Abstract
Persistent human papillomavirus (HPV) infection of the uterine cervix is a risk factor for progression to high-grade squamous intraepithelial lesions. Detection in consecutive genital samples of HPV-16 DNA, a frequently encountered HPV type, may represent persistent infection or reinfection. We undertook a study using PCR–single-strand conformation polymorphism (SSCP) analysis and sequencing of PCR products (PCR-sequencing) to determine if consecutive HPV-16-positive samples contained the same HPV-16 variant. Fifty women (36 human immunodeficiency virus [HIV] seropositive, 14 HIV seronegative) had at least two consecutive genital specimens obtained at 6-month intervals that contained HPV-16 DNA as determined by a consensus L1 PCR assay. A total of 144 samples were amplified with two primer pairs for SSCP analysis of the entire long control region. Fifteen different SSCP patterns were identified in our population, while 22 variants were identified by PCR-sequencing. The most frequent SSCP pattern was found in 75 (53%) samples from 27 (54%) women. The SSCP patterns obtained from consecutive specimens were identical for 46 (92%) of 50 women, suggesting persistent infection. Four women exhibited in consecutive specimens different HPV-16 SSCP patterns that were all confirmed by PCR-sequencing. The additional information on the nature of persistent infection provided by molecular variant analysis was useful for 6% of women, since three of the four women who did not have identical consecutive specimens would have been misclassified as having persistent HPV-16 infection on the basis of HPV typing.
High-risk human papillomavirus (HPV) types are now considered etiological agents of cancer of the uterine cervix (5, 28). HPV infection is the most frequent sexually transmitted disease (STD) worldwide, and up to 60% of sexually active women will be infected by HPV in the genital tract (17). Irrespective of HPV infection status, fewer than 1 in 10,000 women will develop invasive cervical cancer. The fact that most HPV-infected women do not develop cytological anomalies or cancer (7) underlines the importance of factors modulating the progression of cervical disease to cancer in HPV-infected women. These factors may include the HPV genotype and molecular variant, the HPV viral load, persistence of HPV infection, coinfection with other STD agents, and the immune status of the host (7, 12, 26, 29, 30, 39).
Recent studies have demonstrated that infection with high-risk HPV types is usually transient (10, 16, 17, 34). Persistence of HPV infection substantially increases the risk of progression to clinically relevant preinvasive and invasive disease (2, 17, 18, 23, 24, 26). Persistence of HPV infection is often defined as the detection of the same HPV type in consecutive samples obtained at 3- to 6-month intervals. However, when the HPV genotype involved is prevalent in the population of women under study, reinfection with the same genotype may occur, simulating persistent infection. This detection of the same genotype may result in misclassification of consecutive transient HPV infections as persistent infections.
Genomic heterogeneity of HPV-16 isolates, the most prevalent type detected in high-grade and cancerous lesions of the uterine cervix (5, 17, 33), results from a limited number of nucleotide changes, especially in noncoding regions of the HPV genome (19). On this basis, HPV-16 can be classified into more than 40 variants (6, 20, 25, 41) which have a less than 2% variation in DNA sequence (33). Molecular variant analysis of HPV-16 isolates allows us to distinguish reinfection from persistence (11, 32). Sequence analysis of hypervariable regions of the HPV genome (11) remains the gold standard method for molecular variant analysis. However, single-strand conformation polymorphism (SSCP) analysis is an alternative to sequencing for this purpose (38). Only one group has compared the value of HPV-16 variant analysis of PCR-SSCP patterns with that of PCR-sequencing of clinical specimens (32). A simplified dot blot analysis to classify HPV-16 isolates into the major HPV-16 lineages has also been described (35).
The objective of our study was to evaluate the accuracy of PCR-SSCP analysis in classifying women who had consecutive genital samples containing HPV-16 DNA as being persistently infected or being reinfected by HPV-16. We also compared the numbers of variants identified by the SSCP and DNA-sequencing procedures. Using both methods, we investigated if sexually active women considered persistently infected with HPV-16 harbored the same variant over time.
MATERIALS AND METHODS
Patients' protocol.
This study focused on 50 women participating in The Canadian Women's HIV Study Group (13, 14) who had at least two consecutive samples containing HPV-16 DNA sequences as determined by the L1 MY09-MY11 consensus PCR assay. Women were included from the date of the first HPV-16-positive sample to the date of loss of HPV-16, loss to follow-up, or September 1997, whichever came first. All participants provided written informed consent to participate. Ethics committees of each participating institution had approved The Canadian Women's HIV Study protocol.
The Canadian Women's HIV Study has been conducted across Canada since 1993 by 28 STD clinics, human immunodeficiency virus (HIV) clinics, and family practices. It evaluates the relationship between HPV infection, HIV infection, and cervical disease using cross-sectional, cohort, and descriptive methodologies (13, 14). The original study design, the characteristics of the cohort, and HPV typing results have already been described in detail elsewhere (8, 13, 14, 14). Women seropositive for HIV-1 were eligible to participate in The Canadian Women's HIV Study, as were HIV-1-seronegative women who had three or more lifetime male sexual partners or were at risk for STD, if they agreed to have annual serological tests for HIV antibodies (13, 14).
In The Canadian Women's HIV Study, a standardized questionnaire was completed upon study entry and 6 months thereafter to obtain information on sociodemographic characteristics, sexual behavior, history of STD, and opportunistic infections, as well as recent CD4 T-lymphocyte counts (13, 14). For HIV-seronegative women, vaginal tampon specimens were obtained at 6-month intervals while cervicovaginal lavages and Pap smears were collected at 1-year intervals. For HIV-seropositive participants, all three specimens were obtained at 6-month intervals. When a vaginal tampon and a cervicovaginal lavage were obtained at the same annual visit and both contained HPV-16 DNA, the isolate from the cervicovaginal lavage was chosen for molecular variant analyses.
Sample processing.
Participants inserted and immediately withdrew a vaginal tampon before pelvic examination (8). The tampon was placed into a sterile jar containing 50 ml of transport medium composed of 10 mM Tris-HCl (pH 7.5), 50 mM EDTA, and 150 mM NaCl. Cervicovaginal lavages were obtained during pelvic examinations with 10 ml of phosphate-buffered saline (pH 7.4) (8). Specimens were refrigerated within 1 h and transported to a central laboratory on wet ice. Tampons were then squeezed to obtain a suspension of exfoliated cells. Cell suspensions from tampons and cervicovaginal lavages were pelleted after centrifugation at 2,500 rpm (IEC CENTRA-8R) for 10 min at 4°C and resuspended in 500 μl of 10 mM Tris (pH 8.3). Cell suspensions were lysed with Tween 20 at a final concentration of 0.8% (vol/vol), and digested with 250 μg of proteinase K per ml for 2 h at 45°C (8). Cell lysates were boiled for 10 min and stored at −70°C until tested. The delay between sampling and processing was never extended beyond 5 days.
β-Globin and HPV DNA amplification.
Five microliters from each sample was amplified for β-globin DNA with the PC04 and GH20 primers to control for DNA integrity, for the presence of an adequate number of epithelial cells, and for amplification inhibitors (3, 8). β-Globin-negative samples were extracted with phenol-chloroform and precipitated with ethanol (27). Five hundred nanograms of extracted nucleic acids was then reamplified for β-globin. β-Globin-positive samples (lysate or extracted DNA) were then tested for HPV. A volume of 5 μl of each cell lysate was amplified under standard conditions with the MY09, MY11, and HMB01 consensus HPV primers (8, 9, 13, 16). Negative, weakly positive (10 HPV-18 DNA copies), and strongly positive (HPV-6/11, -16, -31, -33, -35, -39, and -45) controls were included in each amplification run. Amplified products were spotted onto nylon membranes and were reacted as described elsewhere under stringent conditions with 32P-labeled oligonucleotide probes for HPV-6, -11, -16, -18, -31, -33, -35, -39, -45, -51, -52, -53, -56, and -58 (3, 8, 16).
Amplification of HPV-16 LCR for molecular variant analyses.
The complete long control region (LCR) of HPV-16, a hypervariable noncoding segment of the HPV-16 genome between nucleotide positions 7109 and 222, was amplified with two sets of primers (32, 37, 38): primers A and B amplified a 418-bp fragment (nucleotide positions 7109 to 7527) designated the AB fragment, while primers C and D generated a 682-bp fragment (nucleotide positions 7445 to 222) designated the CD fragment.
Amplification reactions were performed with 2.5 μl of lysate in a 10-μl reaction volume containing 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, 2.5 U of AmpliTaq or AmpliTaq Gold DNA polymerase (Perkin-Elmer Cetus, Montreal, Quebec, Canada), 0.5 μM each primer, 0.25 mM (each) dCTP, dTTP, and dGTP, 0.075 mM dATP, and 0.5 μl of [α-32P]dATP (3,000 Ci/mmol, 10 mCi/ml; Amersham, Montreal, Quebec, Canada). AmpliTaq Gold DNA polymerase was used when signals generated with AmpliTaq were too weak. Amplification reactions were completed in a 9600 thermal cycler (Roche Diagnostic System, Mississauga, Ontario, Canada) for 35 cycles (37). Each cycle consisted of a denaturation step at 94°C for 25 s, a reannealing step at 62°C for 25 s, and an extension step at 72°C for 50 s. The last cycle was followed by an extension step at 72°C for 10 min. In each set of PCRs, we included negative controls to ensure the absence of contamination. A plasmid containing HPV-16 DNA (pHPV-16) obtained from H. zur Hausen, Heidelberg, Germany, was considered the reference strain for SSCP analysis and was also used as a positive control.
Molecular variant analysis by SSCP analysis.
The SSCP analysis of HPV-16 LCR-amplified products was carried out as described originally by Xi et al. (37, 38). SSCP patterns of single-stranded HPV-16 PCR products from the five fragments of each HPV-16 isolate were compared with the reference patterns generated by pHPV-16. HPV-16 isolates could thus be classified into reference-like and non-reference-like variants. HPV-16 isolates were considered different variants if at least one fragment showed polymorphism. All specimens from a patient were run on the same gel. All representative isolates having SSCP patterns different from the patterns generated by pHPV-16 were compared together on one gel, which permitted comparisons of all SSCP patterns found in our population. Half of the samples were analyzed twice by SSCP analysis, including all samples from participants with isolates exhibiting a change of SSCP pattern over time. Two observers independently interpreted the gels, and both made the same interpretations of SSCP patterns, without knowledge of the results of DNA sequence analysis.
Molecular variant analysis by PCR-sequencing.
Amplifications were performed as described above by replacing [α-32P]dATP by 0.25 mM dATP in the master mix. HPV-16 LCR fragments were generated with the Expand High Fidelity PCR system (Boehringer Mannheim, Laval, Quebec, Canada) which is a mixture of Taq DNA polymerase and Bwo DNA polymerase that has a low rate of misincorporation. When the intensities of the bands generated were too weak, AmpliTaq Gold DNA polymerase was used. PCR-amplified HPV-16 DNA fragments were purified by the QIAquick gel extraction kit protocol as recommended by the manufacturer (Qiagen Inc., Mississauga, Ontario, Canada).
Direct sequencing of PCR products was done initially. Because we often obtained ambiguities at several base positions, we decided to clone PCR products before sequencing for the remaining samples using a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) with the pCR2.1 TOPO vector and competent Escherichia coli TOP10. Clones containing the AB fragment were identified by digestion with the restriction enzymes HindIII and XhoI. Plasmid DNA from transformed clones was then further purified before being sequenced with the QIAprep Spin Miniprep system (Qiagen Inc.) according to the manufacturer's instructions.
Five hundred nanograms of the purified template was sequenced on a Long Reader 4200 DNA sequencing system (Li-Cor, Lincoln, Nebr.). The sequencing M13 primer was labeled with IR800. The data were processed with the software Baseimage IR. PCR-generated fragments were sequenced from both directions. Due to possible misincorporation of bases by the Taq DNA polymerase and to the possibility of multiple variants in one sample, isolates from the same woman for which clones demonstrated a difference in nucleotide sequence were sequenced at least twice. Isolates were sequenced up to five times if differences persisted.
Data analysis.
HPV-16 LCR DNA sequences were compared using the BLAST sequence analysis software from Genetics Computer Group (1) to known HPV sequences contained in the GenBank database (Los Alamos National Laboratories). HPV-16 LCR DNA sequences were compared with the sequence from the HPV-16 prototype strain reported in the HPV compendium from the Los Alamos National Laboratory (25). Isolates with a DNA sequence different from that of the prototype strain were classified as non-prototype-like variants. Sequences from non-prototype-like variants were aligned and compared two by two with the BLAST sequence analysis program to further classify HPV-16 variants. Sequencing analysis was completed without knowledge of SSCP results.
RESULTS
Consecutive detection of HPV-16 in clinical specimens.
From September 1993 to September 1997, 807 women were recruited in The Canadian Women's HIV Study. Four hundred eighty-four participants were HIV seropositive, while 323 were HIV seronegative. Fifty (6.2%) of the 807 women had at least two consecutive specimens containing HPV-16 DNA and were selected for this study. Thirty-six (7.4%) of the 484 HIV-seropositive women and 14 (4.3%) of the 323 HIV-seronegative had persistent HPV-16 infection (P = 0.100). The evaluation of determinants of persistence of HPV infection is part of a global study that will be the subject of future publications. We here consider only the 50 women with persistent HPV-16 infection.
The average ages of 36 HIV-seropositive women and 14 HIV-seronegative women were 33.8 ± 9.9 years (median, 32 years; age range, 20 to 65 years) and 23.9 ± 4.6 years (median, 22.5 years; age range, 19 to 35 years), respectively (P < 0.001, Student's t test). The mean CD4 cell count for 36 HIV-seropositive women at the time of the first HPV-16-positive test was 290 × 106 ± 234 × 106 cells per liter (median of 252 × 106 cells per liter). Of the 36 HIV-seropositive participants, 11 (32.4%) women had CD4 counts below 200 × 106 cells per liter, 19 (55.9%) had CD4 counts between 200 and 499, and 4 (11.8%) had CD4 counts above 500. Each woman had between two and eight consecutive samples that tested positive for HPV-16: 50% of women with persistent infection had two samples, 24% had three samples, 14% had four samples, and 12% had between five and eight consecutive samples containing HPV-16 DNA sequences.
Analysis of persistent HPV-16 infection by the SSCP method.
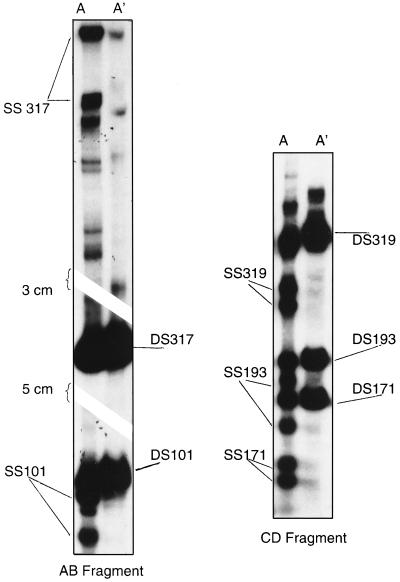
SSCP analysis was used to evaluate the genomic diversity of HPV-16 LCR DNA amplified from 144 clinical specimens. Overall, 141 of the 144 specimens (104 cervicovaginal lavages, 37 tampons) from 50 women with persistent HPV-16 infection gave a positive result for HPV-16 by SSCP analysis when the primer pair A-B or C-D was used. We obtained adequate signals for SSCP analysis for 141 (97.9%) of 144 samples using primers A and B and for 118 (81.9%) of 144 samples using primers C and D. Examples of SSCP patterns are shown in Fig. 1. The three samples for which an interpretable SSCP pattern could not be obtained with the AB fragment did not generate an SSCP pattern with the CD fragment and generated weak signals for HPV-16 DNA by the L1 consensus PCR assay (data not shown).
FIG. 1.
SSCP patterns of fragments AB and CD. Samples were amplified with primers located in the LCR and subjected to nondenaturing gel electrophoresis after digestion with restriction enzymes, as described in Materials and Methods. ds is for double-stranded DNA. A' is for digested PCR products without denaturation, while A is for denatured digested PCR products.
With primer pair A-B, 14 SSCP patterns were found in 141 isolates. With primer pair C-D, six SSCP patterns were found in 118 isolates. Overall, 15 different variants were identified by SSCP analysis if we consider results obtained with both primer pairs. The number of isolates generating reference-like or non-reference-like SSCP patterns with each primer pair is provided in Table 1. With the 66 samples containing non-reference-like HPV-16 variants upon SSCP analysis of the AB fragment, SSCP analysis of the CD fragment showed a pattern suggestive of a reference-like variant in 35 samples. Overall, SSCP testing of the AB fragment identified eight non-reference-like variants that would have been misclassified as reference variants by SSCP analysis of the CD fragment. Of the 50 women with persistent HPV-16 infection, 27 (54%) women were infected with the reference-like HPV-16 variant. Five variants infected more than 84% of women with persistent HPV-16 infection (data not shown).
TABLE 1.
Genetic diversity of 144 HPV-16 isolates from 50 women by SSCP analysis of PCR-amplified LCRsa
| Fragment CD | No. of isolates with SSCP results corresponding to reference or non-reference-like patterns
|
|||
|---|---|---|---|---|
| Reference | Fragment AB non-reference-likeb | NAd | Total | |
| Reference | 62 (1) | 35 (8) | 0 (0) | 97 |
| Non-reference-likec | 0 (0) | 21 (5) | 0 (0) | 21 |
| NA | 13 | 10 | 3 | 26 |
| Total | 75 (1) | 66 (13) | 3 (0) | 144 |
The number of isolates demonstrating reference-like or non-reference-like SSCP patterns are described in the table according to the fragment used for SSCP. The reference-like pattern was that generated by pHPV-16. The SSCP analysis of PCR-amplified fragment AB and fragment CD was accomplished as described in Materials and Methods. Numbers in parentheses are the numbers of different patterns identified by SSCP analysis of the AB fragment when both fragments could be analyzed.
Thirteen non-reference-like patterns were generated with the AB fragment.
Five non-reference-like patterns were generated with the CD fragment.
NA is for not amplified.
The SSCP patterns of the HPV-16 LCR obtained from consecutive specimens were identical for 46 (92%) of 50 women, suggesting persistent HPV-16 infection by the same variant. In samples from five of these women, the presence in one sample of a weaker signal suggested infection with a minor variant. Three of these weak changes were visible on SSCP analysis of the AB fragment, and two changes were visible on SSCP analysis of the CD fragment. No change of signal, either weak or complete, was visible by SSCP analysis of both fragments. Four women had a complete modification of SSCP pattern of the digested AB fragment (Table 2). One of these four women (WM 638) is still considered as having persistent infection since the same variant was detected in the next three consecutive specimens. All changes were confirmed by at least one more round of PCR-SSCP analysis for each sample.
TABLE 2.
Virological characteristics of consecutive isolates from five women who exhibited changes in HPV-16 variants over time
| Participant | Sample | HIV statusa | Sampling time (mo/yr) | SSCP patternb | Sequence typeb |
|---|---|---|---|---|---|
| WM 638 | 1 | Seroneg. | 7/95 | 12 | XXI |
| 2 | Seroneg. | 4/96 | 1 | II | |
| 3 | Seroneg. | 8/96 | 1 | II | |
| 4 | Seroneg. | 3/97 | 1 | II | |
| WQ 505 | 1 | Seroneg. | 5/94 | 1 | II |
| 2 | Seroneg. | 10/94 | 12 | XXII | |
| 3 | Seroneg. | 4/95 | 1 | II | |
| WT 034 | 1 | Seropos. | 9/93 | 13 | IX |
| 2 | Seropos. | 8/94 | 1 | II | |
| WT 085 | 1 | Seropos. | 5/94 | 12 | V |
| 2 | Seropos. | 10/94 | 1 | II |
Seroneg. is for HIV seronegative, and Seropos. is for HIV seropositive.
SSCP pattern 1 corresponds to the pHPV-16 HPV pattern. SSCP types and sequence types are described in more detail in the legend to Fig. 2.
Comparison of results of SSCP analysis and DNA sequencing for analysis of HPV-16 variants.
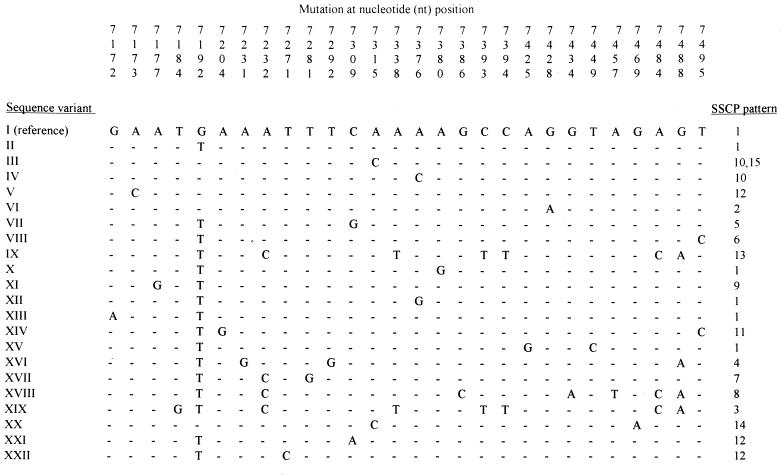
Sequencing of the AB fragment (PCR-sequencing) generated by amplification of the HPV-16 LCR served as the gold standard test to evaluate the ability of PCR-SSCP analysis to identify HPV-16 variants. Mutations defining each variant are displayed in Fig. 2 and compared with results obtained by SSCP analysis. The nucleotide positions that were frequently involved in mutations identifying variants included nucleotide position 7192 (change of a G to a T), found in 16 variants, and nucleotide positions 7232 (change of an A to a C) and 7488 (change of a G to an A), found in 4 variants each (Fig. 2). Overall, PCR-sequencing identified 22 different variants in comparison with only 15 variants identified by SSCP analysis. SSCP analysis of the AB fragment defined as the prototype strain six HPV-16 strains identified by DNA sequence analysis as variants. Other isolates generated SSCP pattern 12 but had a different DNA sequence (sequences V, XXI, and XXII). Variants 10 and 15 generated different SSCP patterns of the CD fragment but had identical AB fragments by SSCP analysis and PCR-sequencing (Fig. 2). Ten of the 28 women for whom SSCP analysis identified the presence of a reference-like variant were infected by nonprototypic variants as determined by DNA sequencing (data not shown).
FIG. 2.
DNA sequence analysis from variants of HPV-16. The sequence variant was determined arbitrarily from the combinations of mutations identified in the amplified AB fragment. The reference sequence was that of the prototype strain reported in the HPV compendium as described in Materials and Methods. The SSCP pattern was determined according to the patterns of bands obtained by SSCP analysis of the AB and CD fragments. Two SSCP patterns were described for sequence variant III on the basis of differences in the migrations of the CD fragment between these two variants, but only the AB fragment was sequenced, illustrating the advantage of testing all the LCR instead of only one section of the LCR.
However, classifications of participants as reinfected versus persistently infected were identical using SSCP analysis or sequencing. The differences in SSCP patterns in consecutive HPV-16 isolates from the same women, which occurred with four participants, were all confirmed by DNA sequencing. The modifications of SSCP patterns corresponded to a base substitution(s) (Table 2).
DISCUSSION
We demonstrate here the usefulness of molecular variant analysis in identifying women with persistent HPV-16 infection. This is the largest published study to date refining the definition of HPV persistence in HIV-infected women using DNA sequencing, and it is also the largest evaluation comparing in parallel SSCP analysis to sequencing for HPV-16 variant analysis of women at risk for STD. Variant analysis has been used to investigate the epidemiology of HPV-16, the phylogenetic analysis of evolution of HPV-16, and determinants of cervical disease progression (4, 6, 11, 19–21, 32, 38–41). HPV-16 nonprototype variants may confer a higher risk than that of the prototype variant for the development of high-grade cervical or anal lesions (36, 39).
Targeting hypervariable regions of the HPV genome increases the probability of finding molecular variants (38). SSCP analysis of the HPV-16 LCR was first described by Xi et al. (38). Using their primers, only 2.1% of HPV-16-positive samples could not be amplified. In comparison to the number of variants identified by PCR-sequencing, the primer pair C-D did not perform as well as the primer pair A-B in terms of the number of variants identified (14 of 22 [63.6%] versus 6 of 22 [27.3%]; P = 0.034, z statistic test) and the number of isolates that generated visible bands (141 of 144 [97.9%] versus 118 of 144 [87.5%]; P = 0.001, McNemar's test). Of the nine women showing a change of SSCP pattern over time or the addition of weak signals suggestive of a minor variant, seven changes were revealed with the AB fragment and two were revealed with the CD fragment only. Another group has also confirmed the superiority of primer pair A-B for SSCP and sequence analyses (32).
The most frequent variant identified by SSCP analysis in our group of persistently infected women was the reference-like strain. Variant analysis is limited by the extent to which HPV variants tend to occur in tight geographical clusters. If a variant occurs frequently in a population, then it becomes an insensitive marker of persistence because of the high probability of reinfection by the same variant. Fortunately, 14 non-reference-like variants identified by PCR-sequencing accounted for nearly 50% of persistent HPV-16 infections in our population. The ethnic diversity of women recruited and the fact that participants in The Canadian Women's HIV Study were sexually active and continuously exposed to STD agents, as well as the national basis for recruitment, reduced the possibility of finding a limited number of variants. PCR-sequencing permitted us to identify 22 variants from 141 HPV-16 isolates detected in 50 sexually active women. A greater number of HPV-16 variants can be found in populations at risk for STD (32).
Although others found that PCR-sequencing and PCR-SSCP analysis gave comparable results for HPV-16 variant analysis, these comparisons were done only on selected specimens exhibiting different SSCP patterns (37–39). Since SSCP analysis and sequencing were conducted only on a subset of samples, these experiments were restricted mainly to the evaluation of the specificity of SSCP analysis by demonstrating that changes in SSCP patterns corresponded to a base substitution(s). Besides our work, only one study compared in parallel sequencing to SSCP analysis of HPV-16 PCR products (32). Those researchers also reached the conclusion that the resolution of the SSCP method for variant analysis was less than that of PCR-sequencing.
We chose to sequence fragment AB, since it exhibits more variability than fragment CD (32) and since nearly all HPV-16 isolates could be amplified with primer pair A-B. In our population of women persistently infected with HPV-16, differences in SSCP patterns were reproducible and could be explained by base substitutions. Although our results demonstrate that SSCP analysis is an adequate alternative to sequencing in identifying HPV-16 variants in consecutive specimens, we found, as reported by others (20, 31, 32, 38), that a more complete analysis of the variable region can be obtained by sequence analysis of PCR-amplified products. We found seven variants defined by sequencing that did not generate different SSCP patterns in our cohort.
In contrast to others (32, 37, 38) who found no change of predominant variant in 344 consecutive specimens from 70 patients, we observed a change of HPV-16 variant for 6% of women with consecutive specimens containing HPV-16 DNA. Three of these four women would have been misclassified as having persistent infection without molecular variant analysis. These differences with our study could be explained by several factors. First, 70% of our study population was HIV seropositive. Second, women in our cohort were exposed to HPV, as demonstrated by the high rate of prevalence of HPV DNA detection (13) and the high risk for STD in participating women, as presented elsewhere (13, 14). Finally, the initial study that used SSCP analysis to study persistence (37) may have underestimated the frequency of reinfection since two variants accounted for up to 70% of all HPV-16 variants identified. It is of interest that another team in Brazil has identified, as we have, a change of variant in consecutive specimens (11). Participant WQ 505 illustrates the complexity of variant analysis. A new variant replaced the first variant but was then replaced in the last sample by the first variant. Since we did not sequence more than five clones of HPV-16 PCR products per sample, we could not exclude the possibility that the first variant was still present in the second specimen at a low copy number. This woman may thus be persistently infected by one variant as well as having been transiently infected by a new variant.
Establishment of infection with multiple variants has been demonstrated (6, 15, 19, 21, 22, 37, 38). We demonstrate here that even in a high-risk population, detection of a new variant is a rare event but does occur. SSCP patterns for five women suggested the presence of more than one variant. Since we did not systematically sequence multiple clones for each sample, we could rely only on PCR-SSCP analysis to estimate the prevalence (10%) of coinfection with more than one variant.
Our experiments confirmed that PCR-SSCP analysis is an alternative in identifying women persistently infected by HPV-16. However, SSCP analysis has a lower resolution in identifying molecular variants than sequencing. The additional information on the nature of persistent infection provided by molecular variant analysis was useful for 6% of women who would have been misclassified as persistently infected by HPV-16. Molecular variant analysis may become more interesting if specific variants are associated with increased risk of disease progression or persistence of HPV infection.
ACKNOWLEDGMENTS
We thank Diane Gaudreault and Diane Bronsard for processing genital samples.
The Medical Research Council of Canada and the National Health Research and Development Programme, Health Canada, supports The Canadian Women's HIV Study. F.C. and M.R. are clinical research scholars supported by the FRSQ.
Appendix
Other members of The Canadian Women's HIV Study Group include the following investigators throughout Canada: John Gill (Calgary); Barbara Romanowski, Stephen Shafran (Edmonton); Rob Grimshaw, David Haase, Lynn Johnston, Wally Schlech (Halifax); Stephan Landis, John Sellors, Fiona Smaill (Hamilton); François Beaudoin, Ngoc Biu, Alena Capek, Marc Boucher, Michel Chateauvert, Manon Coté, Douglas Dalton, Gretty Deutsch, Julian Falutz, Diane Francoeur, Lisa Hallman, Eleanor Hew, Lina Karayan, Marina Klein, Louise Labrecque, Richard Lalonde, Christiane Lavoie, Catherine Lounsbury, John Macleod, Nicole Marceau, Gail Myhr, Grégoire Noel, Robert Piché, Manisha Raut, Chantal Rondeau, Jean-Pierre Routy, Karoon Samikian, Pierre Simard, Christina Smeja, Graham Smith, Paul-Pierre Tellier, and Emil Toma (Montreal); Garry Garber, Garry Victor (Ottawa); Louise Côté, Edith Guilbert, Michel Morissette, Hélène Senay, Sylvie Trottier (Quebec); Phil Berger, Lisa Friedland, Donna Keystone, Joan Murphy, Anne Phillips, Marion Powell, Anita Rachlis, Pat Rockman, Irving Salit, Cheryl Wagner, Sharon Walmsey (Toronto); Kurt Williams (Saskatoon); Ian Bowmer, Rory Windrim (St.-John's); Roger Sandre (Sudbury); Penny Ballem, David Burdge, Brian Conway, Mariane Harris, Deborah Money, Julio Montaner, Deborah Money, and Janice Veenhuizen (Vancouver).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf J J, Dreyfus M, Ritter J, Meyer P, Philippe E, Obert G. A PCR study on the coexistence of herpes simplex virus, cytomegalovirus and human papillomavirus DNAs in cervical neoplasia. Int J Gyn Cancer. 1996;6:389–395. [Google Scholar]
- 3.Bauer H M, Ting Y, Greer C E, Chambers J C, Tashiro C J, Chimera J, Reingold A, Manos M M. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–477. [PubMed] [Google Scholar]
- 4.Bernard H-U, Chan S-Y, Manos M M, Ong C-K, Villa L L, Delius H, Peyton C L, Bauer H M, Wheeler C M. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170:1077–1085. doi: 10.1093/infdis/170.5.1077. [DOI] [PubMed] [Google Scholar]
- 5.Bosch F X, Manos M M, Muñoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V The IBSCC Study Group. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 6.Chan S-Y, Ho L, Ong C-K, Chow V, Drescher B, Durst M, Ter Meulen J, Villa L, Luande J, Mgaya H N, Bernard H-U. Molecular variants of human papillomavirus type 16 from four continents suggest ancient pandemic spread of the virus and its coevolution with humankind. J Virol. 1992;66:2057–2066. doi: 10.1128/jvi.66.4.2057-2066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutlée F, Mayrand M H, Provencher D, Franco E. The future of HPV testing in clinical laboratories and applied virology research. Clin Diagn Virol. 1997;8:123–141. doi: 10.1016/s0928-0197(97)00021-4. [DOI] [PubMed] [Google Scholar]
- 8.Coutlée F, Hankins C, Lapointe N The Canadian Women's HIV Study Group. Comparison between vaginal tampon and cervicovaginal lavage specimens collection for detection of human papillomavirus DNA by the polymerase chain reaction. J Med Virol. 1997;51:42–47. doi: 10.1002/(sici)1096-9071(199701)51:1<42::aid-jmv7>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 9.Coutlée F, Trottier A M, Ghattas G, Leduc R, Toma E, Sanche G, Rodrigues I, Turmel B, Allaire G, Ghadirian P. Risk factors for oral human papillomavirus in adults infected and not infected with human immunodeficiency virus. Sex Transm Dis. 1997;24:23–31. doi: 10.1097/00007435-199701000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Evander M, Edlund K, Gustafsson A, Jonsson M, Karlsson E, Rylander E, Wadell G. Human papillomavirus infection is transient in young women: population-based cohort study. J Infect Dis. 1995;171:1026–1030. doi: 10.1093/infdis/171.4.1026. [DOI] [PubMed] [Google Scholar]
- 11.Franco E L, Villa L L, Rahal P, Ruiz A. Molecular variant analysis as an epidemiological tool to study persistence of cervical human papillomavirus infection. J Natl Cancer Inst. 1994;86:1558–1559. doi: 10.1093/jnci/86.20.1558. [DOI] [PubMed] [Google Scholar]
- 12.Guibinga G H, Coutlée F, Kessous A K, Hankins C, Lapointe N The Canadian Women's HIV Study Group. Role of herpes simplex type 2 virus in genital cancers: review of the evidence. Arch STD/HIV Res. 1995;9:163–179. [Google Scholar]
- 13.Hankins C, Coutlee F, Lapointe N, Simard P, Tran T, Samson J, Hum L The Canadian Women's HIV Study Group. Prevalence of risk factors associated with human papillomavirus infection in women living with HIV. Can Med Assoc J. 1999;160:185–191. [PMC free article] [PubMed] [Google Scholar]
- 14.Hankins C, Lapointe N, Walmsley S The Canadian Women's HIV Study Group. Participation in clinical trials among women living with HIV in Canada. Can Med Assoc J. 1998;159:1359–1365. [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel P A, Chan S Y, Ho L, O'Connor M, Balaram P, Campo M S, Fujinaga K, Kiviat N, Kuypers J, Pfister H. Variation of human papillomavirus type 6 (HPV-6) and HPV-11 genomes sampled throughout the world. J Clin Microbiol. 1995;33:1746–1754. doi: 10.1128/jcm.33.7.1746-1754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildesheim A, Schiffman M H, Gravitt P E, Glass A G, Greer C E, Zhang T, Scott D R, Rush B B, Lawler P, Sherman M E, Kurman R J, Manos M M. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 17.Ho G Y F, Bierman R, Beardsley L, Chang C J, Burk R D. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 18.Ho G Y F, Burk R D, Klein S, Kadish A S, Chang C J, Palan P, Basu J, Tachezy R, Lewis R, Romney S. Persistant genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–1371. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 19.Ho L, Chan S-Y, Chow V, Chong T, Tay S-K, Villa L L, Bernard H-U. Sequence variants of human papillomavirus type 16 in clinical samples permit verification and extension of epidemiological studies and construction of phylogenetic tree. J Clin Microbiol. 1991;29:1765–1772. doi: 10.1128/jcm.29.9.1765-1772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho L, Chan S Y, Burk R D, Das B C, Fujinaga K, Icenogle J P, Kahn T, Kiviat N, Lancaster W, Mavromara-Nazos P. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J Virol. 1993;67:6413–6423. doi: 10.1128/jvi.67.11.6413-6423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho L, Tay S-K, Chan S-Y, Bernard H-U. Sequence variants of human papillomavirus type 16 from couples suggest sexual transmission with low infectivity and polyclonality in genital neoplasia. J Infect Dis. 1993;168:803–809. doi: 10.1093/infdis/168.4.803. [DOI] [PubMed] [Google Scholar]
- 22.Icenogle J P, Laga M, Miller D, Manoka A T, Tucker R A, Reeves W C. Genotypes and sequence variants of human papillomavirus DNAs from human immunodeficiency virus type 1-infected women with cervical intraepithelial neoplasia. J Infect Dis. 1993;166:1210–1216. doi: 10.1093/infdis/166.6.1210. [DOI] [PubMed] [Google Scholar]
- 23.Koutsky L A, Holmes K K, Critchlow C W, Stevens C E, Paavonen J, DeRouen T A, Galloway D A, Vernon D, Kiviat N B. A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. N Engl J Med. 1992;327:1272–1278. doi: 10.1056/NEJM199210293271804. [DOI] [PubMed] [Google Scholar]
- 24.Londesborough P, Ho L, Terry G, Cuzick J, Wheeler C, Singer A. Human papillomavirus genotype as a predictor of persistence and development of high-grade lesions in women with minor cervical abnormalities. Int J Cancer. 1996;69:364–368. doi: 10.1002/(SICI)1097-0215(19961021)69:5<364::AID-IJC2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Myers G, Bernard B H-U, Delius H, Baker C C, Icenogle J, Halpern A L, Wheeler C. Human papillomavirus. A compilation of analysis of nucleic acid and amino acid sequences. Publication LA-UR 95-3675. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 26.Remmink A J, Walboomers J M M, Helmerhorst T J M, Voorhorst F J, Rozendaal L, Risse E K J, Meijer C J L M, Kenemans P. The presence of persistent high-risk HPV genotypes in dysplastic cervical lesions is associated with progressive disease: natural history up to 36 months. Int J Cancer. 1995;61:306–311. doi: 10.1002/ijc.2910610305. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schiffman M H. New epidemiology of human papillomavirus infection and cervical neoplasia. J Natl Cancer Inst. 1995;87:1345–1347. doi: 10.1093/jnci/87.18.1345. [DOI] [PubMed] [Google Scholar]
- 29.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh I M, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 30.Swan D C, Tucker R A, Tortolero-Luna G, Mitchell M F, Wideroff L, Unger E R, Nisenbaum R A, Reeves W C, Icenogle J P. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol. 1999;37:1030–1034. doi: 10.1128/jcm.37.4.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Belkum A. Low-stringency single specific primer PCR, DNA sequencing and single-strand conformation polymorphism of PCR products for identification of genetic variants of human papillomavirus type 16. J Virol Methods. 1995;55:435–443. doi: 10.1016/0166-0934(95)00096-6. [DOI] [PubMed] [Google Scholar]
- 32.van Belkum A, Juffermans L, Schrauwen L, van Doornum G, Burger M, Quint W. Genotyping human papillomavirus type 16 isolates from persistently infected promiscuous individuals and cervical neoplasia patients. J Clin Microbiol. 1995;33:2957–2962. doi: 10.1128/jcm.33.11.2957-2962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa L L. Human papillomaviruses and cervical cancer. Adv Cancer Res. 1997;71:321–341. doi: 10.1016/s0065-230x(08)60102-5. [DOI] [PubMed] [Google Scholar]
- 34.Wheeler C M, Greer C E, Becker T M, Hunt W C, Anderson S M, Manos M M. Short-term fluctuations in the detection of cervical human papillomavirus DNA. Obstet Gyn. 1996;88:261–268. doi: 10.1016/0029-7844(96)00120-2. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler C M, Yamada T, Hildesheim A, Jenison S A. Human papillomavirus type 16 sequence variants: identification by E6 and L1 lineage-specific hybridization. J Clin Microbiol. 1997;35:11–19. doi: 10.1128/jcm.35.1.11-19.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xi L F, Critchlow C W, Wheeler C M, Koutsky L A, Galloway D A, Kuypers J, Hughes J P, Hawes S E, Surawicz C, Goldbaum G, Holmes K K, Kiviat N B. Risk of anal carcinoma in situ in relation to human papillomavirus type 16 variants. Cancer Res. 1998;58:3839–3844. [PubMed] [Google Scholar]
- 37.Xi L F, Demers G W, Kiviat N B, Kuypers J, Beckmann A M, Galloway D A. Analysis of human papillomavirus type 16 variants indicates establishment of persistent infection. J Infect Dis. 1995;172:747–755. doi: 10.1093/infdis/172.3.747. [DOI] [PubMed] [Google Scholar]
- 38.Xi L F, Demers W G, Kiviat N B, Kuypers J, Beckmann A-M, Galloway D A. Sequence variation in the noncoding region of human papillomavirus type 16 detected by single-strand polymorphism analysis. J Infect Dis. 1993;168:610–617. doi: 10.1093/infdis/168.3.610. [DOI] [PubMed] [Google Scholar]
- 39.Xi L F, Koutsky L A, Galloway D A, Kuypers J, Hughes J P, Wheeler C M, Holmes K K, Kiviat N B. Genomic variation of human papillomavirus type 16 and risk for high grade cervical intraepithelial neoplasia. J Natl Cancer Inst. 1997;89:796–802. doi: 10.1093/jnci/89.11.796. [DOI] [PubMed] [Google Scholar]
- 40.Yamada T, Manos M M, Peto J, Greer C E, Munoz N, Bosch F X, Wheeler C M. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J Virol. 1997;71:2463–2472. doi: 10.1128/jvi.71.3.2463-2472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada T, Wheeler C M, Halpern A L, Stewart A C, Hildesheim A, Jenison S A. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J Virol. 1995;69:7743–7753. doi: 10.1128/jvi.69.12.7743-7753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]