Abstract
The primary research approach in pharmacogenetics has been candidate gene association studies (CGAS), but pharmacogenomic genome-wide association studies (GWAS) are becoming more common. We are now at a critical juncture when the results of those two research approaches, CGAS and GWAS, can be compared in pharmacogenetics. We analyzed publicly available databases of pharmacogenetic CGAS and GWAS (i.e., the Pharmacogenomics Knowledgebase [PharmGKB®] and the NHGRI-EBI GWAS catalog) and the vast majority of variants (98%) and genes (94%) discovered in pharmacogenomic GWAS were novel (i.e., not previously studied CGAS). Therefore, pharmacogenetic researchers are not selecting the right candidate genes in the vast majority of CGAS, highlighting a need to shift pharmacogenetic research efforts from CGAS to GWAS.
Keywords: : candidate gene association study, genome-wide association study, pharmacogenetic, pharmacogenomic, research paradigm
Adverse drug reactions are estimated to be the fourth leading cause of death in the USA – ahead of pulmonary disease (before the COVID-19 pandemic), diabetes, AIDS, pneumonia, accidents and automobile deaths [1]. Almost 2.5 million serious adverse drug reactions occur in hospitalized and nursing home patients in the USA every year [1]. In addition to adverse drug reactions, inadequate drug efficacy is a pervasive problem as well. As many as 86% of patients with diabetes, 68% with hypertension and 51% at risk for coronary artery disease fail to meet their drug treatment goals [2–4]. Therefore, there is a critical need to discover effective predictors of drug efficacy and toxicity, or millions of patients will continue to suffer unnecessarily. Multiple factors contribute to drug efficacy and toxicity, such as kidney and liver function, drug–drug interactions and comorbid conditions, but more recent evidence also supports the role of genetics. Heritability is the amount of variation explained by genetics versus environment, and the heritability of different drug responses ranges from 25 to 79% [5]. Therefore, genetics explains a proportion of every drug response and in some cases, genetics explains the majority of drug response.
Pharmacogenetics is the field of research and clinical practice that focuses on the influence of genetics on drug responses. The ultimate goal of pharmacogenetics is ‘precision medicine,’ in which genetic testing is used to individually tailor drug regimens to maximize drug efficacy and/or minimize drug toxicity. The field began over 60 years ago [6], and since then, the primary research approach in pharmacogenetics has been candidate gene association studies (CGAS) [7]. CGAS only analyze a few genes and variants based on a priori knowledge that they have an important role in the drug’s pharmacology. Commonly used candidates in pharmacogenetics include genes encoding the drug’s receptor or pathway downstream from the drug’s receptor (i.e., pharmacodynamic candidates); genes involved in the absorption, distribution, metabolism or excretion of the drug (i.e., pharmacokinetic candidates); or genes involved in the immune system (i.e., hypersensitivity candidates). CGAS have yielded some great successes in pharmacogenetics, such as significantly improving the safety and/or efficacy of the drugs abacavir (gene: HLA-B), clopidogrel (gene: CYP2C19), thiopurines (gene: TPMT) and antidepressants (genes: CYP2C19 and CYP2D6) [6].
By only analyzing a few genes and variants, the major limitation of CGAS is that they can miss many other potentially important genes and variants. Genome-wide association studies (GWAS) have typically overcome that limitation by analyzing all approximately 21,000 protein-coding genes and approximately 1,000,000 independent and common variants in the human genome. A highly successful example of the value of pharmacogenomic GWAS is the discovery of a variant in the gene for nudix hydrolase 15 (NUDT15) that was associated with thiopurine-induced leukopenia. Thiopurines are metabolized by thiopurine methyltransferase (gene: TPMT), and thus decades of CGAS focused on TPMT and identified variants associated with thiopurine-induced myelosuppression [8]. Those variants in TPMT are less common in patients of Asian ancestry than those with European ancestry, but thiopurine-induced leukopenia is still more common in patients with Asian ancestry. Therefore additional, yet undiscovered, factors contributed to thiopurine-induced myelosuppression in Asian ancestry patients. A GWAS of Korean patients discovered a variant in NUDT15 (p.Arg139Cys; rs116855232) that was significantly associated with thiopurine-induced leukopenia (odds ratio = 35.6; p = 4.88 × 10-94) [9]. In Koreans, this variant demonstrated 89% sensitivity and 93% specificity for predicting thiopurine-induced leukopenia. The NUDT15 variant was also strongly associated with thiopurine-induced leukopenia in subjects of European ancestry (odds ratio = 9.50; p = 4.64 × 10-4). As a result, current clinical practice guidelines by the National Comprehensive Cancer Network (NCCN) and the Clinical Pharmacogenetics Implementation Consortium (CPIC) now recommend using NUDT15 variants in addition to TPMT in therapy with thiopurines [8,10]. After decades of CGAS, a GWAS made the discovery of this NUDT15 variant possible.
CGAS were the necessary approach in the first few decades of pharmacogenetic research. The technologies required to perform GWAS were not available until the past two decades (e.g., whole-genome arrays, high-performance computing clusters and GWAS software). With the increased feasibility of GWAS, hundreds of pharmacogenomic GWAS have now been published in addition to the thousands of pharmacogenetic CGAS [11–13]. As a result, we are now at a critical juncture when the results of these two research approaches, GWAS and CGAS, can be compared in pharmacogenetics. Therefore, the purpose of this paper is to provide a perspective on the most appropriate research approach for pharmacogenetic discovery: CGAS versus GWAS. This comparison is critical to determine whether pharmacogenetic researchers are investing their time and effort in the right genes and variants to minimize adverse drug reactions and maximize drug efficacy.
Pharmacogenomic GWAS versus CGAS
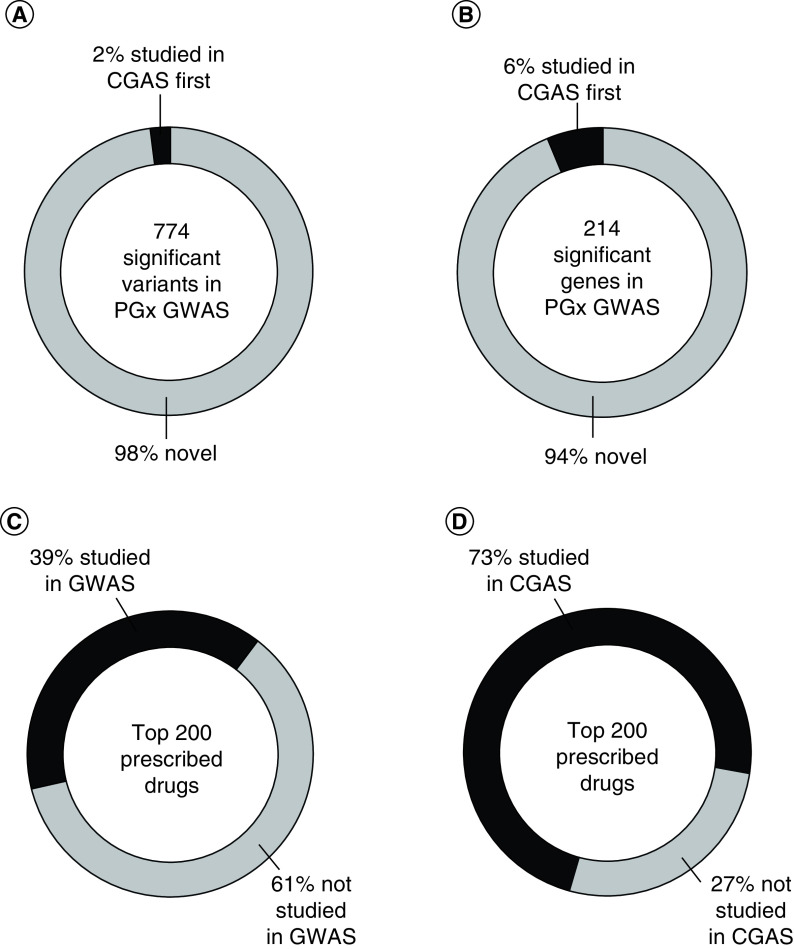
The GWAS catalog, maintained by the National Human Genome Research Institute (NHGRI) and the European Bioinformatics Institute (EBI) [14], curates all GWAS of human data and the results are publicly available for download. Currently in the NHGRI-EBI GWAS catalog (downloaded 25 January 2020), there are 774 variants significantly associated with the response to a drug (with p < 5.0 × 10-8) [14]. The Pharmacogenomics Knowledgebase (PharmGKB®) curates most pharmacogenetic studies (which are mostly CGAS), and its variant annotations are also publicly available for download [15]. Currently in the PharmGKB (downloaded 18 March 2020), there are 6245 variants studied for an association with the response to a drug. We compared the list of 774 significant variants from pharmacogenomic GWAS to the 6245 variants studied in CGAS in PharmGKB, by omitting any GWAS annotated in PharmGKB and matching by drug and response (i.e., efficacy or toxicity). Only 18 of the 774 significant GWAS variants (2%) were previously studied in CGAS for the same drug and response. Therefore, the vast majority (98%) of significant variants discovered in pharmacogenomic GWAS were not previously studied candidate variants (Figure 1A). (Data are available within the Supplementary Material).
Figure 1. . Percentages of novel variants and genes discovered in pharmacogenetic GWAS and the percentage of the top 200 prescribed drugs studied in GWAS.
Percentages of (A) pharmacogenetic variants and (B) genes identified by GWAS with p < 5.0 × 10-8 that were first studied in CGAS and the percentages of the top 200 prescribed drugs studied in (C) GWAS and (D) CGAS.
CGAS: Candidate gene association study; GWAS: Genome-wide association study; PGx: Pharmacogenetic.
Those 774 variants identified in pharmacogenomic GWAS annotate to 214 genes. We compared the list of 214 significant genes from pharmacogenomic GWAS to the 1568 genes annotated in PharmGKB, by omitting any GWAS annotated in PharmGKB and matching by drug and response (i.e., efficacy or toxicity). Only 13 of the 214 significant GWAS genes (6%) were previously studied in CGAS for the same drug and response. Therefore, the vast majority (94%) of significant genes discovered in pharmacogenomic GWAS were not previously studied candidate genes (Figure 1B).
Each variant was categorized into one of three effect categories (see Supplementary Material): pharmacokinetic/pharmacodynamic ([PK/PD]; i.e., the variant or gene likely affects the function or expression of known metabolic enzymes, drug transporters or the drug’s receptor); response (i.e., the mechanism of the variant or gene for affecting drug efficacy is less clear than known PK/PD and it may involve effects downstream of the drug’s receptor or the underlying pathophysiology of the disease the drug is intended to treat); toxicity (i.e., the variant or gene may or may not be related to PK/PD, but the main end point is toxicity as opposed to efficacy). When comparing the types of variants first studied by CGAS versus those first discovered by GWAS, CGAS more often identified variants directly involved in a priori known PK/PD; whereas, GWAS are discovering more variants with less clear mechanisms related to drug response and toxicity (Figure 2).
Figure 2. . Types of variants first studied in (A) pharmacogenetic CGAS versus those discovered first in (B) pharmacogenomic GWAS.
Each variant was categorized into one of three categories (see Supplementary Material): PK/PD; (i.e., the variant or gene likely affects the function or expression of known metabolic enzymes, drug transporters or the drug’s receptor); response (i.e., the mechanism of the variant or gene for affecting drug efficacy is less clear than known PK/PD, and it may involve effects downstream of the drug’s receptor or the underlying pathophysiology of the disease the drug is intended to treat); toxicity (i.e., the variant or gene may or may not be related to PK/PD, but the main end point is toxicity as opposed to efficacy).
CGAS: Candidate gene association study; GWAS: Genome-wide association study; PK/PD: Pharmacokinetic/pharmacodynamic.
The top 200 prescribed drugs
Recognizing the potential of pharmacogenomic GWAS, we compared the current list of the top 200 prescribed drugs [16] to the drugs studied in pharmacogenomic GWAS (in the NHGRI-EBI GWAS catalog) [14] and in pharmacogenomic CGAS (annotated in PharmGKB) [15]. Only 78 of the top 200 prescribed drugs (39%) have been studied in a GWAS; whereas, 145 of the top 200 drugs (73%) have been studied in CGAS (Figure 1C & D). Therefore, the number of the most commonly prescribed drugs that have been studied in GWAS is severely lagging behind the number studied in CGAS.
Discussion
This was a simple analysis and it is subject to limitations, but the results are so dramatic that we cannot deny we are not selecting the right genes or variants in pharmacogenetic CGAS. It seems that selection of the correct candidate genes is no more likely than if by chance. If we want to find the genes and variants that are the most important for decreasing adverse drug reactions and maximizing drug efficacy, then we need to shift the pharmacogenetic research paradigm from CGAS to GWAS. The recent increased feasibility of GWAS has led to more GWAS overall [12], but the number of pharmacogenomic GWAS has severely lagged behind [12]. Ten-times more GWAS focused on diseases and other traits (e.g., height) have been published compared with pharmacogenomic GWAS [12]. Despite this increased feasibility of GWAS, most pharmacogenetic research today still remains as CGAS [15]. A major limiting factor is probably the cost of performing a GWAS is greater than performing CGAS. This is because GWAS usually require larger sample sizes, more extensive genotyping and computational time and resources. It is much more difficult to find sample sizes large enough to perform pharmacogenomic GWAS, in other words, a large sample of patients, that are all treated with the same drug for the same indication and with genomic data available [11]. Another potential explanation is that CGAS are more appropriate for pharmacogenetics because the pharmacology of a drug is typically well characterized. Obvious candidate genes for pharmacogenetics should include pharmacodynamic, pharmacokinetic or hypersensitivity genes. However, when evaluating the types of variants first discovered in GWAS versus those first studied in CGAS, our findings showed that variants directly involved in a priori known PK/PD are actually a very small minority (5%) of the types of variants being discovered in pharmacogenomic GWAS. This suggests that drug response is far more complex than initially thought, and the top genes and variants in pharmacogenomics are usually unpredictable.
In our assessment of the types of studies being performed for the top 200 prescribed drugs, it is not exactly clear why so many more of those drugs have been studied in CGAS than GWAS. The reasons for this could be the same as for why there are fewer pharmacogenomic GWAS in general, for example, GWAS are more expensive to perform than CGAS and it is difficult to find large sample sizes with the necessary data for pharmacogenomic GWAS. We speculate that more scientists with the skills necessary to perform pharmacogenomic GWAS are needed in our workforce. CGAS can typically be performed with traditional statistical approaches (i.e., in most ways, variants in CGAS statistical analyses can be treated similarly as any other variable), software (i.e., CGAS can be performed with traditional statistical software such as SAS, SPSS, etc.) and hardware (i.e., CGAS can be performed on a typical desktop or laptop computer). On the other hand, scientists who perform pharmacogenomic GWAS require a unique combination of skills. In addition to the knowledge and skills necessary in clinical pharmacology, scientists performing pharmacogenomic GWAS also need advanced computational skills, such as the analysis with software and packages specific to GWAS, programming and the ability to run analyses on high performance computing clusters. Pharmacogenomic education programs should focus on the skills necessary to perform GWAS.
The research paradigm in complex disease and trait genomics has already shifted away from CGAS to GWAS. In doing so, disease and trait GWAS have discovered novel genes and variants not previously suspected in CGAS. A successful example of that paradigm shift is with the genomics of coronary artery disease (CAD). Decades of CGAS identified numerous genes and variants associated with CAD [17]. However, those candidate genes and variants could not be validated [18,19]. In contrast, GWAS discovered novel variants associated with CAD, that were not previously suspected in CGAS [19], and those variants have been repeatedly validated [20]. Another example involves CGAS of patient outcomes after blood or marrow transplant (BMT) [21]. For over a decade, researchers conducted CGAS in mostly small and heterogeneous datasets to identify 36 non-HLA candidate genes and 45 variants associated with patient survival after allogenic BMT. However, none of those candidate genes nor variants replicated in the first adequately powered and well-characterized dataset [21]. Acknowledging these examples outside the field of pharmacogenomics, our findings suggest that a similar story applies to pharmacogenetics as well: we are usually not selecting the right genes and variants in CGAS.
Unlike those CAD and BMT examples, in which none of the candidate variants nor genes replicated in GWAS, there are a few examples in which pharmacogenetic candidate variants were replicated in subsequent GWAS. The complete list of these 18 variants can be found in the Supplementary Material (tab ‘PGx_CGAS_First’) and it includes a variety of types of genes (e.g., the drug’s receptor, metabolizing enzyme or transporter). Warfarin is an example for which four different variants in three different genes studied in CGAS for affecting warfarin response and/or toxicity were replicated in subsequent GWAS (i.e., CYP4F2*3, VKORC1 -1639 G >A and CYP2C9*2 and *3). Three different variants studied in CGAS of statin response were replicated in subsequent GWAS (PCSK9 rs11591147, SLCO1B1 rs4149056 [521 T >C] and APOE rs7412 [E2]). When compared at the gene level instead of the variant level, genes first studied in CGAS had somewhat slightly better success at being replicated in subsequent GWAS, but that is still a very small minority of the novel genes discovered by pharmacogenomic GWAS.
GWAS are not perfect, and there will certainly be situations in which CGAS are more appropriate than GWAS in pharmacogenomics. The major limitation of most GWAS is that they have very low statistical power. GWAS analyze all approximately 1,000,000 independent variants in the genome, and that means a GWAS makes approximately 1,000,000 statistical tests. Stringent control of the type I error is needed to limit the number of false positives. Thus, very large sample sizes are usually necessary for GWAS to compensate for that reduction in statistical power. CGAS make much fewer statistical tests and, therefore, do not need as large of sample sizes. Large sample sizes are especially difficult in pharmacogenomics if the drug response is rare (e.g., severe adverse drug reactions like torsades de pointes). Another limitation of GWAS is that they typically identify an associated region of the genome, which can include multiple variants which may or may not be causative, and the causative variant(s) must be identified in follow-up studies. In contrast, CGAS typically analyze variants that are a priori hypothesized to be a causative variant. GWAS are not well suited to detect variants that are not common in the population (i.e., variants present in less than 5% of the population); whereas, CGAS can better target uncommon variants. For example, carboxylesterase 1 (CES1) is the most abundant drug-metabolizing enzyme in human livers and it is responsible for the metabolism of a wide range of drugs [24]. A loss-of-function variant in CES1, G143E (rs71647871), significantly affects the pharmacokinetics and pharmacodynamics of many different drugs [24]. However, even with extensive genotype imputation, G143E is usually not covered by most commercially available arrays because of its low minor allele frequency (∼2–4%) [24]. Thus, the CES1 G143E variant is an example of a pharmacogenetic variant that is better suited for CGAS. Whole-genome arrays that are used for GWAS do not cover some pharmacokinetic and pharmacodynamic variants very well [25,26], that is due to variation in the number of copies of those genes in the population and the presence of similar pseudogenes, whereas, CGAS can use targeted assays to capture variants missed by whole-genome arrays [25,26]. The phenotypes for pharmacogenomic GWAS can be more challenging to collect than for other types of GWAS. For example, to ideally measure response to a drug, the phenotype must be measured both before and the after the drug exposure, and often that type of data are not collected.
Researchers are developing approaches to overcome some of these challenges of pharmacogenomic GWAS. Approaches to increase the likelihood of identifying the causative variants in GWAS include exome-based GWAS [22] (i.e., GWAS of only the protein coding regions of the genome) and statistical fine mapping [23]. One approach to overcome the challenge of the large sample sizes required for GWAS is to form large consortia [12]. Consortia focused on selective serotonin reuptake inhibitors, metformin, warfarin and clopidogrel have made pharmacogenomic GWAS for those drugs possible [12]. Another approach is to leverage large-scale genomic biobanks that can link prescription, genomic and outcomes data in large samples of participants [13]. Although, it is worth noting that large-scale genomic biobanks have limitations that can still make pharmacogenomic analyses challenging. For example, large-scale biobanks typically enroll broad populations regardless of their health status and drug therapy. Thus, the sample size for a specific pharmacogenomic analysis for a particular drug and/or indication can be still be limited. Moreover, pharmacogenomic analyses of large-scale biobanks are usually dependent on prescription data, which does not accurately represent drug exposure or adherence (e.g., in a 12 month period, actual medication exposure is estimated to be 43% of the prescribed exposure) [27]. On such a large scale, diagnosis codes are usually relied upon for phenotype identification, but diagnosis codes are not always accurate (accuracy can range from approximately 50 to 98%) [28]. Biobanks usually have protocols in place in order for scientists to have access to the data, but the process of cleaning prescription data to perform pharmacogenomic GWAS is a challenging task (i.e., filtering out the drug(s) of interest from extensive prescription histories, standardizing drug doses across the same drugs within a class and filtering out multiple other drugs that can be involved in drug–drug interactions).
Pharmacogenomic GWAS may have an advantage over disease and trait GWAS in regards to power. Some pharmacogenomic GWAS have been able to detect significant associations with very small sample sizes, due to the variants having very large effect sizes [11]. For example, a GWAS of flucloxacillin-induced liver injury was able to detect a significant association with the HLA-B*5701 variant in only 51 cases and 282 controls [29]. This was possible because the effect size was extremely large (odds ratio = 80.6; p = 9.0 × 10-19). Thousands of years of human evolution have led variants with large effects on disease or traits to be rare among the human population. However, most drugs have only been invented in the past 100 years. Thus, unlike disease genomics, it is possible that variants with strong effects on drug response can still be common in the population; and therefore, detectable in relatively smaller GWAS. Another approach that pharmacogenomic researchers are using to overcome the sample size limitations for GWAS is the use of polygenic scores [30]. Polygenic scores aggregate the individually small effects of multiple variants from GWAS into a single score with a cumulatively larger effect size, and thus the effects of polygenic scores are more detectable in small sample sizes [30].
Conclusion
The vast majority of pharmacogenomic GWAS are discovering novel genes and variants that were not previously studied in pharmacogenetic CGAS. This suggests that we are not selecting the right genes and variants in pharmacogenetic CGAS. Many more of the most commonly prescribed drugs have been studied in CGAS than GWAS. Acknowledging the limitations of GWAS, we need to dramatically shift our discovery efforts in pharmacogenomics from CGAS to GWAS. There are some situations in which CGAS may still be necessary or appropriate (e.g., the variant or phenotype is rare), but in the vast majority of cases, CGAS should only be used as follow-up investigation of GWAS findings. Current data shows that we are almost guaranteed to discover novel genes and variants compared with the candidates that are currently being studied.
Future perspective
In 5–10 years, we expect that the research paradigm in pharmacogenetics will shift even further to pharmacogenomics, in other words, less CGAS will be performed and more GWAS will be performed. The field of pharmacogenetics has had several major successful examples discovered in the past via CGAS, in other words, single genes or variants with large enough effect sizes to be clinically meaningful. However, those early successes were the ‘low hanging fruit’, and thus genomic approaches, such as GWAS coupled with polygenic scores, will become the norm in the future for pharmacogenomics. Many of these successful pharmacogenomic examples have now been published [13], but genomic approaches will become more widespread.
Executive summary.
Background
The primary research approach in pharmacogenetics has been candidate gene association studies (CGAS), but pharmacogenomic genome-wide association studies (GWAS) are becoming more common.
We are now at a critical juncture when the results of those two research approaches, CGAS and GWAS, can be compared in pharmacogenetics.
Pharmacogenomic GWAS versus CGAS
The vast majority of variants (98%) and genes (94%) discovered in pharmacogenomic GWAS were novel (i.e., not previously studied CGAS).
The top 200 prescribed drugs
Only 78 of the top 200 prescribed drugs (39%) have been studied in a GWAS; whereas, 145 of the top 200 drugs (73%) have been studied in CGAS.
Discussion
Pharmacogenetic researchers are not selecting the right candidate genes and variants in the vast majority of CGAS.
The research paradigm in disease and trait genomics has already shifted successfully from CGAS to GWAS.
Pharmacogenetic researchers have developed strategies for overcoming the limitations of GWAS.
Conclusion
We need to dramatically shift our discovery efforts in pharmacogenomics from CGAS to GWAS.
There are some situations in which CGAS may still be necessary or appropriate, but in the vast majority of cases, CGAS should only be used as follow-up investigation of GWAS findings.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/pgs-2021-0108
Financial & competing interests disclosure
JA Luzum is supported by a NHLBI K08 award (K08HL146990). HL McLeod is on the Board of Directors of Vyant Bio (Nasdaq: VYNT) and is a co-Founder of Clariifi and Interpares Biomedicine. HL McLeod is a medical advisor to EviCORE and Pharmazam, as well as an expert consultant to Illumina. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.US FDA. Preventable adverse drug reactions: a focus on drug interactions (2018). https://www.fda.gov/drugs/drug-interactions-labeling/preventable-adverse-drug-reactions-focus-drug-interactions#ADRs:%20Prevalence%20and%20Incidence
- 2.Ali MK, Bullard KM, Saaddine JB, Cowie CC, Imperatore G, Gregg EW. Achievement of goals in US diabetes care, 1999–2010. N. Engl. J. Med. 368(17), 1613–1624 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Chow CK, Teo KK, Rangarajan S et al. Prevalence, awareness, treatment, and control of hypertension in rural and urban communities in high-, middle-, and low-income countries. JAMA 310(9), 959–968 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Akyea RK, Kai J, Qureshi N, Iyen B, Weng SF. Sub-optimal cholesterol response to initiation of statins and future risk of cardiovascular disease. Heart 105(13), 975–981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roden DM, Wilke RA, Kroemer HK, Stein CM. Pharmacogenomics: the genetics of variable drug responses. Circulation 123(15), 1661–1670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirmohamed M. Pharmacogenetics: past, present and future. Drug Discov. Today 16(19–20), 852–861 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat. Rev. Genet. 4(12), 937–947 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Relling MV, Schwab M, Whirl-Carrillo M et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin. Pharmacol. Ther. 105(5), 1095–1105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang SK, Hong M, Baek J et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat. Genet. 46(9), 1017–1020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Despite decades of candidate gene association studies focusing on TPMT in thiopurine-induced leukopenia, this genome-wide association study (GWAS) discovered that variants in NUDT15 are associated with thiopurine-induced leukopenia.
- 10.National Comprehensive Cancer Netword. Pediatric acute lymphoblastic leukemia. (2020) https://www.nccn.org/professionals/physician_gls/pdf/ped_all.pdf
- 11.Daly AK. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 11(4), 241–246 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Giacomini KM, Yee SW, Mushiroda T, Weinshilboum RM, Ratain MJ, Kubo M. Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat. Rev. Drug Discov. 16(1), 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describes the challenges of pharmacogenomic GWAS and compares them to disease and trait GWAS.
- 13.McInnes G, Yee SW, Pershad Y, Altman RB. Genomewide association studies in pharmacogenomics. Clin. Pharmacol. Ther. 110(3), 637–648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper is an in-depth and current evaluation of pharmacogenomic GWAS.
- 14.Buniello A, MacArthur JAL, Cerezo M et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47(D1), D1005–d1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whirl-Carrillo M, McDonagh EM, Hebert JM et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92(4), 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ClinCalc.com. ClinCalc DrugStats Database (2021). https://clincalc.com/DrugStats/Top200Drugs.aspx
- 17.Casas JP, Cooper J, Miller GJ, Hingorani AD, Humphries SE. Investigating the genetic determinants of cardiovascular disease using candidate genes and meta-analysis of association studies. Ann. Hum. Genet. 70(Pt 2), 145–169 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA 297(14), 1551–1561 (2007). [DOI] [PubMed] [Google Scholar]; • Shows that previously studied candidate genes for coronary artery disease could not be validated.
- 19.Samani NJ, Erdmann J, Hall AS et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357(5), 443–453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• After years of candidate gene association studies focused on other genes and variants, this genome-wide association study discovered a novel loci, chromosome 9p21.3, as associated with the development of coronary artery disease.
- 20.Schunkert H, Götz A, Braund P et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation 117(13), 1675–1684 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows that the chromosome 9p21.3 loci that was discovered by GWAS is robustly replicated.
- 21.Karaesmen E, Rizvi AA, Preus LM et al. Replication and validation of genetic polymorphisms associated with survival after allogeneic blood or marrow transplant. Blood 130(13), 1585–1596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandi MT, Williams MS, van der Spek P, Koromina M, Patrinos GP. Exome-wide analysis of the DiscovEHR cohort reveals novel candidate pharmacogenomic variants for clinical pharmacogenomics. Genes (Basel) 11(5), 561–572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19(8), 491–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Her L, Zhu HJ. Carboxylesterase 1 and precision pharmacotherapy: pharmacogenetics and nongenetic regulators. Drug Metab. Dispos. 48(3), 230–244 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters EJ, McLeod HL. Ability of whole-genome SNP arrays to capture ‘must have’ pharmacogenomic variants. Pharmacogenomics 9(11), 1573–1577 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Gamazon ER, Skol AD, Perera MA. The limits of genome-wide methods for pharmacogenomic testing. Pharmacogenet. Genomics 22(4), 261–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowan CG, Flory J, Gerhard T et al. Agreement and validity of electronic health record prescribing data relative to pharmacy claims data: a validation study from a US electronic health record database. Pharmacoepidemiol. Drug Saf. 26(8), 963–972 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Burns EM, Rigby E, Mamidanna R et al. Systematic review of discharge coding accuracy. J. Public Health (Oxf.) 34(1), 138–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daly AK, Donaldson PT, Bhatnagar P et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 41(7), 816–819 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Lanfear DE, Luzum JA, She R et al. Polygenic score for β-blocker survival benefit in European ancestry patients with reduced ejection fraction heart failure. Circ. Heart Fail. 13(12), e007012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Successful example of how pharmacogenetic researchers can use polygenic scores to overcome the low power of GWAS which typically assess the individual effects of genetic variants.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.