, especially those within DAGs, which may lead to differential activity of kinase-directed signaling transduction pathways between the two races, thereby contributing to the racial disparity in prostate cancer.
Differential activities of disparity associated transcription factors between AA and CA
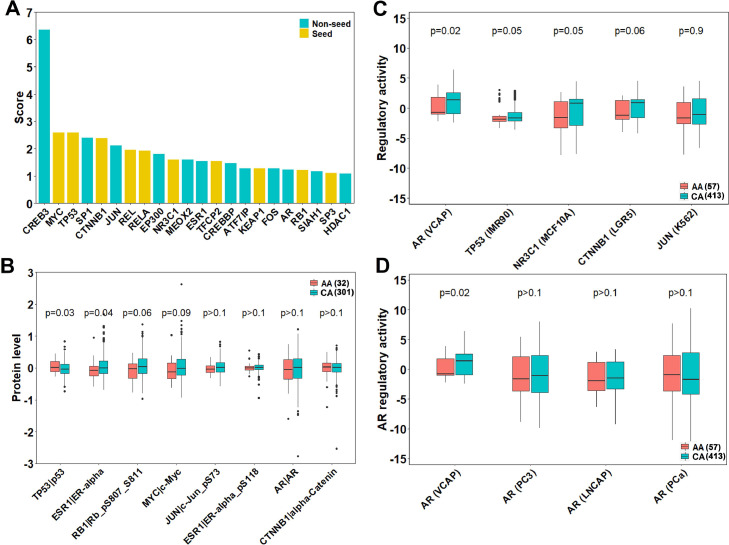
Transcription factors regulate the expression of many genes, which in turn affects cellular growth, division and apoptosis. Dysregulation of key transcription factors (AR, ERG) has been reported to contribute to tumorigenesis and progression of prostate cancer [57,58]. Therefore, we also examined potential roles of transcription factors within the DAGs in racial disparity. A total of 22 transcription factors were found within the DAGs, with 10 seeds and 12 non-seeds (Fig. 6A). Using the TCGA RPPA dataset, we compared protein levels of 7 transcription factors within DAGs between AA and CA. AA was found with higher protein levels of TP53 (p = 0.03) and lower protein levels of ESR1 (p = 0.04), RB1 (p = 0.06) and MYC (p = 0.09) compared to CA (Fig. 6B). Interestingly, phosphorylated ESR1 along with JUN, AR and CTNNB1 had comparable protein levels between AA and CA (Fig. 6B). After adjusting for the same clinical variables with a generalized linear model, TP53 still remained significantly different in protein levels between AA and CA (Table S6).
Fig. 6.
Transcription factors within DAGs exhibited differential regulatory activities between AA and CA. (A): Propagation score of 22 transcription factors within DAGs. (B): Protein levels of the transcription factors within DAGs between AA and CA. Protein names of corresponding genes were after “|”. For phosphorylated proteins, positions of the phosphorylated amino acids were after “_p”. (C): Regulatory activities of the transcription factors between AA and CA. Names of cell lines from which the binding profile used to calculate the regulatory activity were indicated in parentheses. (D): AR regulatory activities calculated by different binding profiles from multiple prostate cancer cell lines between AA and CA. P values were calculated by Wilcoxon rank-sum tests.
In addition to protein expression levels, mutations and alternative splicing can also affect the regulatory activity of transcription factors [59], [60], [61], which in turn impacts a number of downstream pathways. Hence, we subsequently compared regulatory activities of transcription factors within DAGs between AA and CA. Using a previously developed deconvolutional algorithm [45], we calculated transcription factor regulatory activities based on TCGA PRAD gene expression profile and transcription factor binding profiles from the ChEA dataset [44]. In contrast to protein levels, TP53 exhibited significantly higher regulatory activities in AA instead of CA (Figs. 6C and S5), indicating different mechanisms in dysregulation of TP53 downstream pathways between the two races. While the TCGA RPPA data didn't include the protein level of NR3C1, its regulatory activity did differ significantly between the two races (Fig. 6C). Furthermore, while AR, CTNNB1, and JUN did not differ in protein levels between the two races, we still observed significant or weakly significant differences in their regulatory activities (Fig. 6C). Specifically, the differences in AR regulatory activities between AA and CA remained significant even after adjusting for the same clinical variables as previously described (Table S7).
As a key driver in tumorigenesis and development of prostate cancer [46], AR has been investigated in multiple prostate cancer cell lines and tumor tissues in terms of its binding profiles [62], [63], [64], [65]. The ChEA dataset includes four AR binding profiles measured from different prostate cancer cell lines or tissues (VCAP, PC3, LNCAP and tumor tissue). We calculated and compared regulatory activities in AA and CA with the four different profiles. As shown, only AR regulatory activities calculated using the corresponding binding profile from VCAP were significantly different between AA and CA (Fig. 6D). Out of the three cell lines, VCAP was the only one that can express AR-V7 protein, a splice variant of AR, which was rare in primary prostate cancer but common in CRPC and can render resistance to anti-androgen drugs [66,67]. Based on this result, there may be differences in alternative splicing events between AA and CA, resulting in differential delays to advanced prostate cancer stages.
Discussion
To investigate potential DAGs underlying racial disparity in prostate cancer, we integrated differential molecular alterations between AA and CA from different data types and applied a network-based approach to identify 130 DAGs. Subsequent characterizations revealed these DAGs were highly associated with patient prognosis as well as tumorigenesis, development and progression in prostate cancer. Furthermore, kinases and transcription factors that act as DAGs differed in activity state or regulatory activity between AA and CA, although some of them were comparable at the protein level. These results suggested that, with the network-based integration method, we detected some new racial disparity associated genes in prostate cancer that weren't detectable by common comparative analysis due to their similar expression levels.
We observed AA had significantly lower protein levels of activated AKT1, MAPK1 and GSK3B but higher levels of their inactivated state proteins compared to CA. Indeed, an elevated PI3K/AKT/mTOR signaling pathway, containing GSK3B, is observed in many prostate cancer cases [68,69]. An activated MAPK pathway is often found in CRPCs and is associated with resistance to anti-androgen drugs [69,70]. Based on these results, it might be more important to also consider kinase activity instead of only comparing mRNA or protein expression. Similarly, AR has significantly different regulatory activities regardless of similar protein levels between the two races. More importantly, we can observe the difference only by calculating regulatory activity based on the AR-binding profiles of the VCAP cell line that expresses AR-V7 isoform [66]. AR-V7 is highly expressed in CRPC and associated with resistance to anti-androgen therapy [67]. This result highlights the potential different alternative splicing of AR between the two races, which in turn leads to racial disparity.
Interestingly, we observed significant different levels of unphosphorylated ESR1 but similar levels of its phosphorylated form between AA and CA. As ESR1 exhibits different allele frequencies between AA and CA [71], there may already be effects of differential variants within races, in which case dysregulation of associated downstream pathways at phosphorylation level might not be necessary.
It should be noticed that both AA and CA populations might have diverse genetic backgrounds. Simply categorizing patients into AA and CA may overlook admixture from multiple genetic ancestries. Different proportion of other ancestries within AA population will also confound current results. Besides, the sample sizes between AA and white are imbalanced, which may reduce the statistical power. Therefore, we used meta-analysis to combine multiple datasets and network-based methods to integrate differentially altered genes between AA and CA from different data types. Although these methods can compensate to some extent for the relatively low statistical power, a larger and more balanced dataset would be more desirable. Similarly, due to low mutation frequencies of the majority of genes, we used unadjusted p values without considering other confounding variables to identify differential mutated genes. Such an approach improves sensitivity, but it also leads to the identification of some genes that are not differentially mutated. In addition, we showed the DAGs were highly associated with prognosis. However, there's no race information within that dataset. So differential association of DAGs with survival between the two races will need to be further examined.
Most importantly, our results were generated based on solely computational method. Even though the DAGs identified can be validated with high essentiality and association with prostate cancer incidence using cell line and GWAS data, experimental validation will be needed to evaluate the specific role they play in prostate cancer. For example, knock-out animal studies can be performed to examine role of these DAGs in vivo. The clinical value of DAGs can be explored through the use of drugs that target these DAGs in cell line or animal models. Besides, by using a network-based approach, we lost the direction of differential molecular alterations. Considering the identified DAGs alone, we are unable to identify their roles in prostate cancer or the direction they contributed to racial disparity, whether they increased risk for AAs or decreased risk for CA. Furthermore, the label propagation method only considered the presence of the differential molecular alterations and overlooked how much they differ between AA and CA. Although such method allows integrating results from different data types with different distributions, it will somewhat amplify the effect of relatively less significant molecular alterations and reduce the effect of extremely significant molecular alterations. Last, the ChEA dataset [44] contains a limited number of transcription factor binding profiles, so many of the profiles we used to calculate regulatory activity were not from prostate cancer cell lines. These results would be verified and consolidated using profiles directly from prostate cancer cell lines.
Conclusion
In summary, we applied a network-based algorithm to integrate differential molecular alterations between AA and CA and identified 130 DAGs. By characterizing these DAGs, we observed significant differential downstream pathway activities of MAPK, AKT and AR. Targeting these pathways may advance precision medicine, and ultimately benefits AA as well as other ethnicities.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Data availability
The data that support the findings of this study are openly available from Gene Expression Omnibus repository (GSE6956, GSE17356, GSE41969, GSE91037 and GSE16560).
CRediT authorship contribution statement
Baoyi Zhang: Formal analysis, Investigation, Validation, Visualization, Writing – original draft. Kevin Yao: Writing – review & editing. Chao Cheng: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgements
This work is supported by the Cancer Prevention Research Institute of Texas (CPRIT) (RR180061 to CC) and the National Cancer Institute of the National Institutes of Health (1R21CA227996 to CC). CC is a CPRIT Scholar in Cancer Research.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101327.
Appendix. Supplementary materials
Reference
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C.E., Miller K.D., Goding Sauer A., Jemal A., Siegel R.L. Cancer statistics for African Americans, 2019. CA Cancer J. Clin. 2019;69:211–233. doi: 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 3.Ravery V., Dominique S., Hupertan V., Ben Rhouma S., Toublanc M., Boccon-Gibod L., et al. Prostate cancer characteristics in a multiracial community. Eur. Urol. 2008;53:533–538. doi: 10.1016/j.eururo.2007.04.048. [DOI] [PubMed] [Google Scholar]
- 4.Powell I.J., Bock C.H., Ruterbusch J.J., Sakr W. Evidence supports a faster growth rate and/or earlier transformation to clinically significant prostate cancer in black than in white American men, and influences racial progression and mortality disparity. J. Urol. 2010;183:1792–1796. doi: 10.1016/j.juro.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weprin S.A., Parker D.C., Jones J.D., Kaplan J.R., Giusto L.L., Mydlo J.H., et al. Association of low socioeconomic status with adverse prostate cancer pathology among African American men who underwent radical prostatectomy. Clin. Genitourin. Cancer. 2019;17:e1054–e1059. doi: 10.1016/j.clgc.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Mahal B.A., Ziehr D.R., Aizer A.A., Hyatt A.S., Sammon J.D., Schmid M., et al. Getting back to equal: the influence of insurance status on racial disparities in the treatment of African American men with high-risk prostate cancer. Urol. Oncol. 2014;32:1285–1291. doi: 10.1016/j.urolonc.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Ziehr D.R., Mahal B.A., Aizer A.A., Hyatt A.S., Beard C.J., D Amico A.V., et al. Income inequality and treatment of African American men with high-risk prostate cancer. Urol. Oncol. 2015;33:18.e7–18.e13. doi: 10.1016/j.urolonc.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Khani F., Mosquera J.M., Park K., Blattner M., O'Reilly C., MacDonald T.Y., et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin. Cancer Res. 2014;20:4925–4934. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Zheng S.L., Na R., Wei L., Sun J., Gallagher J., et al. Distinct genomic alterations in prostate tumors derived from African American men. Mol. Cancer Res. 2020;18:1815–1824. doi: 10.1158/1541-7786.MCR-20-0648. [DOI] [PubMed] [Google Scholar]
- 10.Koga Y., Song H., Chalmers Z.R., Newberg J., Kim E., Carrot-Zhang J., et al. Genomic profiling of prostate cancers from men with African and European ancestry. Clin. Cancer Res. 2020;26:4651–4660. doi: 10.1158/1078-0432.CCR-19-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W., Zheng S.L., Na R., Wei L., Sun J., Gallagher J., et al. Distinct genomic alterations in prostate tumors derived from African American men. Mol. Cancer Res. 2020;18:1815–1824. doi: 10.1158/1541-7786.MCR-20-0648. [DOI] [PubMed] [Google Scholar]
- 12.Faisal F.A., Sundi D., Tosoian J.J., Choeurng V., Alshalalfa M., Ross A.E., et al. Racial variations in prostate cancer molecular subtypes and androgen receptor signaling reflect anatomic tumor location. Eur. Urol. 2016;70:14–17. doi: 10.1016/j.eururo.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaston K.E., Kim D., Singh S., Ford O.H., Mohler J.L. Racial differences in androgen receptor protein expression in men with clinically localized prostate cancer. J. Urol. 2003;170:990–993. doi: 10.1097/01.ju.0000079761.56154.e5. [DOI] [PubMed] [Google Scholar]
- 14.Shuch B., Mikhail M., Satagopan J., Lee P., Yee H., Chang C., et al. Racial disparity of epidermal growth factor receptor expression in prostate cancer. J. Clin. Oncol. 2004;22:4725–4729. doi: 10.1200/JCO.2004.06.134. [DOI] [PubMed] [Google Scholar]
- 15.Yuan J., Kensler K.H., Hu Z., Zhang Y., Zhang T., Jiang J., et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang F.W., Mosquera J.M., Garofalo A., Oh C., Baco M., Amin-Mansour A., et al. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discov. 2017;7:973–983. doi: 10.1158/2159-8290.CD-16-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavin R., Pettersson A., Hendrickson W.K., Fiorentino M., Finn S., Kunz L., et al. SPINK1 protein expression and prostate cancer progression. Clin. Cancer Res. 2014;20:4904–4911. doi: 10.1158/1078-0432.CCR-13-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang K.-C., Evans A., Donnelly B., Bismar T.A. SPINK1 overexpression in localized prostate cancer: a rare event inversely associated with ERG expression and exclusive of homozygous PTEN deletion. Pathol. Oncol. Res. 2017;23:399–407. doi: 10.1007/s12253-016-0119-9. [DOI] [PubMed] [Google Scholar]
- 19.Faisal F.A., Kaur H.B., Tosoian J.J., Tomlins S.A., Schaeffer E.M., Lotan T.L. SPINK1 expression is enriched in African American prostate cancer but is not associated with altered immune infiltration or oncologic outcomes post-prostatectomy. Prostate Cancer Prostatic Dis. 2019;22:552–559. doi: 10.1038/s41391-019-0139-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Yin X., Shen P., Sun G., Yang Y., Liu J., et al. The association between SPINK1 and clinical outcomes in patients with prostate cancer: a systematic review and meta-analysis. Onco. Targets Ther. 2017;10:3123–3130. doi: 10.2147/OTT.S127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M.H., Ross A.E., Alshalalfa M., Erho N., Yousefi K., Glavaris S., et al. SPINK1 defines a molecular subtype of prostate cancer in men with more rapid progression in an at risk, natural history radical prostatectomy cohort. J. Urol. 2016;196:1436–1444. doi: 10.1016/j.juro.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Song Y., Ye M., Dai X., Zhu X., Wei W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat. Rev. Urol. 2020;17:339–350. doi: 10.1038/s41585-020-0314-z. [DOI] [PubMed] [Google Scholar]
- 23.Wang B.-D., Yang Q., Ceniccola K., Bianco F., Andrawis R., Jarrett T., et al. Androgen receptor-target genes in African American prostate cancer disparities. Prostate Cancer. 2013;2013 doi: 10.1155/2013/763569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neslund-Dudas C.M., McBride R.B., Kandegedara A., Rybicki B.A., Kryvenko O.N., Chitale D., et al. Association between cadmium and androgen receptor protein expression differs in prostate tumors of African American and European American men. J. Trace Elem. Med. Biol. 2018;48:233–238. doi: 10.1016/j.jtemb.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leiserson M.D.M., Vandin F., Wu H.-.T., Dobson J.R., Eldridge J.V., Thomas J.L., et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimitrakopoulos C., Hindupur S.K., Häfliger L., Behr J., Montazeri H., Hall M.N., et al. Network-based integration of multi-omics data for prioritizing cancer genes. Bioinformatics. 2018;34:2441–2448. doi: 10.1093/bioinformatics/bty148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace T.A., Prueitt R.L., Yi M., Howe T.M., Gillespie J.W., Yfantis H.G., et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 28.Timofeeva O.A., Zhang X., Ressom H.W., Varghese R.S., Kallakury B.V.S., Wang K., et al. Enhanced expression of SOS1 is detected in prostate cancer epithelial cells from African-American men. Int. J. Oncol. 2009;35:751–760. [PMC free article] [PubMed] [Google Scholar]
- 29.Powell I.J., Dyson G., Land S., Ruterbusch J., Bock C.H., Lenk S., et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol. Biomarkers Prev. 2013;22:891–897. doi: 10.1158/1055-9965.EPI-12-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann B., Acosta A.M., Richards Z., Deaton R., Sapatynska A., Murphy A., et al. Association of high miR-182 levels with low-risk prostate cancer. Am. J. Pathol. 2019;189:911–923. doi: 10.1016/j.ajpath.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. limma Powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347 doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csardi, G., Nepusz, T. The igraph software package for complex network research:9.
- 34.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., et al. Defining a cancer dependency map. Cell. 2017;170:564–576. doi: 10.1016/j.cell.2017.06.010. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DepMap 20Q2 public [internet]. figshare; 2020 [cited 2021 Oct 22]. Available from: https://figshare.com/articles/dataset/DepMap_20Q2_Public/12280541/4.
- 39.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sboner A., Demichelis F., Calza S., Pawitan Y., Setlur S.R., Hoshida Y., et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med. Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng C., Yan X., Sun F., Li L.M. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinform. 2007;8:452. doi: 10.1186/1471-2105-8-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huggins C., Hodges C.V. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin. 1972;22:232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 47.Bitting R.L., Goodman M., George D.J. Racial disparity in response to prostate cancer systemic therapies. Curr. Oncol. Rep. 2020;22:96. doi: 10.1007/s11912-020-00966-z. [DOI] [PubMed] [Google Scholar]
- 48.Yu C.-C., Chen L.-C., Lin W.-H., Lin V.C., Huang C.-Y., Lu T.-L., et al. Genetic association analysis of cell cycle regulators reveals YWHAZ has prognostic significance in prostate cancer. Cancer Genomics Proteomics. 2020;17:209–216. doi: 10.21873/cgp.20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lage-Vickers S., Bizzotto J., Valacco M.P., Sanchis P., Nemirovsky S., Labanca E., et al. The expression of YWHAZ and NDRG1 predicts aggressive outcome in human prostate cancer. Commun. Biol. 2021;4:103. doi: 10.1038/s42003-020-01645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murata T., Takayama K., Urano T., Fujimura T., Ashikari D., Obinata D., et al. 14-3-3ζ, a novel androgen-responsive gene, is upregulated in prostate cancer and promotes prostate cancer cell proliferation and survival. Clin. Cancer Res. 2012;18:5617–5627. doi: 10.1158/1078-0432.CCR-12-0281. [DOI] [PubMed] [Google Scholar]
- 51.Bossy-Wetzel E., Bakiri L., Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asim M., Siddiqui I.A., Hafeez B.B., Baniahmad A., Mukhtar H. Src kinase potentiates androgen receptor transactivation function and invasion of androgen-independent prostate cancer C4-2 cells. Oncogene. 2008;27:3596–3604. doi: 10.1038/sj.onc.1211016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carver B.S., Chapinski C., Wongvipat J., Hieronymus H., Chen Y., Chandarlapaty S., et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maddika S., Ande S.R., Wiechec E., Hansen L.L., Wesselborg S., Los M. Akt-mediated phosphorylation of CDK2 regulates its dual role in cell cycle progression and apoptosis. J. Cell Sci. 2008;121:979–988. doi: 10.1242/jcs.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X.Q., Sun P., Paller A.S. Inhibition of integrin-linked kinase/protein kinase B/Akt signaling: mechanism for ganglioside-induced apoptosis. J. Biol. Chem. 2001;276:44504–44511. doi: 10.1074/jbc.M106563200. [DOI] [PubMed] [Google Scholar]
- 56.Beenstock J., Mooshayef N., Engelberg D. How do protein kinases take a selfie (autophosphorylate)? Trends Biochem. Sci. 2016;41:938–953. doi: 10.1016/j.tibs.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 57.Culig Z., Santer F.R. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. 2014;33:413–427. doi: 10.1007/s10555-013-9474-0. [DOI] [PubMed] [Google Scholar]
- 58.Adamo P., Ladomery M.R. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2016;35:403–414. doi: 10.1038/onc.2015.109. [DOI] [PubMed] [Google Scholar]
- 59.Mäkinen N., Heinonen H.-.R., Moore S., Tomlinson I.P.M., van der Spuy Z.M., Aaltonen L.A. MED12 exon 2 mutations are common in uterine leiomyomas from South African patients. Oncotarget. 2011;2:966–969. doi: 10.18632/oncotarget.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.-P., et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Wang Y., Rao X., Wang Y., Feng W., Liang H., et al. Roles of alternative splicing in modulating transcriptional regulation. BMC Syst. Biol. 2017;11:89. doi: 10.1186/s12918-017-0465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei G.-H., Badis G., Berger M.F., Kivioja T., Palin K., Enge M., et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin B., Wang J., Hong X., Yan X., Hwang D., Cho J.-.H., et al. Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS ONE. 2009;4:e6589. doi: 10.1371/journal.pone.0006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urbanucci A., Sahu B., Seppälä J., Larjo A., Latonen L.M., Waltering K.K., et al. Overexpression of androgen receptor enhances the binding of the receptor to the chromatin in prostate cancer. Oncogene. 2012;31:2153–2163. doi: 10.1038/onc.2011.401. [DOI] [PubMed] [Google Scholar]
- 65.Guseva N.V., Rokhlin O.W., Bair T.B., Glover R.B., Cohen M.B. Inhibition of p53 expression modifies the specificity of chromatin binding by the androgen receptor. Oncotarget. 2012;3:183–194. doi: 10.18632/oncotarget.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharp A., Coleman I., Yuan W., Sprenger C., Dolling D., Rodrigues D.N., et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Invest. 2019;129:192–208. doi: 10.1172/JCI122819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C., et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hermida M.A., Dinesh Kumar J., Leslie N.R. GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv. Biol. Regul. 2017;65:5–15. doi: 10.1016/j.jbior.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 69.Shorning B.Y., Dass M.S., Smalley M.J., Pearson H.B. The PI3K-AKT-mTOR pathway and prostate cancer: at the crossroads of AR, MAPK, and WNT signaling. Int. J. Mol. Sci. 2020;21:E4507. doi: 10.3390/ijms21124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bluemn E.G., Coleman I.M., Lucas J.M., Coleman R.T., Hernandez-Lopez S., Tharakan R., et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489. doi: 10.1016/j.ccell.2017.09.003. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernández J., Balic I., Johnson-Pais T.L., Higgins B.A., Torkko K.C., Thompson I.M., et al. Association between an estrogen receptor alpha gene polymorphism and the risk of prostate cancer in black men. J. Urol. 2006;175:523–527. doi: 10.1016/S0022-5347(05)00240-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available from Gene Expression Omnibus repository (GSE6956, GSE17356, GSE41969, GSE91037 and GSE16560).