Abstract
Amplified-fragment length polymorphism (AFLP) analysis with the endonucleases BglII and MfeI was used to genotype 91 Campylobacter jejuni subsp. jejuni strains from outbreaks and sporadic cases. AFLP-generated fragments were labeled with fluorescent dye and separated by capillary electrophoresis. The software packages GeneScan and GelCompar II were used to calculate AFLP pattern similarities and to investigate phylogenetic relationships among the genotyped strains. The AFLP method was compared with two additional DNA-based typing methods, pulsed-field gel electrophoresis (PFGE) using SmaI and restriction fragment length polymorphism analysis on PCR products (PCR-RFLP) of the flaA and flaB genes. We found that AFLP analysis of C. jejuni strains is a rapid method that offers better discriminatory power than do both PFGE and PCR-RFLP. AFLP and, to a lesser extent, PCR-RFLP could differentiate strains within the same PFGE profiles, which also makes PCR-RFLP an alternative to PFGE. We were able to clearly distinguish 9 of 10 recognized outbreaks by AFLP and to identify similarities among outbreak and sporadic strains. Therefore, AFLP is suitable for epidemiological surveillance of C. jejuni and will be an excellent tool for source identification in outbreak situations.
Bacteria belonging to the genus Campylobacter are regarded worldwide as the most frequent cause of food-borne human gastroenteritis (19, 36).
For epidemiologic characterization of Campylobacter isolates, two serotyping methods are commonly used (Penner and Lior methods) (25, 33). However, additional methods are needed for outbreak investigation and phylogenetic studies since the majority of Campylobacter isolates belong to a limited number of serotypes (30) and many strains remain nontypeable.
Typing procedures based on DNA analysis have gained considerable interest in recent years. It has, however, proved difficult to combine speed and simplicity with high discriminative power and reproducibility. Often there are considerable difficulties in interpreting the results of these methods. The pulsed-field gel electrophoresis (PFGE) method, which comprises agarose gel separation of large endonuclease-digested fragments, has proven to be both discriminatory and reproducible. PFGE has high discriminatory power (26) and has proven useful in outbreak situations (14, 31). However, PFGE is rather time-consuming compared to other DNA-based methods. The randomly amplified polymorphic DNA (RAPD) assay is based on random PCR amplification of DNA fragments by short (typically 10-bp) primers. The location and number of annealing sites for the RAPD primers will vary between different strains, thus creating different patterns when strains are analyzed by gel electrophoresis. The RAPD technique is fast and easy to perform, but it has proven difficult to reproduce stable RAPD patterns between experiments (15). Decreased reproducibility compromises the discriminatory power (16) and makes the exchange of data between laboratories difficult. The flagellin genes of several strains of Campylobacter have been sequenced, and all of the studied strains contain two flagellin genes names flaA and flaB, which share about 92% homology (2, 9, 10) and are both necessary to produce a fully active flagellar filament (9). These genes have proven to be suitable targets for genotyping involving PCR and restriction fragment length polymorphism (RFLP) analysis of the resulting fragments (2, 3, 29), which are rapid and low-cost genotyping assays.
The amplified-fragment length polymorphism (AFLP) method is a promising typing technique for several bacterial species (1, 5–7, 18–22, 24). The AFLP method generates fingerprints from DNA of both eukaryotic and prokaryotic origin without any prior knowledge of the sequence. The method is reviewed by Janssen et al. (17) and Savelkoul et al. (35). The AFLP method is comparatively rapid, and with samples run on capillary electrophoresis or sequencing instruments with internal size markers, fragments can be separated with a 1-bp size difference (24). The resulting fingerprint patterns are stored in digital form, which means that they can easily be exported to analysis software or be exchanged with other laboratories. AFLP fingerprinting of Campylobacter spp. was first reported by Kokotovic and On (22) and later by Duim et al. (6). Recently, a computer-assisted analysis of genotyping methods for Campylobacter jejuni and Campylobacter coli including AFLP was published (4).
We compared the discriminatory power of AFLP, PFGE, and PCR-RFLP for genotyping a selection of sporadic and outbreak-related C. jejuni strains. In addition, we built a searchable database for AFLP fingerprint profiles from C. jejuni isolates.
MATERIALS AND METHODS
Bacterial strains.
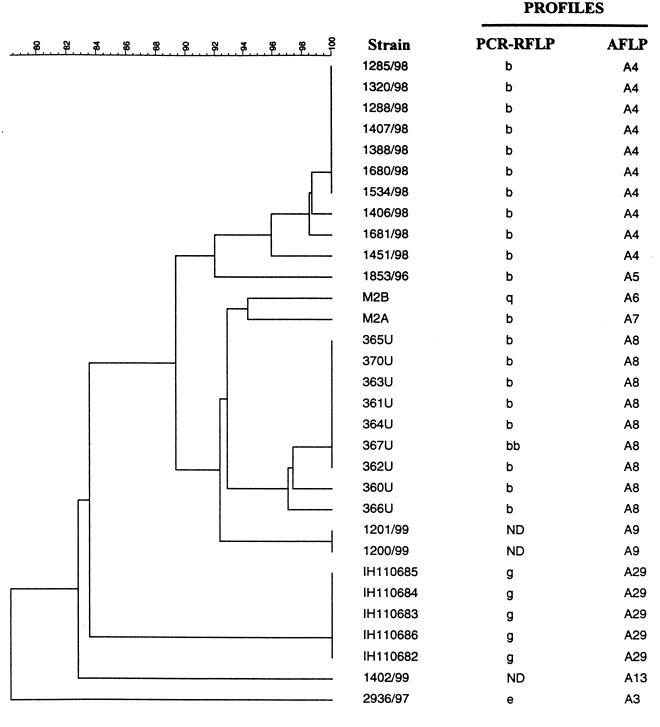
In all, 91 Campylobacter jejuni subsp. jejuni isolates, where 85 strains belong to biotype 1 and 6 strains (142/96, 911/96, 14368U, 14363/81U, 14360U, and 956/97) belong to biotype 2, were used. Isolates include 58 outbreak and 30 sporadic strains with 87 human clinical isolates and 4 isolates from other sources (Fig. 1). Strains were obtained from the strain collection at the National Reference Laboratory for Enteropathogenic Bacteria at the National Institute of Public Health, Oslo, Norway. Furthermore, strains from one suspected and nine recognized outbreaks in Norway, as well as one Finnish outbreak, were included (12, 23, 27, 28).
FIG. 1.
Dendrogram made from the AFLP fragment patterns of all the analyzed strains with their corresponding PFGE, PCR-AFLP, and AFLP profile names. Different outbreaks are numbered and are from different regions and years as follows: 1, southwest Norway, 1998; 2, north Norway, 1988; 3, Lake Mjøsa annual bicycle race, 1999; 4, north Norway, 1981; 5, lake Mjøsa annual bicycle race, 1997; 6, central Norway, 1998; 7, south Norway, 1997; 8, central Norway, 1995; 9, south Norway, 1997; 10, Finland, 1998. ND, not done.
AFLP fingerprinting.
Bacterial genomic DNA was extracted using a commercial kit (Easy-DNA; Invitrogen BV, Leek, The Netherlands). The AFLP reaction was performed as detailed previously (22, 38) using the BglII and MfeI restriction endonucleases (21). This enzyme combination and AFLP assay should theoretically give 22 fragments in the range of 50 to 500 bp according to the published sequence of C. jejuni NCTC11168 (32) and analysis with Lasergene software (DNASTAR Inc., Madison, Wis.). PCRs were performed as described elsewhere (22). The PCR product was diluted 1:2, and 1 μl was used for capillary electrophoresis on an ABI-310 Genetic Analyzer (PE Biosystems, Foster City, Calif.) with POP4-polymer and GeneScan TAMRA-500 as internal standard in each sample (PE Biosystems).
PFGE genotyping.
Standard methods for SmaI macrorestriction and PFGE were used (37). Bacterial cells were treated with formaldehyde to inhibit DNase activity (8). The DNA fragments were separated in 1% SeaKem GTG agarose (FMC, Rockland, Maine) with 0.25× modified Tris-borate-EDTA buffer for 25 h at 350 V and 12°C, with pulse times from 1 to 16 s using a Beckman Gene Line II electrophoresis unit (Beckman, Fullerton, Calif.).
PCR-RFLP genotyping.
PCR-RFLP was performed essentially as described by Ayling et al. (3) from DNA isolated by the Easy-DNA kit. The method is based on RFLP analysis of PCR products of the C. jejuni flaA and flaB genes. The PCR primers were as previously published (3). The PCR was carried out on a Perkin-Elmer GeneAmp 9700 PCR system (PE Biosystems). The temperature profile was 94°C denaturation for 1 min followed by 45 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2 min and then finally 58°C for 90 s and a 5-min extension step at 72°C. PCR products were then ethanol precipitated, washed with 70% ethanol, and digested with 10 U of the restriction enzyme DdeI at 37°C overnight. The digested PCR products were separated on a 1.8% Metaphor agarose gel (FMC) in 1× Tris-borate-EDTA buffer for 4 h at 120 V.
Data analysis.
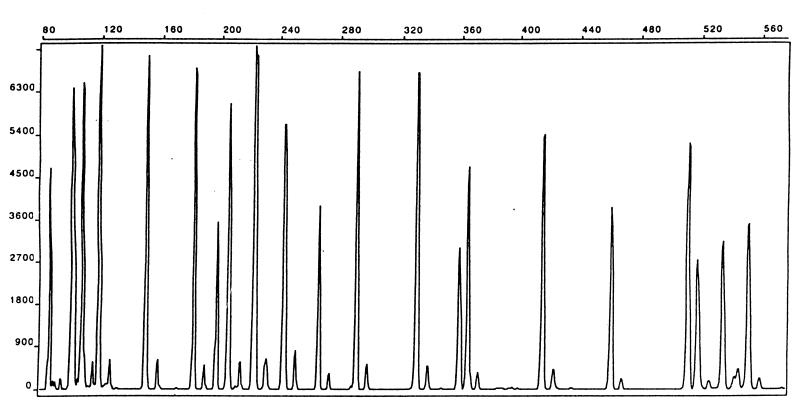
The PFGE and PCR-RFLP gels were stained with ethidium bromide and photographed under UV illumination. The resulting photographs were visually inspected and assigned to different fingerprint patterns. The resulting electropherograms generated by capillary electrophoresis of the AFLP fragments were compared using GeneScan (PE Biosystems) software with separate colors given to individual strain patterns. The patterns were superimposed and were visually inspected for polymorphous peaks. The internal standard was also overlaid in all compared samples to ensure a correct interpretation and alignment of the band patterns. A gel image was then constructed by GelCompar II (Applied Maths BVBA, Kortrijk, Belgium) from the Applied Biosystems Inc. (ABI) trace files. Both software packages were used for identifying specific patterns. GenScan allowed easy correction for differences arising as a consequence of slightly different termination of individual runs and to ensure that all runs had approximately the same peak intensities and peak shapes before import of files to GelCompar. The GelCompar II software, on the other hand, had far more analysis tools for quantifying strain differences than did GeneScan, and therefore the fine analysis of similarities was performed within GelCompar II. The fragment peaks were high, with a relative fluorescence up to about 7,000, and well defined (Fig. 2). This allowed us to use a relative fluorescence value of 900 as threshold. AFLP fragment peaks with fluorescence values less than 900 were not included in the analysis. A phylogenetic tree was constructed with GelCompar II using Dice coefficients and cluster analysis with the unweighted pair group method with arithmetic averages from the ABI trace files.
FIG. 2.
AFLP fragment pattern of a C. jejuni strain. The horizontal scale is fragment sizes in base pairs, and the vertical scale is relative fluorescence. One of the PCR primers was labeled with the dye FAM (5′-carboxyfluorescein).
RESULTS
AFLP.
Genotyping of 91 C. jejuni strains by AFLP allowed 40 distinct patterns to be distinguished (Fig. 1). The fingerprints generated by the BglII-MfeI enzyme combination had sharp and easily distinguishable peaks. About 20 to 25 fragments in the range of 50 to 500 bp were produced (Fig. 2) in accordance with the number of fragments calculated from the C. jejuni genome sequence (32). The patterns were noncomplex and suitable for visual comparison of fingerprints. The electropherograms sometimes showed a variation in the height of the peaks, but less variation was found in the present study than with EcoRI-MseI AFLP performed on Salmonella enterica subsp. enterica strains examined previously (24). It was evident that outbreak strains clustered together with a similarity in the range of 96 to 100%, with the majority showing a homology of more than 98% (Fig. 1). From the knowledge of our outbreak strains and allowing for small errors that may arise between different AFLP runs, we designated all strains within a window of similarity of between 95 and 100% homology as being identical. This limit gave rise to the 40 different AFLP patterns named A1 to A40 (Fig. 1). All outbreaks could be distinguished except outbreaks 5 and 6 (Fig. 1), which were clustered together with 97% similarity. Outbreaks 5 and 6 were from separate geographic locations and from different years; thus, we had expected more divergent patterns between these outbreaks. We used a window of similarity of between 90 and 95% homology to designate strains into families which are highly related but not identical. This allowed us to identify relationships between different outbreaks and with sporadic strains (Fig. 1).
Macrorestriction and PFGE.
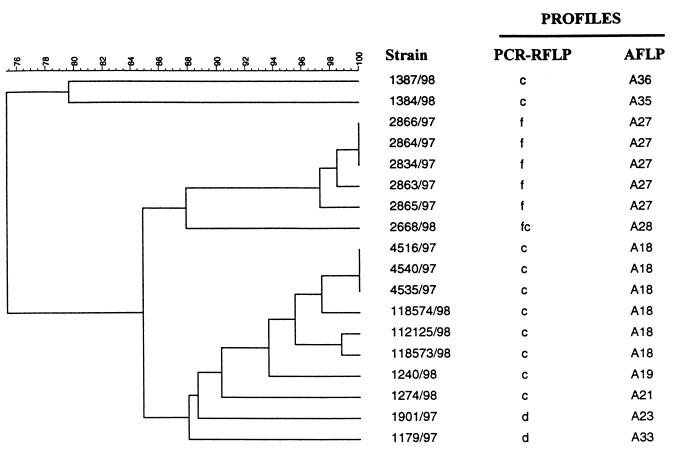
By SmaI macrorestriction and PFGE of 85 C. jejuni biotype 1 strains comprising 36 AFLP profiles, we observed 20 different PFGE profiles (Fig. 1). In Fig. 3, a representative PFGE gel is shown. Some of the patterns were, however, quite similar, especially with minor variations of the B, E, and G PFGE profiles, usually with one band absent or present or one size-shifted band. These variations are treated as separate PFGE profiles in this study. The AFLP method had a higher discriminative power than did PFGE, and several strains with identical PFGE profiles could be distinguished with AFLP. The two large PFGE profile groups named E and B could both be subdivided into nine AFLP profiles. Figure 4 shows the PFGE E profile subdivided by AFLP. The AFLP profiles clearly distinguish the three outbreaks in this group, one outbreak from Finland (12), one from southwestern Norway in 1998, and one from north Norway in 1980. Figure 5 shows the PFGE profile B subdivided by AFLP. This group (PFGE B) comprises strains from three different outbreaks. AFLP can distinguish two of the three outbreaks on the 95% similarity level. Outbreak 5 and outbreak 6 are also indistinguishable by AFLP at the 95% similarity level for identical strains, and we must conclude that they were caused by the same strain. The above-mentioned variations in the B and E profiles were also seen with AFLP. The variation within the G profile was not discovered by AFLP. The BII and BI profiles are clustered together in two separate families distinct from other strains with the B profile with AFLP (Fig. 1).
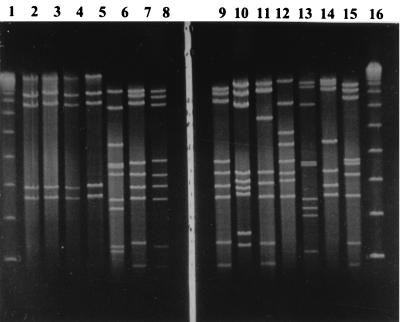
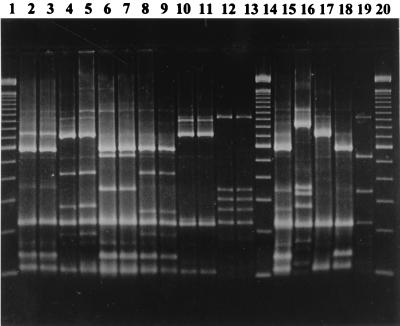
FIG. 3.
PFGE gel with the following profiles; lane 1, λ size marker; lane 2, E profile from outbreak 1; lane 3, E profile from outbreak 10; lane 4, E profile from outbreak 2; lane 5, E profile from outbreak 3; lane 6, G profile from outbreak 7; lane 7, B profile from outbreak 5; lane 8, B profile from outbreak 9; lane 9, B profile from outbreak 6; lane 10, P profile from outbreak 8; lane 11, sporadic strain with V profile; lane 12, sporadic strain with N profile; lane 13, sporadic strain with O profile; lane 14, sporadic strain with C profile; lane 15, sporadic strain with L profile; lane 16, λ size marker.
FIG. 4.
Dendrogram showing AFLP subdivision of strains which all have the PFGE E profile. Nine AFLP profiles are resolved at the 95% similarity level for designating a unique profile. Four or five PCR-RFLP profiles are resolved (four profiles if the variable bands within PCR-RFLP profile b are disregarded). ND, not done.
FIG. 5.
Dendrogram showing AFLP subdivision of strains which all have the PFGE B profile. Nine AFLP profiles are resolved at the 95% similarity level for designating a unique profile. Four PCR-RFLP profiles are resolved.
PCR-RFLP.
By DdeI PCR-RFLP of the flaA and flaB genes from 84 strains comprising 35 AFLP profiles, we found 18 or 19 different PCR-RFLP patterns. On repeated PCR amplifications and restriction cutting, we discovered that two bands at 185 and 325 bp were unstable in the strains with the PCR-RFLP b profile. These bands, when absent, made the PCR-RFLP profile b identical to the g profile (Fig. 6). In Fig. 4, it is shown that PCR-RFLP can subdivide the PFGE E profile into five profiles, or four profiles when the unstable bands were disregarded, of 28 tested strains. In Fig. 5, it is shown that PCR-RFLP can subdivide the PFGE B profile into four profiles of 18 tested strains. The PCR-RFLP method clustered outbreaks 1 and 2 together with the exception of one strain (367U). Outbreaks 1 and 2 also appeared identical with PFGE and could be separated only with AFLP. When the above-mentioned unstable bands were absent, strains from outbreak 10 became identical to strains from outbreaks 1 and 2. Outbreaks 1 and 2 belong to two neighboring families according to their AFLP patterns and are 89% similar but originate from geographic locations distant from each other. Isolates of outbreak 1 were from southwestern Norway in 1998, and isolates from outbreak 2 (27, 29) were from north Norway in 1988. PCR-RFLP and PFGE can, however, distinguish outbreaks 5 and 6 from outbreak 7, which is 92% similar and belongs to the same family according to AFLP. Thus, one cannot predict directly from the similarity index in AFLP when a new pattern would arise in PCR-RFLP or PFGE. PCR-RFLP could distinguish the PFGE BII profile from the other strains with the B profile, but it clustered the strains with PFGE BI profiles together with the B strains of outbreak 9 (Fig. 1). One strain (2791/97) has a variation of the E pattern (EI), and this strain is not grouped together with any other E strains by AFLP, nor is it included in any family (i.e., ≥90% similarity with other strains). By PCR-RFLP, this strain is grouped with a strain that has the PFGE B profile (2668/98). The variations within the G profile were not recognized by PCR-RFLP.
FIG. 6.
PCR-RFLP gel with the following profiles: lane 1, 100-bp ladder; lanes 2 and 3, profile a from outbreak 7; lanes 4 and 5, profile b from outbreak 1; lanes 6 and 7, profile c from outbreak 5; lanes 8 and 9, profile f from outbreak 9; lanes 10 and 11, profile g from outbreak 10; lanes 12 and 13, profile s from outbreak 8; lane 14, 100-bp ladder; lane 15, profile d; lane 16, profile e; lane 17, profile ga; lane 18, profile h; lane 19, profile k; lane 20, 100-bp ladder. The two bands at approximately 185 and 325 bp in profile b, lanes 4 and 5, are unstable and do not appear on all gels when repeated PCR-RFLP analyses are performed on the same DNA.
PFGE combined with PCR-RFLP.
When we combined the results for the PFGE and PCR-RFLP methods, 31 different profiles, or 30 profiles with unstable PCR-RFLP bands absent, could be separated, versus 32 for AFLP for 81 strains examined. The combination of these two methods did not offer any time or labor reductions compared to AFLP, nor did it give a higher power of discrimination than AFLP. Thus, even with the combination of two different genotyping methods, the resolution was still less than what was obtained with AFLP.
DISCUSSION
We have genotyped 91 strains of C. jejuni by AFLP. Most isolates were also genotyped by PFGE (85 strains) and PCR-RFLP (84 strains). The strain collection consisted of outbreak and sporadic isolates (Fig. 1). The strains were isolated throughout Norway with a large geographic distribution, and a time span from 1980 to 1999. This relatively large collection of outbreaks gave a good basis for evaluating the performance of the AFLP genotyping method. It is advantageous to include strains from several known outbreaks because this will aid in the assessment of similarity indices and finding a similarity value above which strains can be regarded as identical. In the present study, this value was set at 95% similarity.
We have demonstrated that AFLP is well suited for genotyping outbreak strains by precisely sizing DNA restriction fragments unique for those strains. We found that the discriminative power of AFLP was higher than that of both PFGE and PCR-RFLP. AFLP could distinguish strains that were identical by PFGE. It was speculated that strains from outbreak 1 in southwestern Norway could have been carried to Finland and caused an outbreak (outbreak 10) there later the same year. Outbreak 1 took place at a Nordic sport event in Norway, and some participants at this event lived in the community where the Finnish outbreak (outbreak 10) occurred later the same year. By PFGE, the two outbreaks had identical profiles, but AFLP genotyping showed that they were separate and distinct outbreaks (Fig. 1). By PCR-RFLP, these two outbreaks were sometimes identical or different on repeated analyses due to two unstable bands in the PCR-RFLP b profile. A number of cases of campylobacteriosis were reported among participants in an annual bicycle race in east Norway in 1997 (outbreak 5) and again in 1999 (outbreak 3). We examined whether these two outbreaks were caused by the same strain. The AFLP analysis showed that the outbreaks (outbreaks 3 and 5) were caused by genetically different strains (Fig. 1). Outbreak 3 was assigned to two new genotypes by AFLP (A9 and A13), but with PFGE, only one profile, which was displayed by epidemiologically unrelated strains as well, could be resolved (E). PFGE, however, confirmed that outbreaks 3 and 5 were caused by different strains (Fig. 1). All three genotyping methods gave the same fingerprint for outbreak 5 and outbreak 6. We concluded that these outbreaks have the same clonality even though they are separated in time and geographic region (southeast Norway in 1997 and central Norway in 1998). The fingerprinting results from outbreaks 5 and 6 point out the advantage of typing techniques, which possess the power to link some outbreaks with different time and place distributions. Of the observed variations within the PFGE B, E, and G patterns, only the G pattern variation was not detected by AFLP typing. This could reflect instability in a region of the C. jejuni genome that affects an SmaI restriction site and was discovered only by PFGE. C. jejuni genomic instability has previously been reported with PFGE typing (11, 39). The numbers of fragments generated by AFLP for analysis and genotype definition are about two to five times higher than the number of SmaI PFGE fragments, which probably can explain some of the increased discriminative power of AFLP versus PFGE. The discriminative power of PFGE can be increased by macrorestriction with other endonucleases, and this has been recommended for PFGE typing of C. jejuni (31). Macrorestriction with several restriction endonucleases would, however, further increase the time and labor used versus those for the AFLP protocol. We observed that the PCR-RFLP method also offered good discrimination but, like PFGE, failed to separate strains from some of the different outbreaks. PCR-RFLP could, however, separate the two outbreaks within the PFGE B profile. We discovered run-to-run variations within the PCR-RFLP b profile, which resulted in the presence or absence of two specific bands. These bands when absent gave rise to the PCR-RFLP g profile. The existence of this profile is thus questionable, but it is included as a distinct profile in this study for reasons of comparison, since the main objective was to evaluate the AFLP assay, but if PCR-RFLP is used as the primary genotyping method, the two unstable bands should be disregarded. The removal of the PCR-RFLP g or b profile will increase the discriminatory power of AFLP versus that of PCR-RFLP. The use of PCR-RFLP analysis of the flaA and flaB genes for long-term monitoring of C. jejuni strains has additionally been questioned because of both intragenomic and intergenomic recombination between the flagellin genes (13). The PCR-RFLP procedure is easy to perform, and typing polymorphisms in other genes could possibly increase its discriminative power. The gyrA and pflA genes are two additional genes which may be included in PCR-RFLP genotyping (34). Our results indicate that multiple runs of PCR-RFLP analysis on the same samples should be performed routinely in order to localize variant bands. When PCR-RFLP and PFGE were combined for our strain collection, the discriminatory power was still less than that for AFLP.
The present data document that the AFLP method used in this study gives a resolution for discriminating C. jejuni species that can be matched only by combining two other genotyping methods. The AFLP method is, however, flexible and can most likely be further optimized for higher resolution. The multicolor system of the ABI apparatus also allows for several AFLP reactions, e.g., with different enzyme or primer combinations to be analyzed in the same run. This is at the moment unfeasible for both the PFGE and the PCR-RFLP genotyping methods.
Our results are in agreement with results from other groups that have used AFLP for fingerprinting of Campylobacter (4, 6, 22). Although using different restriction endonucleases and different protocols, all studies, including ours, report AFLP to be a highly discriminatory method, which displays a relationship between strains that corresponds well with epidemiological data. The discriminatory power of AFLP is reported to be comparable to or higher than that of analysis by PFGE (4, 22), which is also seen in the present study. A higher discriminatory power of AFLP than of flagellin typing with the flaA gene has additionally been reported elsewhere (4). We observed that AFLP had a higher power of discrimination than did combined flaA and flaB typing.
Results from our laboratory indicate that, among the various AFLP protocols and species tested, the protocol described in this article has the highest discriminatory power versus PFGE. AFLP genotyping of S. enterica subsp. enterica serovars was found to achieve comparable discriminative power versus XbaI PFGE (24), while XbaI PFGE was clearly the best method for genotyping Shiga toxin-producing Escherichia coli isolates compared to AFLP (14a). The discriminative power of AFLP appears to be highly influenced by the species under study, and optimization of the method is needed for each species as is optimization even within serovars of the same species as previously noted for S. enterica (24).
In conclusion, we have presented data which showed a higher discriminative power of AFLP than of both PFGE and PCR-RFLP in typing outbreak-related and sporadic C. jejuni strains. We have further analyzed the AFLP data in a software package and shown its usefulness in epidemiological studies and outbreak investigations. The different outbreaks could easily be distinguished, and the genetic similarity between all strains could be quantified. The different profiles are stored in a database that will be used as a tool for continuous surveillance of C. jejuni strains and for tracing the source of future outbreaks in Norway. The digital nature of the data makes them easy to share, but standardization of the AFLP typing method will be needed.
ACKNOWLEDGMENTS
We gratefully acknowledge A. Siitonen, National Public Health Institute, Helsinki, Finland; L. Bevanger, Trondheim Regional Hospital, Trondheim, Norway; E. Wahl, Regional Food Inspection Authority, Trondheim, Norway; and M. Varslot, Innherrad Hospital, Levanger, Norway, for submitting strains to the Norwegian National Reference Laboratory for Enteropathogenic Bacteria.
REFERENCES
- 1.Aarts H J, Hakemulder L E, Van Hoef A M. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int J Food Microbiol. 1999;49:95–102. doi: 10.1016/s0168-1605(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Guerry P, Trust T J. Distribution and polymorphism of the flagellin genes from isolates of Campylobacter coli and Campylobacter jejuni. J Bacteriol. 1993;175:3051–3057. doi: 10.1128/jb.175.10.3051-3057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayling R D, Woodward M J, Evans S, Newell D G. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry campylobacters for epidemiological investigations. Res Vet Sci. 1996;60:168–172. doi: 10.1016/s0034-5288(96)90013-2. [DOI] [PubMed] [Google Scholar]
- 4.de Boer P, Duim B, Rigter A, van der Plas P J, Jacobs-Reitsma W F, Wagenaar J A. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000;38:1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai M, Efstratiou A, George R, Stanley J. High-resolution genotyping of Streptococcus pyogenes serotype M1 isolates by fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol. 1999;37:1948–1952. doi: 10.1128/jcm.37.6.1948-1952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duim B, Wassenaar T M, Rigter A, Wagenaar J. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 1999;65:2369–2375. doi: 10.1128/aem.65.6.2369-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geornaras I, Kunene N F, von Holy A, Hastings J W. Amplified fragment length polymorphism fingerprinting of Pseudomonas strains from a poultry processing plant. Appl Environ Microbiol. 1999;65:3828–3833. doi: 10.1128/aem.65.9.3828-3833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson J R, Sutherland K, Owen R J. Inhibition of DNAse activity in PFGE analysis of DNA from Campylobacter jejuni. Lett Appl Microbiol. 1994;19:357–358. doi: 10.1111/j.1472-765x.1994.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 9.Guerry P, Alm R A, Power M E, Logan S M, Trust T J. Role of two flagellin genes in Campylobacter motility. J Bacteriol. 1991;173:4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerry P, Logan S M, Thornton S, Trust T J. Genomic organization and expression of Campylobacter flagellin genes. J Bacteriol. 1990;172:1853–1860. doi: 10.1128/jb.172.4.1853-1860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hänninen M L, Hakkinen M, Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:2272–2275. doi: 10.1128/aem.65.5.2272-2275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hänninen M L, Pajarre S, Klossner M L, Rautelin H. Typing of human Campylobacter jejuni isolates in Finalnd by pulsed-field gel electrophoresis. J Clin Microbiol. 1998;36:1787–1789. doi: 10.1128/jcm.36.6.1787-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington C S, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington C S, Thomson-Carter F M, Carter P E. Molecular epidemiological investigation of an outbreak of Campylobacter jejuni identifies a dominant clonal line within Scottish serotype HS55 populations. Epidemiol Infect. 1999;122:367–375. doi: 10.1017/s0950268899002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Heir, E., B. A. Lindstedt, T. Vardund, Y. Wasteson, and G. Kapperud. Genomic fingerprinting of shigatoxin producing Escherichia coli (STEC) strains: comparison of pulsed-field gel electrophoresis (PFGE) and fluorescent amplified fragment length polymorphism (FAFLP). Epidemiol. Infect., in press. [DOI] [PMC free article] [PubMed]
- 15.Hernandez J, Fayos A, Ferrus M A, Owen R J. Random amplified polymorphic DNA fingerprinting of Campylobacter jejuni and C. coli isolated from human faeces, seawater and poultry products. Res Microbiol. 1995;146:685–696. doi: 10.1016/0923-2508(96)81065-5. [DOI] [PubMed] [Google Scholar]
- 16.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 18.Janssen P, Maquelin K, Coopman R, Tjernberg I, Bouvet P, Kersters K, Dijkshoorn L. Discrimination of Acinetobacter genomic species by AFLP fingerprinting. Int J Syst Bacteriol. 1997;47:1179–1187. doi: 10.1099/00207713-47-4-1179. [DOI] [PubMed] [Google Scholar]
- 19.Karras D J. Incidence of foodborne illnesses: preliminary data from the foodborne diseases active surveillance network (FoodNet) Ann Emerg Med. 2000;35:92–93. [PubMed] [Google Scholar]
- 20.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh J M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokotovic B, Friis N F, Jensen J S, Ahrens P. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J Clin Microbiol. 1999;37:3300–3307. doi: 10.1128/jcm.37.10.3300-3307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokotovic B, On S L. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol Lett. 1999;173:77–84. doi: 10.1111/j.1574-6968.1999.tb13487.x. [DOI] [PubMed] [Google Scholar]
- 23.Lind L, Sjögren E, Melby K, Kaijser B. DNA fingerprinting and serotyping of Campylobacter jejuni isolates from epidemic outbreaks. J Clin Microbiol. 1996;34:892–896. doi: 10.1128/jcm.34.4.892-896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindstedt B A, Heir E, Vardund T, Kapperud G. Fluorescent amplified-fragment length polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J Clin Microbiol. 2000;38:1623–1627. doi: 10.1128/jcm.38.4.1623-1627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lior H, Woodward D L, Edgar J A, Laroche L J, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–162. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 27.Melby K, Holmen L A, Svendby J G, Eggebø T, Andersen B M. Epidemiologic outbreak of Campylobacter infection. Tidsskr Nor Laegeforen. 1991;111:1530. [PubMed] [Google Scholar]
- 28.Melby K, Storvold G, Congi R V, Penner J L. Serotyping of Campylobacter jejuni isolated from sporadic cases and outbreaks in northern Norway. Acta Pathol Microbiol Immunol Scand B. 1985;93:83–86. doi: 10.1111/j.1699-0463.1985.tb02856.x. [DOI] [PubMed] [Google Scholar]
- 29.Nachamkin I, Bohachick K, Patton C M. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J Clin Microbiol. 1993;31:1531–1536. doi: 10.1128/jcm.31.6.1531-1536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen E M, Engberg J, Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol Med Microbiol. 1997;19:47–56. doi: 10.1111/j.1574-695X.1997.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 31.On S L, Nielsen E M, Engberg J, Madsen M. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol Infect. 1998;120:231–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, Jagels K, Karlyshev A V, Moule S, Pallen M J, Penn C W, Quail M A, Rajandream M A, Rutherford K M, van Vliet A H, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 33.Penner J L, Hennessy J N. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragimbeau C, Salvat G, Colin P, Ermel G. Development of a multiplex PCR gene fingerprinting method using gyrA and pflA polymorphisms to identify genotypic relatedness within Campylobacter jejuni species. J Appl Microbiol. 1998;85:829–838. doi: 10.1046/j.1365-2672.1998.00597.x. [DOI] [PubMed] [Google Scholar]
- 35.Savelkoul P H, Aarts H J, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker J L, Schouls L, Lenstra J A. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999;37:3083–3091. doi: 10.1128/jcm.37.10.3083-3091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skirrow M B. Diseases due to Campylobacter, Helicobacter and related bacteria. J Comp Pathol. 1994;111:113–149. doi: 10.1016/s0021-9975(05)80046-5. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki Y, Ishihara M, Funabashi M, Suzuki R, Isomura S, Yokochi T. Pulsed-field gel electrophoretic analysis of Campylobacter jejuni DNA for use in epidemiological studies. J Infect. 1993;27:39–42. doi: 10.1016/0163-4453(93)93628-h. [DOI] [PubMed] [Google Scholar]
- 38.Vos P, Hogers R, Bleeker M, Reijans M, van-de-Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]