Abstract
Two strains of a previously undescribed Actinomyces-like bacterium were recovered in pure culture from infected root canals of teeth. Analysis by biochemical testing and polyacrylamide gel electrophoresis of whole-cell proteins indicated that the strains closely resembled each other phenotypically but were distinct from previously described Actinomyces and Arcanobacterium species. Comparative 16S rRNA gene-sequencing studies showed the bacterium to be a hitherto unknown subline within a group of Actinomyces species which includes Actinomyces bovis, the type species of the genus. Based on phylogenetic and phenotypic evidence, we propose that the unknown bacterium isolated from human clinical specimens be classified as Actinomyces radicidentis sp. nov. The type strain of Actinomyces radicidentis is CCUG 36733.
The genus Actinomyces embraces a heterogeneous group of anaerobic and facultatively anaerobic, asporogenous, gram-positive, non-acid-fast, rod-shaped organisms (10) many of which occur as inhabitants of mucosal surfaces, particularly the oral cavity, of humans and some other homeothermic animals. In addition, some Actinomyces species are long-established pathogens of humans and animals, and in recent years several new Actinomyces species and related taxa (e.g., Arcanobacterium and Actinobaculum) associated with human disease have been described (3, 4, 5, 7, 8, 13). At present, the identification of clinical isolates of Actinomyces and close relatives to the species level using conventional phenotypic methods can be very difficult, which has hampered knowledge of their relationships, natural habitats, prevalence, and pathogenicity. Much of this problem stems from poor test reproducibility, lack of discriminatory power of tests, and possible heterogeneity within some taxa. In recent years the implementation of molecular genetic techniques, in particular 16S rRNA gene sequencing, in concert with molecular chemical methods (e.g., whole-cell protein profiling) of analysis has resulted in much improved classification and species identification (3, 7, 8). As a result of these investigations, a plethora of new Actinomyces and related species have been described from human and animal sources. Despite this increase in the number of new Actinomyces species from humans, there is evidence that many isolates from clinical material do not correspond to described species and that the use of these more precise molecular diagnostic methodologies will inevitably result in the recognition of even more new taxa (6). In this article, we report the results of a polyphasic taxonomic study on two strains of an unusual Actinomyces species isolated in pure culture from infected root canals of teeth. Based on the presented findings, another new species of the genus Actinomyces, Actinomyces radicidentis, is described.
MATERIALS AND METHODS
Cultures and phenotypic characterization.
Strains CCUG 36733T and CCUG 42377 were isolated from infected root canals of human teeth. Strain CCUG 36733T was isolated in 1996, from the upper right canine of an 80-year-old Swedish woman. The tooth had been treated by a dentist on earlier repeated occasions and finally had been root filled. Due to persistent symptoms, the patient was referred to a specialist in endodontics who removed the root filling and took a bacteriological sample from which strain CCUG 36733T was the only organism recovered. The second strain, CCUG 42377, was isolated in 1999, from two upper incisors of a 25-year-old Swedish man. The teeth were treated by a dentist, but, due to persistent symptoms, permanent root filling was avoided during a 7-year period. The patient was finally referred to a specialist in endodontics, who filled the teeth. Three years later, a periapical abscess developed in this region. Penicillin was administered, and the root fillings were removed under aseptic conditions. Bacteriological analysis of samples from the two teeth revealed the growth of strain CCUG 42377 in pure culture. The unidentified isolates were cultured on Columbia agar (Difco, Detroit, Mich.) supplemented with 5% horse blood at 37°C in air. Forty-eight hours was needed for the development of colonies suitable for biochemical testing. The strains were characterized by using the API rapid ID32Strep, API Zym, and API Coryne systems according to the manufacturer's instructions (API bioMërieux, Marcy l'Etoile, France). Polyacrylamide gel electrophoretic analysis of whole-cell proteins was performed as described by Pot et al. (9). For the protein profiling, young cultures produced under optimal conditions were used. For densitometric analysis, normalization, and interpretation of protein patterns, the GCW 3.0 software package (Applied Maths, Kortrijk, Belgium) was used. The similarity between all pairs of traces was expressed by the Pearson product moment correlation coefficient, converted for convenience to a percentage similarity. Generated profiles were compared with a comprehensive database maintained by the Culture Collection of the University of Göteborg (CCUG), Göteborg, Sweden.
16S rRNA gene sequencing and phylogenetic analyses.
The 16S rRNA genes of the two isolates were amplified by PCR and directly sequenced using a Taq Dye-Deoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and an automatic DNA sequencer (model 373A; Applied Biosystems). The closest known relatives of the new isolates were determined by performing database searches. These sequences and those of other known related strains were retrieved from the GenBank or Ribosomal Database Project libraries and aligned with the newly determined sequences using the program PILEUP (1). The resulting multiple-sequence alignment was corrected manually, and a distance matrix was calculated using the programs PRETTY and DNADIST (using the Kimura-2 correction parameter) (2). A phylogenetic tree was constructed by the neighbor-joining method with the program NEIGHBOR (2). The stability of the groupings was estimated by bootstrap analysis (500 replications) using the programs DNABOOT, DNADIST, NEIGHBOR, and CONSENSE (2). Parsimony analysis was also performed using the same package (2).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain CCUG 36733T has been deposited in GenBank under accession number AJ251986.
RESULTS AND DISCUSSION
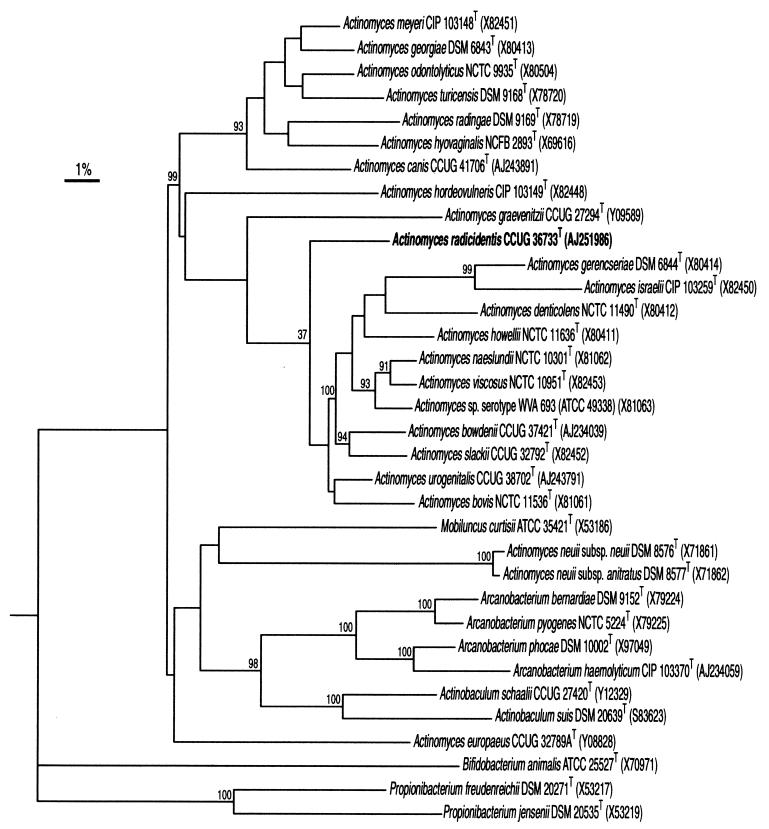
The two isolates consisted of gram-positive coccoid-shaped cells which were non-acid fast and non-spore forming. The strains were catalase positive and grew on Columbia blood agar under aerobic and anaerobic conditions. Using the commercial biochemical kits, both strains were unidentified. Phenotypically they closely resembled each other, producing acid from glucose, maltose, mannitol, melibiose, melezitose, lactose, d-raffinose, ribose, sucrose, and trehalose. Neither of the isolates produced acid from d-arabitol, l-arabinose, cyclodextrin, glycogen, pullulan, sorbitol, or tagatose. Alanine phenylalanine proline arylamidase, acid phosphatase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-galacturonidase, leucine arylamidase, and pyrazinamidase were produced by the isolates, but tests for arginine dihydrolase, alkaline phosphatase, α-fucosidase, β-glucuronidase, glycyl tryptophane arylamidase, lipase C14, α-mannosidase, β-mannosidase, N-acetyl-β-glucosaminidase, chymotrypsin, and trypsin were negative. Variable reactions were observed for urease production and nitrate reduction. The biochemical reactions of the isolates were consistent with their assignment to the genus Actinomyces. To assess the phenotypic resemblance of the two isolates to each other and to reference Actinomyces species, a comparative analysis of whole-cell protein profiles by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) was performed. The two isolates clustered together and formed a distinct group with a within-group correlation level of 85%. The nearest taxa to the unknown isolates on the protein-profiling analysis corresponded to Actinomyces howellii and Arcanobacterium phocae, although this association was loose, joining the aforementioned cluster at a correlation level of approximately 50% (Fig. 1). The PAGE results therefore confirmed that the two unidentified strains represent a phenotypically homogeneous group of organisms and that they are distinct from all Actinomyces species and close relatives described to date. To ascertain the phylogenetic relationships of the clinical isolates, their 16S rRNA genes were sequenced and subjected to a comparative analysis. The almost complete gene sequences (>1,400 nucleotides) of the two strains were determined, and pairwise analysis showed these to be almost identical (99.8% sequence similarity). Sequence database searches confirmed that the unknown bacterium was most closely related to species of the genus Actinomyces (results not shown). Highest sequence relatedness was shown with Actinomyces bovis (95.6% sequence similarity), Actinomyces bowdenii (95%), Actinomyces naeslundii (95%), Actinomyces viscosus (95.2%), and Actinomyces slackii (96.8%). The results of neighbor-joining analysis are shown in Fig. 2 and confirmed the association of the unknown clinical bacterium (as exemplified by strain CCUG 36733T) with A. bovis and related species, with the unknown organism branching at the periphery of the A. bovis rRNA cluster.
FIG. 1.
Similarity dendrogram based on whole-cell protein patterns of A. radicidentis sp. nov. and related species. Levels of correlation are expressed as percentages of similarity for convenience.
FIG. 2.
Unrooted tree showing the phylogenetic relationships of A. radicidentis sp. nov and some other high-G+C content gram-positive bacteria. The tree constructed using the neighbor-joining method was based on a comparison of approximately 1,327 nucleotides. Bootstrap values, expressed as percentages of 500 replications, are given at branching points. Bar, 1% sequence divergence.
It is now recognized that the genus Actinomyces is not monophyletic and consists of several rRNA lineages worthy of separate generic status (7, 8). It is evident from the present 16S rRNA study that the novel bacterium reported here forms a distinct subline branching proximal to the base of a cluster of species which includes the type species A. bovis; therefore, it can be regarded as an authentic Actinomyces species. Sequence divergence values of 3.2% to 9.0% with other members of the A. bovis cluster are clearly indicative of a new species. Although there is no precise correlation between percentage 16S rRNA divergence values and species delineation, it is recognized that organisms that differ by ≥3% do not belong to the same species (11). The observed >3% sequence divergence between the unknown clinical isolates and currently described Actinomyces species is therefore consistent with separate species status. Support for the separateness of the unknown bacterium from infected teeth also comes from phenotypic evidence. The unknown bacterium can be biochemically readily distinguished from all other described catalase-positive or catalase-variable Actinomyces species (Table 1). Similarly the two clinical isolates formed a very distinct cluster upon PAGE analysis of whole-cell proteins, which was far removed from all members of the Actinomyces species group (Fig. 1). It is now firmly established that this molecular chemical approach is extremely reliable for comparing closely related strains and shows excellent correlation with DNA-DNA pairing (12). Furthermore, PAGE protein profiling has been used with great success in Actinomyces systematics (see, e.g., references 3 and 7). The presented PAGE protein profiling results unequivocally demonstrate that the two unknown isolates represent a separate species from all described Actinomyces species. Therefore, based on the distinct phenotypic characteristics of the unknown bacterium and the use of molecular chemical and molecular genetic evidence in concert, we conclude that the two clinical isolates merit classification as a new species of the genus Actinomyces, for which we propose the name A. radicidentis. It seems likely that A. radicidentis may be part of the normal indigenous oral microflora of humans. The recovery of this bacterium in pure culture from infected root canals of teeth indicates that it is possibly a hitherto unrecognized facultative pathogen. We consider that the formal description of this new species will facilitate its identification in the clinical laboratory, thereby permitting a future evaluation of its distribution and clinical significance.
TABLE 1.
Characteristicsa useful in differentiating A. radicidentis from other catalase-positive or catalase-variable Actinomyces speciesb
| Species | Catalase | Nitrate reduction | Hydrolysis of:
|
Fermentation of:
|
Production ofc:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aesculin | Gelatin | Urea | d-Ara | l-Ara | Glu | Man | Suc | d-Xyl | Mel | Gly | Pull | Rib | Tre | PAL | PYZ | β-GLU | β-NAG | |||
| Actinomyces radicidentis | + | V | + | W | V | − | − | + | + | + | − | + | − | − | + | + | − | + | + | − |
| Actinomyces bowdenii | + | + | + | − | − | − | − | + | − | + | − | + | − | − | + | + | V (−) | V | + (W) | − |
| Actinomyces canis | + | − | − | − | − | − | + | + | − | V | + | − | + | + | + | − | − | + (W) | − | + |
| Actinomyces hordeovulneris | + (W) | − | + | W | − | − | V | + | − | + | + | − | V | + | V | V | − | V | V | + |
| Actinomyces howellii | + | − | + | − | − | − | − | + | − | + | + | V | − | − | − | V | + | + | + | + |
| Actinomyces naeslundii (serotype I) | V (−) | + | V (+) | V (−) | W | − | − | V (+) | V (−) | V (+) | V (−) | V (−) | − | − | − | + | − | + | + | − |
| Actinomyces naeslundii (serotype II) | V (−) | + | V | − | W | − | − | + | − | + | − | + | − | − | − | + | + (W) | + | + | − |
| Actinomyces neuii subsp. anitratus | + | − | − | − | − | + | − | + | + | + | V (+) | + | − | − | + | + | V | + | − | − |
| Actinomyces neuii subsp. neuii | + | + | − | − | − | + | − | + | + | + | + | V | − | − | V | V | − | + | − | − |
| Actinomyces slackii | + | + | + | V | V (−) | − | − | + | − | V | + | + | − | − | − | + | − | V | + | − |
| Actinomyces viscosus (serotype I) | + | + | + | − | V | − | − | + | − | + | − | − | − | − | − | V | + | − | + | − |
| Actinomyces viscosus (serotype II) | + | + | + | − | V | − | − | + | − | + | − | V | − | − | − | V | + | + | + | − |
| Arcanobacterium phocae | V (+) | − | − | − | − | − | − | + | V (+) | + | V (−) | V (+) | + | − | + | V | + | + | − | − |
Tests performed using commercial API systems, as described in the text. +, positive; −, negative; W, weak reaction; V, variable; V (+), most strains positive; V (−), most strains negative.
d-Ara, d-arabitol; l-Ara, l-arabinose; Glu, glucose; Man, mannitol; Mel, melezitose; Suc, sucrose; d-Xyl, d-xylose; Gly, glycogen; Pull, pullulan; Rib, ribose; Tre, trehalose.
PAL, alkaline phosphatase; PYZ, pyrazinamidase; β-GLU, β-glucosidase; β-NAG, β-N-acetylglucosaminidase.
Description of A. radicidentis sp. nov.
Actinomyces radicidentis (L. n. radix root, L. gen. masc. n. dentis of the tooth, L. gen. masc. n. radicidentis of the root of the tooth) cells are coccoid, stain gram positive, and are non-acid fast and nonmotile, facultatively anaerobic, and catalase positive. Using API systems, acid is produced from d-glucose, maltose, mannitol, melibiose, melezitose, methyl-β-d-glucopyranoside, lactose, d-raffinose, ribose, sucrose, and trehalose. Acid is not produced from d-arabitol, l-arabinose, cyclodextrin, glycogen, pullulan, sorbitol, or tagatose. Esculin and gelatin (weak) are hydrolyzed, but hippurate is not. Alanine phenylalanine proline arylamidase, acid phosphatase, α-galactosidase, β-galactosidase, β-galacturonidase, α-glucosidase, β-glucosidase, leucine arylamidase, and pyrazinamidase are produced. Arginine dihydrolase, alkaline phosphatase, cystine arylamidase, α-fucosidase, ester lipase C8, β-glucuronidase, glycyl tryptophane arylamidase, α-mannosidase, β-mannosidase, N-acetyl-β-glucosaminidase, valine arylamidase, chymotrypsin, trypsin, and pyroglutamic acid arylamidase are not produced. Activities for urease are variable. Acetoin production is weakly positive. Nitrate reduction is variable. Organisms have been isolated from infected root canals of teeth. Habitat is not known. The type strain is CCUG 36733 (= CIP 106352T).
ACKNOWLEDGMENTS
We are grateful to Hans Truper for help in coining the species name and to Lena Dahl for performing PAGE analysis.
Footnotes
Corresponding author. Mailing address: Department of Food Science and Technology, Whiteknights, Reading, RG6 6AP, United Kingdom. Phone: 44 1189357226. Fax: 44 1189310080. E-mail: m.d.collins@reading.ac.uk.
REFERENCES
- 1.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 3.Funke G, Alvarez N, Pascual C, Falsen E, Akervall E, Sabbe L, Schouls L, Weiss N, Collins M D. Actinomyces europaeus sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:687–692. doi: 10.1099/00207713-47-3-687. [DOI] [PubMed] [Google Scholar]
- 4.Funke G, Pascual Ramos C, Fernandez-Garayzabal J, Weiss N, Collins M D. Description of human-derived Centers for Disease Control coryneform group 2 bacteria as Actinomyces bernardiae sp. nov. Int J Syst Bacteriol. 1995;45:57–60. doi: 10.1099/00207713-45-1-57. [DOI] [PubMed] [Google Scholar]
- 5.Funke G, Stubbs S, von Graevenitz A, Collins M D. Assignment of human-derived CDC group 1 coryneform bacteria and CDC group 1-like coryneform bacteria to the genus Actinomyces as Actinomyces neuii subsp. neuii sp. nov., subsp. nov., and Actinomyces neuii subsp. anitratus subsp. nov. Int J Syst Bacteriol. 1994;44:167–171. doi: 10.1099/00207713-44-1-167. [DOI] [PubMed] [Google Scholar]
- 6.Hall V, O'Neill G L, Magee J T, Duerden B I. Development of amplified 16S ribosomal DNA restriction analysis for identification of Actinomyces species and comparison with pyrolysis-mass spectrometry and conventional biochemical tests. J Clin Microbiol. 1999;37:2255–2261. doi: 10.1128/jcm.37.7.2255-2261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawson P A, Falsen E, Akervall E, Vandamme P, Collins M D. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schaalii sp. nov. Int J Syst Bacteriol. 1997;47:899–903. doi: 10.1099/00207713-47-3-899. [DOI] [PubMed] [Google Scholar]
- 8.Pascual C, Falsen E, Akervall E, Sjoden B, Collins M D. Actinomyces graevenitzii sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:885–888. doi: 10.1099/00207713-47-3-885. [DOI] [PubMed] [Google Scholar]
- 9.Pot B, Vandamme P, Kersters K. M. Goodfellow and A. G. O'Donnell (ed.), Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, United Kingdom: J. Wiley and Sons Ltd.; 1994. Analysis of electrophoretic whole-organism protein fingerprints; pp. 493–521. [Google Scholar]
- 10.Schaal K P. Genus Actinomyces. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1383–1418. [Google Scholar]
- 11.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 12.Vandamme P, Pot B, Gillis M, DeVos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wüst J, Stubbs S, Weiss N, Funke G, Collins M D. Assignment of Actinomyces pyogenes-like (CDC coryneform group E) bacteria to the genus Actinomyces as Actinomyces radingae sp. nov. and Actinomyces turicensis sp. nov. Lett Appl Microbiol. 1995;20:76–81. doi: 10.1111/j.1472-765x.1995.tb01290.x. [DOI] [PubMed] [Google Scholar]