Abstract
The Automated Tape-Collecting Ultramicrotome (ATUM) is tape-reeling device that is placed in a water-filled diamond knife boat to collect serial sections as they are cut by a conventional ultramicrotome. The ATUM can collect thousands of sections of many different shapes and sizes, which are subsequently imaged by a scanning electron microscope. This method has been used for large-scale connectomics projects of mouse brain, and is well suited for other smaller-scale studies of tissues, cells, and organisms. Here, we describe basic procedures for preparing a block for ATUM sectioning, handling of the ATUM, tape preparation, post-treatment of sections, and considerations for mapping, imaging, and aligning the serial sections.
Keywords: ATUM, serial section EM, tape collector, 3D EM, SEM
I. INTRODUCTION
New ways to obtain 3-dimensional ultrastructural data are opening exciting frontiers in biology. In the ATUM (Automated Tape Collecting Ultramicrotome) method, sections are cut in a traditional way using an ultramicrotome and diamond knife with a water boat. The sections are picked up on moving tape and then imaged with a scanning electron microscope (SEM). The method was developed by Ken Hayworth, Richard Schalek (sectioning device) and Bobby Kasthuri, and Juan Carlos Tapia (tape imaging method) in the Lichtman lab and was first used to image all the cellular constituents including every synapse, axon and dendrite in a 50-micron cube of mouse cerebral cortex (Kasthuri et al., 2015). This effort involved imaging ~2000 sections (30 nm thick) at an xy resolution of 3 nm per pixel (16,384 × 16,384 pixels). Much of the analysis was done using a computer-assisted manual tracing and segmentation tool (VAST, Berger et al., 2018)
This early effort has been followed by reconstructing neuronal circuits in ~1000-fold larger volumes (cubic millimeter scale). The sectioning approach is quite fast, so the bottleneck has been image acquisition time as the volume size has increased. This limitation has stimulated significant progress on several fronts, such as the Multibeam SEM combining 61 (or 91) single beams (Eberle et al., 2015), supercomputer alignment (Morgan et al., 2016; Hildebrand et al., 2017), and automated segmentation (Ai-Awami et al., 2016; Januszewski et al., 2018).
The underlying procedures of the ATUM method are also still under development. These include fixation/processing, cutting, collecting, protein localization (Fang et al., 2018; Norris et al., 2017), and imaging. These are all being explored extensively, and improvements can be expected.
Our goal in this chapter is to discuss aspects of the ATUM method that will first concern a new lab that is setting up the technique. Also, because these basic procedures are often sufficient for projects in cell biology, which may involve smaller volumes compared to connectomics, we do not emphasize the special challenges related to extremely large petabyte-scale volumes being contemplated in neuroscience projects.
II. METHODS
A. Specimen preparation
Preparing a biological sample for ATUM-based imaging with a SEM involves many of the same chemicals and procedures commonly used for conventional imaging with a transmission electron microscope (TEM), with the only notable difference that more heavy-metal staining is usually needed for ATUM-based SEM imaging compared to TEM imaging. This need is related to the requirement to generate images quickly with the intrinsically slower point by point scanning approach than is required in the widefield transmission electron microscopy strategy. If the electron beam hovers over one pixel briefly, there needs to be a lot of stain (i.e., osmium) to get a recordable signal.
Like any electron microscopy method, ATUM-based projects rely on proper initial sample fixation and subsequent processing. There are many excellent articles on the subject of fixing and processing biological specimens for electron microscopy. A good introductory source is found in the textbook Bozzola and Russel, 1999, and there are many protocols and methods available for specific tissues and animal models, most of which are well suited for ATUM-based SEM imaging (Tapia, et al., 2012; McDonald, 2010). Most protocols are optimized for the preservation and staining of membranes, while compromising other cellular components such as the cytoskeleton. The solutions used for the primary fixation and heavy metal staining can be adjusted to highlight other cellular components as desired but will not be discussed here.
Ideally, the sample to be examined will be processed with a protocol optimized for ATUM-based SEM imaging (Tapia, et al., 2012; Baena and Terasaki, 2019), however, if the sample has already been processed for conventional TEM imaging, it can also be used for ATUM-based SEM imaging. In such case, additional heavy-metal contrast is applied to the serial sections after they have been cut, collected on tape, and the tape has been affixed to a wafer (see section II.H for post-staining sections).
The resin formulation used for tissue embedding is important for ATUM sectioning since it needs to have uniform cutting properties for the hundreds or thousands of serial sections. We typically use epon resin, although other resins have been found to be appropriate. The components used to make resin can be combined in different ratios to control the hardness of the polymerized block. A block made with a hard resin formulation will make cutting ultrathin sections (30–60 nm) much easier, but the sections may be more prone to form wrinkles at the interface between the tissue and the resin. To aid in preventing this, it is recommended to keep the ratio of tissue-to-resin on the section high (discussed in more detail in section II.C.). For cutting and collecting thick sections (200–500 nm), we recommend using a soft resin formulation to further reduce the probability of wrinkle formation.
B. Ultramicrotome
For the beginner, the ultramicrotome can present some exceptional challenges. In particular, the user has to understand the complex geometrical relationships between six degrees of freedom (X and Y axes of the knife holder, rotational angle of the knife holder, clearance angle of the knife holder, and the pitch and rotation of the block). These parameters are set by viewing the knife holder and block in three dimensions using the binocular stereomicroscope that is equipped with the ultramicrotome. Successful positioning of the knife and block requires manipulating a number of very small adjustments, which becomes less difficult with practice and training. Importantly, the tape-based approach does not in any way eliminate the need for a skilled practitioner of ultramicrotomy, thereby we recommend spending time learning the basics of ultramicrotomy and getting used to using diamond knives and an ultramicrotome. Excellent introductory material can be found in Bozzola and Russell, 1999; Hagler, 2007; Maunsbach and Afzelius, 1998.
Our own serial sections have been generated and collected using both RMC Powertome (Boeckeler Instruments, Inc. Tucson, Arizona) and Leica UC6 ultramicrotomes (Leica Microsystems Inc., Buffalo Grove, IL). The ultramicrotomy table and the room environment are important for the successful collection of ultrathin serial sections and are discussed below (see section II.F).
C. Trimming the block
Appropriate trimming of the block face is critical for the continuous and reliable collection of serial sections on tape. Though many of the physical and mechanical requirements of blocks used in ATUM-based serial electron microscopy are similar to blocks used in conventional electron microscopy techniques, the number of serial sections cut and collected by tape usually exceeds the number of serial sections in traditional electron microscopy by hundreds or thousands. The up-scaling in the number of serial sections requires that the tissue blocks have uniform cutting properties and that the cutting face (i.e. block face) is well trimmed on its sides and to an appropriate depth (also known as the mesa).
The block face size and shape is generally dictated by the tissue geometry and by the width of the diamond knife, which rarely exceeds 4 mm. The tape conveyor (ATUM) can collect tissue sections of any size or shape, and the sections can be picked up independently or as a ribbon. Very often, the block face shape and the smoothness of the trimmed edges will dictate the ease of cutting and collecting sections. Therefore, we highly encourage the ATUM practitioner to optimize each sample block face carefully and individually. Below, we discuss useful considerations for trimming a block for ATUM-based SEM imaging.
Block face size
It is generally valuable to trim the block face such that the amount of non-relevant surface is minimized. A useful rule is to trim away most, or if possible, all of the resin that does not contain the tissue being studied. Trimming away empty resin leads to a high tissue-to-resin ratio on the block face, which will make cutting and collecting sections easier than when the ratio is low. This approach reduces the possibility of wrinkles forming on the sections and also helps to extend the lifetime of the diamond knife. Creating a block face with a high tissue-to-resin ratio is not always possible to accomplish when working with very small samples as this would result in a very small block face. In these cases, small samples can be embedded with supporting tissue (such as brain or liver) around them in order to create a larger block face with a high tissue-to-resin ratio (Hildebrand et al., 2017).
The dimensions of our typical bock faces vary greatly depending on the sample being studied. Generally, they tend to be no larger than 4 mm × 2 mm (length × width) or smaller than 0.5 mm × 0.5 mm. A large block face will usually provide sections that get picked-up independently from each other on the tape, making their collection easier. A disadvantage of working with a large block face is that the sections are more likely to have sectioning artifacts such as compression from passing through the knife edge and wrinkles. The smaller the block face, the less likely it is for the sections to acquire sectioning artifacts, however, if the sections are small enough, then two or more sections will collect in the space between the knife edge and the tape collector before they are picked up. It is problematic if they become separated and drift because this may cause the sections to get picked up out of order or drift past the side of the collector and not get picked up at all. If the small sections are trimmed so that they adhere to each other to form a ribbon, the tape collector is usually able to pick them up successfully (Fig. 1A).
Figure 1. Schematic of typical block face shapes and a prediction on how they are picked up by the tape.
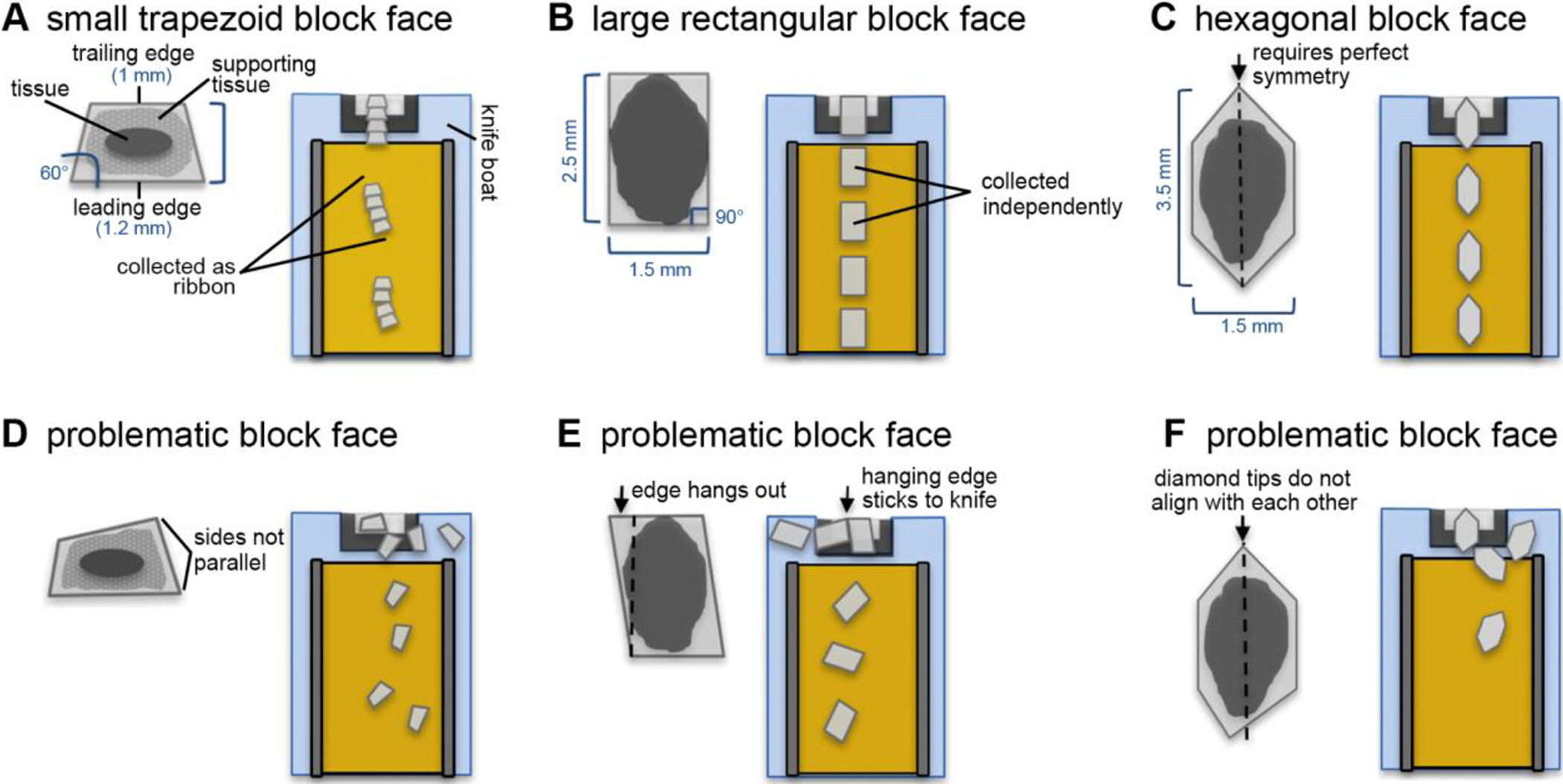
(A) A properly trimmed trapezoid block face tends to give sections that form ribbons. This shape is desirable when the block face is small. (B) A properly trimmed rectangular block face tends to give sections that are picked up independently. (C) An elongated hexagonal-shaped block face eliminates the formation of ribbons. (D) Example of an inappropriately trimmed block face. The leading and trailing edges are not parallel to each other. This leads to an abrupt rotation of the section after the trailing edge is cut, which may lead to some sections becoming stalled on the knife boat. (E) Example of an inappropriately trimmed block face. The trailing edge is longer on one side than the leading edge. The hanging edge tends to stick to the diamond knife edge, impeding the section from moving towards the tape. (F) Example of an inappropriately trimmed block face. The tips of the hexagonal-shaped block face do not align with each other, causing the sections to rotate in the knife boat.
Figure 1A–B illustrates the behavior of a small and a large block face shape, and consecutive section uptake by the tape. The tradeoff between section size, ease of pick-up, and tendency of artifact formation should be evaluated by the microtomist for each sample and will become more intuitive after considerable practice.
Block face shape
For post-imaging stitching and alignment purposes, it is useful to give the block face a shape that facilitates the collection of sequential sections that remain aligned during the cutting procedure. We recommend using a rectangular or trapezoid-shaped block face almost exclusively. A trapezoid block face consists of parallel leading (bottom) and trailing (top) edges, and side edges that form 45–60° angles with the leading edge, resulting in a trailing edge that is shorter than the leading edge. Trapezoid block faces provide the most stable sections, which often remain attached to each other as ribbons (Fig. 1A). A rectangular block face has 90° angles on all corners and the trailing and leading edges are of equal length. Rectangular sections can be collected as ribbons or independently depending on the length of the side edges. Side edges of ~4 mm allow for a direct-to-tape collecting approach, in which the leading edge is picked up by the tape conveyor while the rest of the section is being cut (Fig. 1B). This creates a gap between adjacent sections, which becomes helpful during wafer mapping (see section II.I).
For both trapezoid and rectangular block face shapes, it is crucial to keep the leading and trailing edges parallel to each other, otherwise sections will rotate when the following section is cut, and may stall on the knife boat (Fig. 1D). Similarly, a trailing edge that does not match (or is longer than) the leading edge may cause sections to rotate due to the unmatched portion of the trailing edge becoming stuck to the diamond knife (Fig. 1E). The rotation of sections by itself is not fatal as this can be easily corrected during the wafer mapping. It only becomes a problem if the sections float away from the tape conveyor in the knife boat and become disordered or separated.
In certain cases, the microtomist will want to avoid collecting sections as ribbons because it may become difficult to determine where one section ends and another one starts on the tape, especially when the ribbons consist of sections that are small and rectangular or squared. For this, we have used an elongated hexagon-shaped block face (Morgan et al. 2016; Hildebrand et al., 2017). In this approach, the user begins with a rectangular block face (~4 mm × 2 mm), but then the trailing and leading edges are trimmed into half diamonds that are symmetrical to each other (Fig. 1C). With this block face, the trailing edge does not stick to the diamond knife edge or to the following section, thereby eliminating the formation of ribbons. This block face shape requires perfectly symmetrical trimming so that the ends of the diamond align to each other on adjacent sections (Fig. 1F).
There are many ways to collect serial sections which do not require any of the block face shapes mentioned here. In many cases, non-conventional block face shapes will be needed for particular tissue geometries. We highly encourage the microtomist to experiment with different block face shapes, and to adjust the trimming as needed for each block and for the number of sections wished to be collected.
Trimming tools
For block trimming, several types of tools can be used: razor blades, glass knives, and diamond trimming knives. We have extensively tested these trimming tools for the purpose of providing block faces that are useful for the cutting and collection of serial sections.
Standard single-edged razor blades work well for an initial rough trimming of the block face. The blades are inexpensive, but practice is required to produce evenly trimmed sides and care must be taken not to cut one’s fingers.
Glass knives have a long history in the preparation of biological tissue for electron microscopy (Bozzola and Russell, 1999). Glass knives are fabricated using a glass knife breaker, which produces 45 degree knives from 6.4, 8 and 10 mm thick glass (Hagler, 2007). Traditionally, glass knives are used for generating sections but they dull quickly and so are not ideal for serial sections. They can be used to trim blocks, but caution must be exercised if using both glass and diamond knives intermittently because glass shards embedded into the resin block can chip the diamond knife.
Diamond trimming tools are used exclusively in our laboratories. Though much more expensive than the aforementioned tools, diamond knife tools provide consistently smooth and straight edges. The Trim 45 diamond knife tool (Diatome, Hatfield, PA) and the Trim 90 tool (Diatome, Hatfield, PA) (Fig. 2A) produce the best-finished surfaces (Fig. 2B). The Trim 90 tool has an additional polished facet that produces a smoother mesa sidewall surface compared to the Trim 45. A smoother mesa sidewall increases the likelihood that an ultrathin section will cut and move smoothly across the knife edge.
Figure 2. Trimming a block for the ATUM.
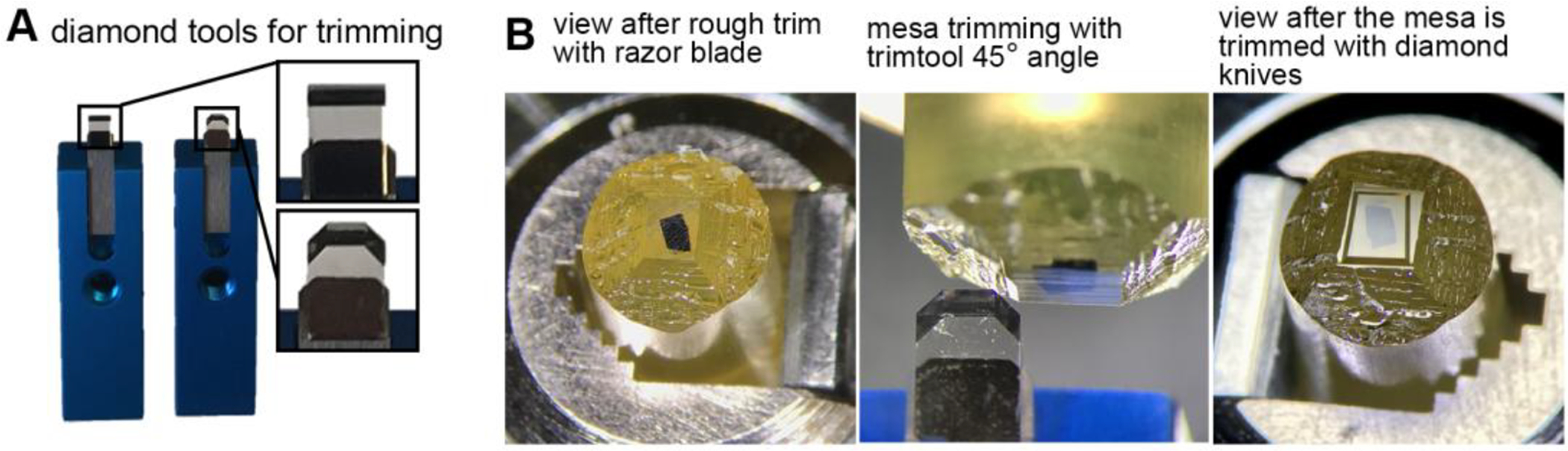
(A) Common diamond trimming knives used for shaping the block face and mesa. A single edge-razor blade is used for an initial rough trimming, then, diamond trimming knives such as Trimtool 45° (left) and Trimtool 90° (right) (Diatome-US, Hatfield, PA) are used to shape and smoothen the block face and mesa. (B) Pictures of a block face during the trimming procedure. Left: A block face after rough trimming with a razor blade. Middle: trimming of the mesa with the side of a Trimtool 45° (Diatome-US, Hatfield, PA). Right: view of a trapezoid-shaped block face and mesa after trimming with diamond knives.
The trimming speed and size is important for creating a smooth mesa. For trimming the block face and sidewalls with a diamond trimming knife, we set the ultramicrotome feed size to 300–500 nm at a speed of 50–80 mm/sec.
D. ATUM
The automated tape-collecting ultramicrotome (ATUM) is a device that facilitates the collection of hundreds to thousands of sections on tape while they are simultaneously cut with an ultramicrotome. The ATUM works by reeling tape similarly to a conveyor belt. The tape collects the sections as the cutting proceeds. The speed at which the tape moves is adjusted such that it is the same or slightly faster than the cutting speed of the ultramicrotome.
Ideally, the ATUM is set up so that as a section is cut, it pushes the previously cut section over the moving tape, which is in turn pulling some of the water with it. The section floats on the water over the tape and is pulled away from the diamond knife edge by the action of the water being pulled by the moving tape. Eventually, the water recedes and the section attaches to the tape, at which point it begins to dry on its way towards the take-up reel.
Although most of our data was acquired using a custom-built ATUM, a commercial version is available through RMC-Boeckeler (Tucson, AZ) called “ATUMtome” when combined with their PowerTome ultramicrotome. We will use the terms “ATUM” to refer to the tape-collecting device by itself, and “ATUMtome” when it is combined with the ultramicrotome.
ATUM parts
Figure 3A shows some of the primary components of the RMC-Boeckeler version of the ATUM. These include a supply reel for the tape, a take-up reel, a tape tensioner and slip clutch to maintain tension, the tape snout which is placed inside the knife boat, bottom and top pinch rollers, Z-height control, and a syringe pump to maintain the water level in the knife, which is controlled automatically by a camera that looks at the water meniscus. In addition, the ATUM may be equipped with a Static Ionizer used to eliminate static charge which builds-up on the block face during sectioning (Diatome Static Line Ionizer II, Electron Microscopy Sciences, Hatfield PA). This may be placed approximately 5 cm away, facing the side of the block face (not shown). In addition to the hardware, the ATUM requires control software, which is provided by the manufacturer.
Figure 3. Components of the ATUM and path of the tape through the ATUM.
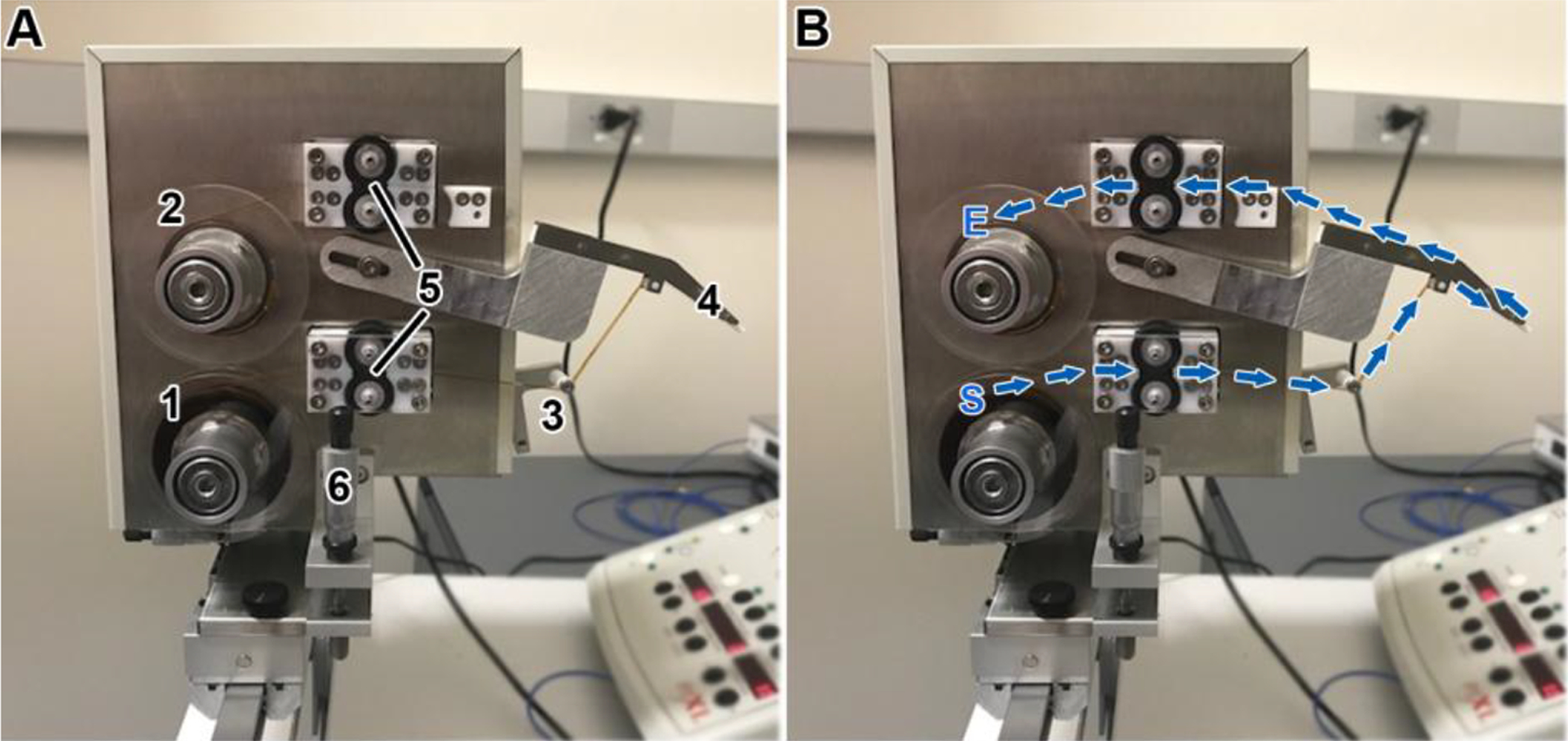
(A) Picture of the ATUM indicating its main components. (1) supply reel for the tape, (2) a take-up reel, (3) tape tensioner, (4) snout, (5) bottom and top pinch rollers, and (6) Z-height control. (B) The path of the tape begins at the supply reel (S), goes through the bottom pinch rollers (set in the “on” position), around the tensioner arm roller, around the snout, through the top pinch rollers, and ends at the take-up reel (E) to which it is attached with adhesive tape.
Setting up the ATUM: tape feeding
The path of the tape through the ATUM starts at the supply reel, moves through the lower tape guides and lower pinch rollers, past the tensioner arm, into the diamond knife boat, over the tip of the tape snout, out of the diamond knife boat, through the upper tape guides and the upper pinch rollers, and onto the take-up reel (Fig. 3B). The tape is moved along this path by the lower and upper pinch roller motors, while tension before the lower pinch roller and after the upper pinch roller are supplied by feed and take-up reel tensioner motors attached to the slip clutches. The tension is adjusted to a certain value in the ATUM software following the manufacturer’s recommendations.
Gloves should be worn when handling tape to avoid contaminating it with grease that would in turn contaminate the SEM chamber. The tape is fed to the ATUM manually by taking the end of the tape from the supply reel and feeding it to the bottom pinch rollers which are set in the “on” position in the ATUM software. As the tape moves through the bottom pinch rollers, the operator directs it around the tensioner arm roller, around the snout, through the top pinch rollers, and then it is adhered to the take-up reel with adhesive tape. After this stage, the snout can be positioned in the diamond knife water boat.
Setting up the ATUM: positioning of the snout and starting a cutting run.
Once the microtomist is satisfied with the shape and size of the block face, the sides of the mesa are trimmed and smooth, and the cutting face has the sample area of interest, the ATUM can be positioned for the collection of sections. Note that ultrathin (30–60 nm) cutting of serial sections (> 100 sections) is performed only with diamond knives. Our serial sections are cut using either an Ultra 35° or Ultra 45° diamond knife (Diatome, Hatfield, PA) with widths up to 4 mm. Diamond knives with a larger boat (Ultra Jumbo, Diatome, Hatfield, PA), especially useful for accommodating the ATUM, are now sold commercially.
Begin by overfilling the knife boat with water. This is advised because the water level on the knife boat is lowered when the ATUM is dipped in, causing the diamond knife edge to become dry. For a beginner, it is sometimes easier to lower the snout into an empty knife boat so that the clearance between the boat and the snout can be easily seen. The knife boat can be filled with water after the snout has been positioned.
Place the ATUM perpendicularly to the diamond knife edge over the knife boat (Fig. 4A).
Slowly lower the snout into the water until it is ~2 mm deep (Fig. 4B) being cautious not to touch the walls of the boat while lowering the snout.
- Advance the snout towards the knife edge until it is ~4 mm away from it (Fig. 4C).
- Take caution while doing this because if the snout is placed too deep in the water boat, the metal sides of the snout may touch the front curved wall of the knife boat, which may in turn push the knife closer to the block, resulting in a thick section being cut the next time the microtome wheel is turned. The operator can look at the snout and assess its depth and proximity to the diamond knife edge by looking both from the top and from the side intermittently.
When the snout is at the appropriate depth and distance from the diamond knife edge, turn the ATUM on using the software (the cutting speed should be set to 0.6–0.3 mm/sec at this stage). The water will initially travel upwards with the tape, and then recede until it reaches an equilibrated level.
After the water has equilibrated, adjust the level by using a 25-gauge syringe to add or remove water as needed. The correct water level can be estimated by looking at the reflection of the ultramicrotome lights on the water with the ultramicrotome binoculars (Fig. 4D). The ATUM snout should sink ~2 mm deep in the water and it should not make contact with any of the knife boat walls.
- With the ATUM “on” (tape moving), cut a few sections by manually turning the ultramicrotome wheel. Adjust the water level again if needed and move the snout closer (if possible) or further from the diamond knife edge such that it is positioned at ~1.5 lengths of a section.
- Note that a distance of 1.5 lengths of a section is only a suggestion for beginners, and may not be achievable if working with small sections. As the ATUM user becomes experienced, they may choose to adjust the snout distance differently for every block.
- After successfully cutting and collecting a few sections manually, start the automatic cutting with the ultramicrotome at a thickness of 60 nm (if the ultimate goal is to cut ultrathin sections), or 500 nm (if the ultimate goal is to cut thick sections) at a cutting speed of 0.3–0.6 mm/sec. Allow enough time for the ATUMtome to cut and collect ~20 sections.
- Try not to touch the ATUMtome or the table it rests on, disturb the sections with your breath, or make jerky movements around the area when the ATUMtome is automatically cutting. Even small vibrations can affect the cutting procedure and cause the ultramicrotome to cut thick-and-thin sections, skip cutting a section, or create chatter on the sections (see section II.F for consideration on the room environment).
- While the ATUMtome is automatically cutting, pay close attention to the way the sections are collected by the tape and assess whether it is “reliable”. A reliable cutting round should theoretically allow the operator to leave the room without worrying about whether the sections are being collected in the right order or if they are becoming lost in the knife boat. However, we encourage the beginner ATUM user to stay and observe the cutting round (usually via a camera set up on the microtome binoculars), again exercising caution as to not touch the ATUMtome or disturb the environment.
- If the sections are rotating severely and floating away in the water boat, stop the cutting run while the ultramicrotome is in the advance position, then try to move the ATUM snout closer to the diamond knife edge so that the tape can collect the sections before they float away. Often times, however, section rotation is caused by the way in which the block face was trimmed, especially when the leading and trailing edges are not parallel to each other. If necessary, remove the ATUM from the knife boat and re-trim the block to fix any irregularities.
- If the initial cutting and collection of sections is successful, begin to decrease the thickness of the sections in 5–10 nm intervals cutting ~5 consecutive sections on each interval until the desired thickness is achieved. Adjust the cutting speed accordingly based on the section thickness. The cutting speed we work at ranges from 0.6 to 0.1 mm/sec for section thicknesses from 500 to 20 nm. The thickness and cutting speed can be adjusted while the microtome is in the advance position.
- Note that the thinner the sections are, the more difficult the cutting run becomes. In our experience, ≥60 nm is generally easy, 50–40 nm is feasible with the right block face shape, and ≤30 nm is difficult, requiring perfect trimming of the block face and the right resin hardness (harder rather than softer resin formulation).
The automatic water level control system equipped with the ATUMtome can be set up at this point following the manufacturers’ instructions.
Figure 4. Procedure to position the ATUM snout in the knife boat.
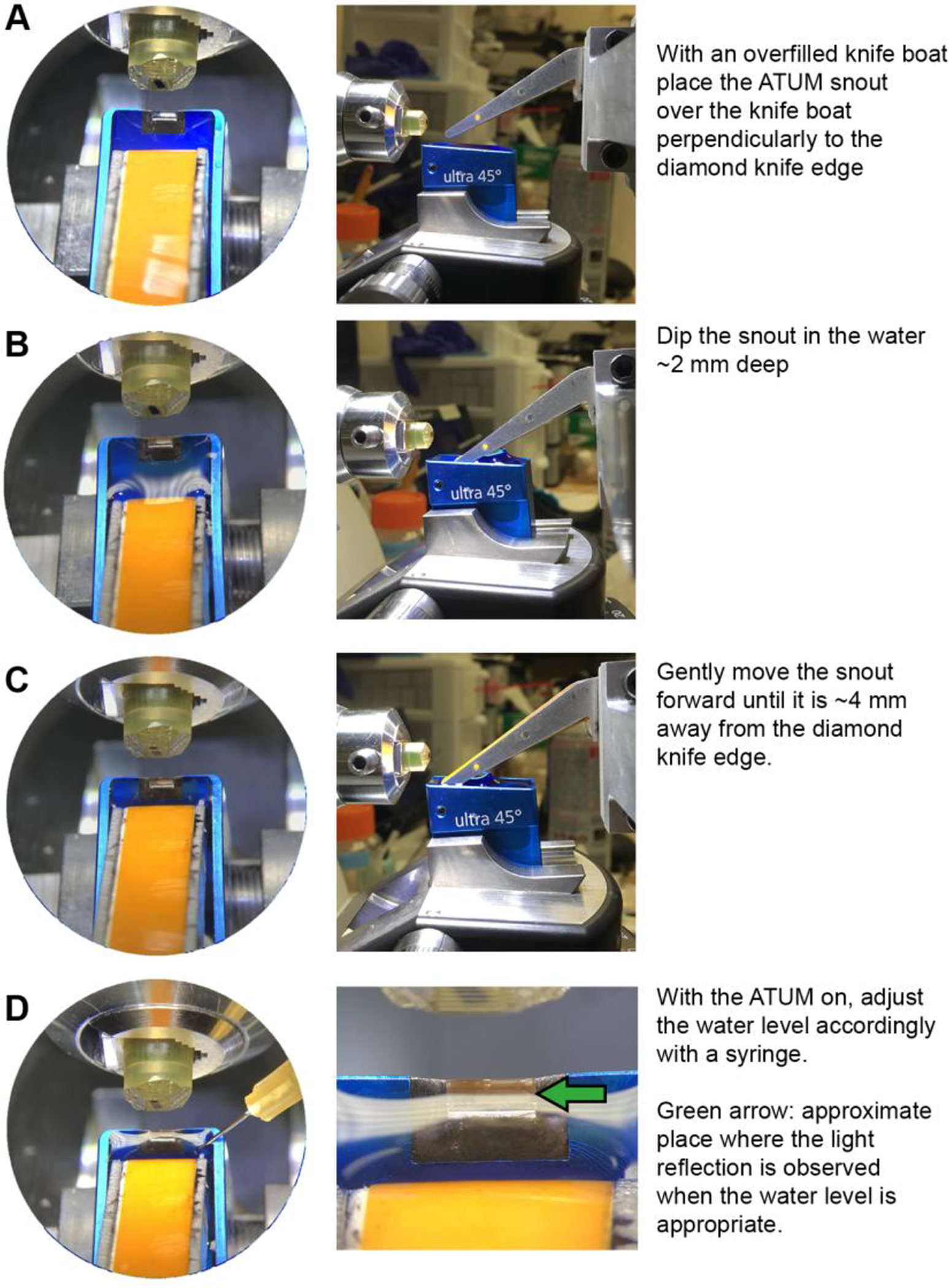
(A-D) Panels on the left show view as seen from the ultramicrotome binoculars. Panels on the right show view as seen from the side (except for D, which shows view from binoculars). Text provides summarized procedure for placing the ATUM snout in the knife boat.
Finishing the cutting run
When enough sections have been collected, stop the automatic cutting while the ultramicrotome is in the advance mode. Raise the nose of the ATUM above the height of the knife boat and move it backwards and to the side. At this point, the tape can be reeled using the ATUM software until the last section is on the take-up reel. Cut the tape with scissors at a point between the top pinch rollers and the take-up reel.
E. Collection tape preparation
Collection tapes
A number of plastic tapes have been used to collect sections, including: Kapton HN, aluminum-coated Kapton HN, polyether ether ketone (PEEK; Sheldahl, Northfiled, MN), and carbon nanotube (CNT)-coated polyethylene terephthalate (PET) tape (Kubota et al., 2018). Kapton tape (8 mm wide) is the most commonly used and widely available tape for ATUM projects.
The physical properties of the tape affect how the sections are picked up from the knife water boat. It is very valuable to make the tape more hydrophilic by treating it with a glow discharge system prior to the collection of sections. Additionally, the tape must provide a path to ground for the imaging electrons in the microscope. Without this path, electrons accumulate in the section and repel later-arriving electrons, thereby disrupting the imaging process. Except for the aluminum-coated Kapton and the CNT tape, the user must treat the tape to make it conductive, either by sputter coating with a metal or by depositing a thin film of carbon. Below we describe the suggested treatments for Kapton tape: plasma/glow discharge system and carbon coating.
Plasma/glow discharge system for tape
The polymer tapes in use are intrinsically hydrophobic, and thus not very wettable. By passing the tape through a glow discharge plasma system (before collecting sections), the ionized atoms in the plasma create polar functional groups on the tape surface. This makes the surface more hydrophilic, minimizing water beading and the concomitant stresses that occur as the water trapped between the tape and section evaporates. These stresses may cause wrinkles in the sections, therefore, we suggest always treating the tape with the glow discharge system before collecting sections.
There are many commercial glow discharge systems available for treating TEM grids, but the chamber that comes with them is typically not large enough for a source and a take-up reel for the tape plus a turning mechanism. A common solution to this is to construct a larger, custom Plexiglas chamber although a commercial product designed specifically for glow discharging tape is expected to become available from Zeiss.
In the system described in Figure 5, a chamber was designed to fit on a Pelco EasiGlow system (Ted Pella, Redding, CA). The parts listed in the Materials section were used to build the chamber and tape transport mechanism. A continuously turning motor or a stepper motor coupled by a drive belt is used to move the tape.
Figure 5. Glow discharge system for tape.
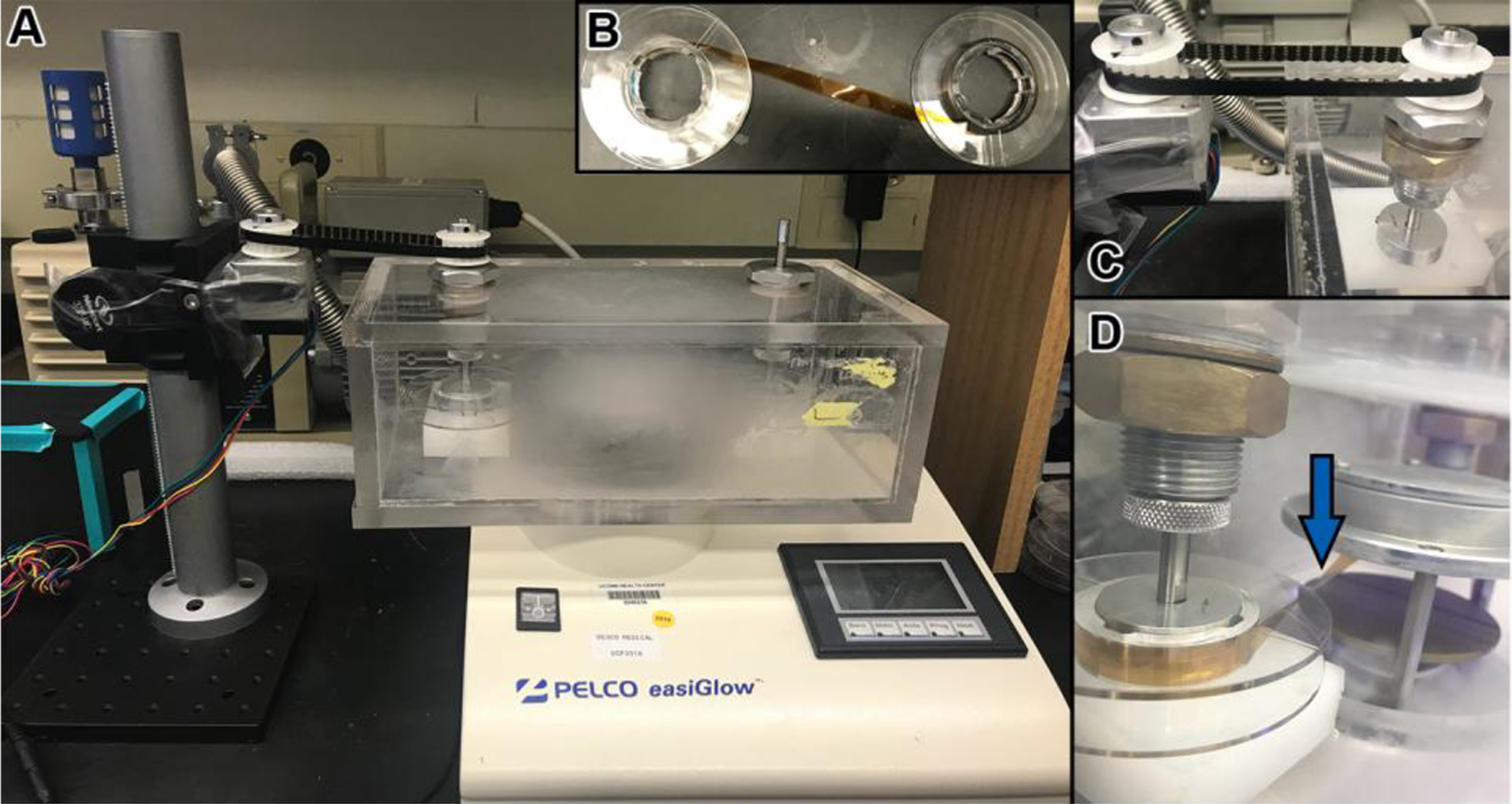
(A) Picture showing the PELCO easiGlow discharge unit (Ted Pella, Inc.) with a custom-made chamber. Within the chamber, the tape is passed from a source reel, through the glow discharge area, and is then collected on a take-up reel. The take-up reel is turned by a motor and a belt system (left). (B) Before placing the tape in the chamber, it is attached from the source reel to the take-up reel with adhesive tape. (C) Motor and belt system for turning reels with tape. (D) One of the reels is turned accordingly so that the tape runs parallel to the discharge plates and the underside of the tape (the side which will collect the sections) faces up as it runs in between the plates (blue arrow).
Carbon coating
If the user intends to image the specimen by secondary electron emission, the conductive carbon layer must be deposited on the tape before collecting the sections because a conductive layer on top of the section would interfere with the secondary electron emission. For backscatter imaging, the conductive layer can be applied before collecting sections or after the sections are collected and the tape is affixed to a wafer.
For carbon coating sections on tape affixed to a wafer, carbon evaporators commonly found in electron microscopy facilities can be used since their chambers are usually large enough for a wafer to fit in. In this approach, the wafer is placed in the carbon evaporation chamber and a film of carbon is deposited under vacuum.
F. Considerations for the ultramicrotomy room
Air currents from ventilation systems, windows, or fans can have a detrimental effect in the way sections are cut. Vibrations from the building, pedestrian traffic, and numerous other sources can disrupt the cutting and collection of sections, and can diminish the diamond knife lifetime. The room temperature and humidity should be controlled for consistent results.
Useful considerations to minimize disturbances:
Ideally, the room should be located on the lowest possible floor of the building, such as the basement.
The room should be small and only used for the purpose of ultramicrotomy. Pedestrian traffic and opening/closing of doors should be minimal.
Strong air vents should be covered or a plastic enclosure should be placed over the ATUMtome to shield it from air currents.
The ATUMtome should be set on an ultramicrotomy table or an air table that is freely-floating using house air or a small compressor. The sides of the table should not touch the walls of the room.
A room humidifier can be used to keep the humidity constant in order to achieve consistent results. The ideal relative humidity is 40–60% to avoid static electricity and the temperature stability should be about +/− 1°C per hour.
G. Wafer fabrication
Wafer Specifics
The primary purpose of the wafer is to provide a flat, stable and electrically conductive surface for adhering the tape with sections, which can then be mounted on an SEM stage. Generally, silicon wafers are specified by diameter, dopant, orientation, resistivity, thickness, polish, and grade. We typically use 100 mm (diameter) silicon wafers (University Wafer, South Boston, MA). We have found that wafers with a thickness of at least 500 μm and with a single side polish (the top surface) are sufficient for most ATUM projects. Wafers with thicknesses less than 500 μm tend to be too fragile and are not advised. While a double sided polished wafer can be used, the wafer cost increases by ~60% and does not provide a significant advantage. To our knowledge, both N- or P-doped wafers, with a low resistivity ranging from 0–100 ohm-cm can be used for ATUM-based projects.
Mounting tape on a wafer
The space used for wafer fabrication should be clean and as dust-free as possible. Gloves should be worn when handling tape and wafers in order to aid in maintaining a clean SEM chamber.
Remove the take-up reel from the ATUM and place it on a reel holder (Fig. 6A) with the sections facing up. Placing the reel holder on a cutting board facilitates unreeling and cutting the tape.
Lay down strips of double-sided carbon tape on the wafer parallel to each other (Fig. 6B, left). Some space should be allotted on the wafer surface to allow for labeling and placement of fiducials (if required for wafer mapping).
Unreel the tape with the sections facing up and cut it with a scalpel into strips that are the same length as the double-sided carbon tape strips that were laid down on the wafer. It is helpful to use the paper/plastic cover of the double-sided tape as a reference for the length of the Kapton tape strips.
Using two forceps, take the strips of Kapton tape with sections and lay it over the adhesive carbon tape (Fig. 6B, middle). Avoid touching the sections with the forceps. A seam roller can be used to press the Kapton tape against the adhesive tape.
Continue to lay the rest of the Kapton tape strips over the carbon tape guides. Use caution to keep the order and orientation of the sections consistent throughout the wafer (e.g. the order of the sections goes from top to bottom and from left to right) (Fig. 6B, right).
Copper tape or carbon adhesive tape can be used to provide additional grounding of the tape to the wafer.
Label the wafer and attach fiducial TEM grids, if needed (Fig. 6B, middle). At this point, the wafer can be coated with carbon if the sample will be used for backscattered electron imaging.
Use a dust-blowing device to clean the surface of the wafer and sections before placing the wafer in the SEM chamber. Wafers with sections can be placed in petri dishes or compact-disc boxes and stored in airtight boxes indefinitely.
Figure 6. Mounting tape with sections on a wafer.
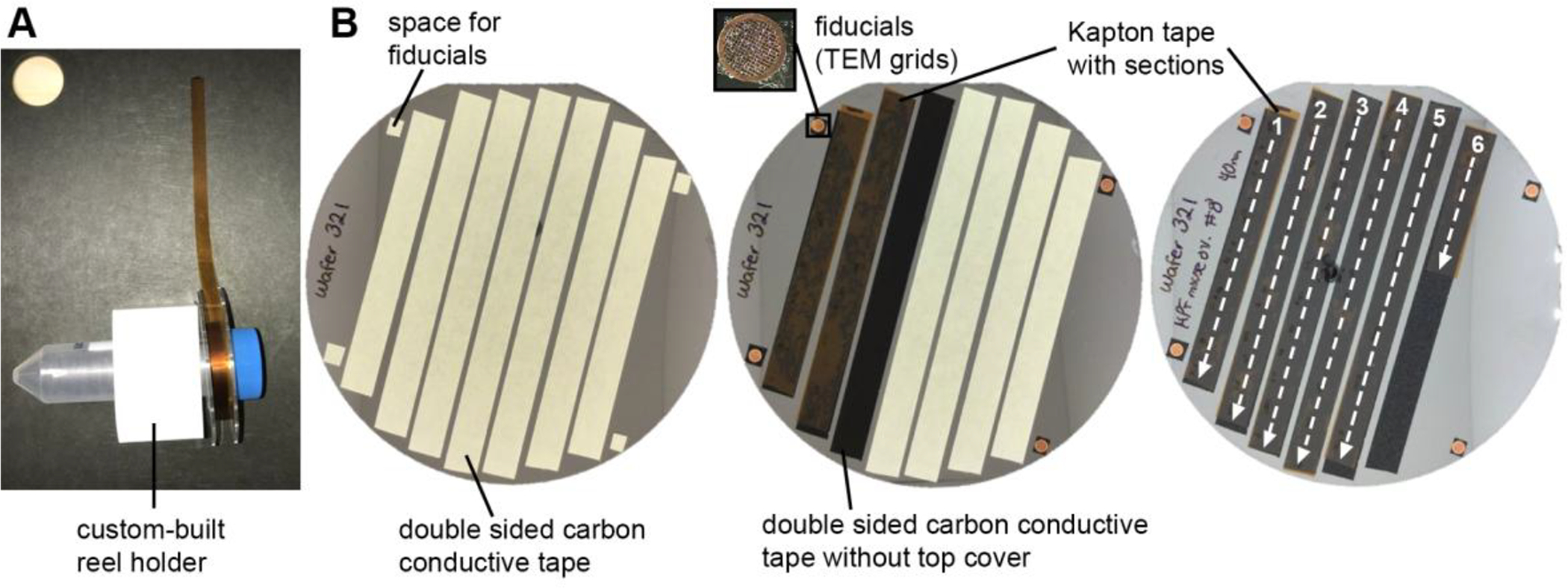
(A) A custom-built reel holder holing a reel with tape with sections. The tape has been spread over a cutting board so that it can be cut into strips. (B) Procedure showing how a wafer is made. Left: strips of double sided carbon conductive tape are laid down along the wafer surface. Small squares for adhering fiducials are placed where appropriate. Middle: the Kapton tape with sections is cut into strips that are then placed over the adhesive carbon tape guides. Right: The orientation and order of the strips of tape should be kept consistent throughout the wafer. In this case, the order of the sections goes from top to bottom and from left to right. The wafer will be mapped in this order.
H. Post-staining sections on wafers
Processing biological samples for ATUM-based SEM imaging usually requires more heavy metal staining than samples processed for TEM imaging. Good heavy metal contrasting can be easily obtained by staining the tissue en-bloc as described in section II.A. If the contrast is still limited, additional membrane contrast can be obtained by post-staining sections on tape, in a similar way to post-staining sections on TEM grids.
After adhering the Kapton tape with sections to the wafer, we recommend placing the wafer in a high vacuum for a few minutes before starting the post-staining procedure. This will allow the Kapton tape to adhere tightly to the carbon tape, reducing the risk of trapping aqueous solutions between the two tapes during the post-staining procedure. The vacuum used for carbon evaporation or the SEM chamber vacuum work well for this purpose. Treating the wafer with affixed tape with a glow discharge system prior to the post-staining procedure will help by making the tape more wettable and allowing the reagents to spread across the sections more easily.
Typical post-staining reagents include 4% uranyl acetate (aqueous or in methanol) and stabilized lead citrate. These solutions are usually drawn into a 10 ml syringe and applied through a 0.22 μm filter directly on the sections. The solutions are washed off by submerging the wafer in double distilled water in 150 mm petri dishes. Additional post-stain solutions may be applied to the wet sections and the washing steps are repeated.
The following post-stain solutions have provided us good results for sections on Kapton tape, but not aluminum tape. Any other post-staining protocol for TEM grids should work as well.
Uranyl acetate:
1–4% in water for 3 minutes.
Water washes:
3 × 3 minutes each with double distilled water in 150 mm petri dishes with gentle agitation.
Lead:
0.5% stabilized lead citrate in water for 3 minutes.
Water washes:
3 × 3 minutes each with double distilled water in 150 mm petri dishes with gentle agitation.
As with TEM grids, the water washes should be generous and the post-stain reagents should never be allowed to dry on the sections as this may cause heavy metal precipitates that will affect the quality of the images. After the final water wash, use a hair drier or dust-blowing device along the tape to blow off the water and avoid accumulations of large drops of water. The wafer is then placed in a 60°C oven for 5–10 minutes or until it is completely dry. It is critical to remove all the water before placing the wafer in the SEM chamber.
I. Imaging and wafer mapping
Imaging is often the most problematic step in the ATUM workflow, and it seems to be particularly true for new labs. This section discusses why imaging can be challenging. Because multiple brands of microscopes can be used (i.e., Zeiss and Thermo Fisher/FEI) as well as several software programs, we will deal more with generalities than with detailed procedures.
Microscope parameters
One important choice is whether to use secondary electron emission or backscatter electron imaging. Acquisition by secondary electron emission can be ten times faster, because the detector signal chain in the electron detector is faster and more efficient than backscatter detection. On the other hand, backscatter imaging produces images of larger dynamic range, which leads to higher contrast. In addition the scanned electron voltage in backscattered mode is higher (~10kV) than in secondary mode (~2kV) meaning the images will be sharper. The choice of detector will dictate when the carbon layer is added to the sample. As discussed in section II.E, secondary electron emission imaging requires the carbon layer to be deposited on the tape underneath the sections while for backscatter electron imagining, it can be deposited either underneath or on top of the sections.
Automated imaging
It is very possible to manually collect serial section images. A microscope operator with no time pressure and infinite patience will make perfectly centered, in-focus data. So, for any project, there is a fail-safe way to get the data! The problem is that manual collection is so time-consuming and is beset by errors as fatigue and tediousness set in. In practice, manual serial imaging is not a viable solution except for small series (<100 sections) or other unusual circumstances that require it.
Fortunately, modern SEMs have excellent automation capabilities. The microscope will reliably go to a specified location, perform an auto-focus / auto-stigmation operation, and take a picture with the specified pixel resolution and pixel number.
A typical large size for a section is 2000 microns × 2000 microns square (i.e., 2 mm × 2 mm). The sections are on a silicon wafer which is held securely on a moveable microscope stage within the SEM chamber. The sections are sometimes picked up in different orientations (see Figure 1). Every point in every section has a unique x-y coordinate on the microscope stage. In order to automate image collection, one must therefore determine, for each section, the points corresponding to the center of the biological structure of interest.
Pixel Math
The user should learn “pixel math”. This is somewhat akin to learning “solution math” for making solutions in the lab. This only involves arithmetic, but an error can have big consequences. It is not difficult to be off by thousand-fold or more due to the wrong use of units or by multiplying instead of dividing. In general, some basic calculations are necessary to optimize image collection; usually one is balancing the quality of the data (resolution, noise level), which necessarily slows the imaging, against the time involved in obtaining the image.
Three key quantities are the number of pixels, resolution, and field of view (FOV; which can also be called ROI = region of interest, both refer to the height and width of the area of the image). In survey mode, the microscope is typically set to obtain 1024 × 1024 pixel images, whereas in data collection mode, this can be 16,384 × 16,384 or even higher on some SEMs. At 1 nanoAmp of current, the beam resolution (spot size) is ~2–3 nanometers (depending on the voltage). (The SEM range of pixel resolution is between ~2 to ~2000 nanometers per pixel). These three quantities are related by an equation: the field of view is equal to the number of pixels multiplied by the pixel size. For instance, a low resolution image of 1024 pixels at 2000 nanometers per pixel has a field of view of 2048 nm = 2 mm.
A fourth key quantity is the pixel dwell time. This is how long the beam takes to scan each pixel. A typical value might be in the range of 100ns to 1 μs per pixel. Longer dwell times result in less noisy images but correspondingly longer acquisition times. A 1024 × 1024 pixel image with 1 μs dwell time is obtained in 1 second. A 4096 × 4096 pixel image with the same dwell time will take 16 seconds.
As an example of how to use these quantities, one can consider how the accuracy of the stage motor can affect automated imaging. For the standard stage on a Zeiss SEM, this accuracy is ±4 microns (more accurate and correspondingly more expensive stages are available). A targeting map that is more accurate than this is superfluous. This limitation also has consequences for the field of view of automated collection. For a high-resolution image at 4 nm per pixel, this error corresponds to 1000 pixels. In other words, the field of view can be off by 1000 pixels due to stage positioning error. It would be necessary to take ~10k × 10k pixel images in order to reduce this error to 10% of the field of view. The large image guarantees that the center of the field of view is present in each section. A 10k × 10k pixel image with 1 μs dwell time takes 100 sec (10,000 × 10,000 × 1 usec), so that a high-resolution imaging run of 200 sections would take 5.5 hours.
Software for automated image collection
A computer program is required for automated image collection. This program is installed on a second computer that can bypass the main computer to control functions of the SEM. This third party or custom in-house software keeps track of each section and its rotation. Moreover, it allows the operator to specify the imaging conditions: pixel dimensions, nm per pixel, pixel dwell time, brightness and contrast, stigmation, scan rotation as well as adjusting parameters for auto-focus.
Several software options exist, but the Atlas 4/ Atlas 5 Array Tomography (AT) software for the Zeiss SEM is the most commonly used commercially available software. The non-commercial program Wafer Mapper (Hayworth et al., 2014) was used for Kasthuri et al (2015) and is available for download online (https://software.rc.fas.harvard.edu/lichtman/LGN/WaferMapper.html). The Matlab wrapper that connects to the Atlas API (application programming interface) has not yet been modified for the new Atlas 5 AT software. For FEI microscopes, we use a Matlab-based custom software program called SEM Navigator written by Daniel Berger of the Lichtman lab.
Initial mapping
One approach is to use the SEM to take a low-resolution image of the wafer. This takes ~2 hours because the largest field of view of the SEM is about 2 mm × 2 mm, which is small compared to the 100 mm diameter silicon wafer. There are faster ways to take an initial image outside the microscope. One solution is to use a Zeiss Axio Imager microscope with a motorized stage to take a mosaic of images. This takes an image at a resolution of ~4 μm. Another solution is to take a picture of the wafer with a digital camera. The Terasaki lab uses the camera of an iPod touch with a custom-made photobox; the resolution of this image is ~200 μm. The photo box is a 6-inch cube made of white artist’s cardboard. The bottom is open and lies on a black cardboard on the table. The wafer is placed on guidelines on the black cardboard and an iPod touch (or cell phone camera) lies on the top surface of the photobox, so that the wafer and camera are in the same relative position each time. Because the camera is directly above the wafer, the camera takes a reflected image of itself. To minimize the size of this reflection, the photobox has a small hole to accommodate just the lens of the iPod touch. The position of the photobox is adjusted so that the reflection does not lie on top of a section (Figure 6B shows wafer images taken with this photobox). To provide uniform lighting inside the box, a single LED strip light is taped along the four walls about an inch above the bottom of the box.
The wafer image must then be aligned with the microscope stage coordinates. This is done by placing two to four special EM grids (fiducials) at corners of the wafer (see Figure 6B). These grids have a center hole that is 10 um in diameter. In the photograph, pixel coordinates of the centers of the grids are determined. Then, in the SEM, the microscope stage x and y coordinates of these holes are determined. The photo and SEM coordinates are matched and some math and trigonometry is done to scale and rotate the coordinate systems so that photograph coordinates of each section can be converted into microscope stage coordinates.
Mapping
A map (also called region of interest target map) consists of a coordinate system for each section: an origin (x and y coordinates of a specific point on the wafer) and the orientation of the x and y axes. The x-y axes of the section are perpendicular to each other, but depending on the section, are rotated at an angle relative to the x and y axes of the stage.
The Atlas 5 AT program uses one of the corners as the origin of the coordinate system for each section. The user makes a template of the first section, and then aligns this template on each section.
There are two factors that introduce error into the map. One is the significant variation from section to section in the appearance of the corner at high magnification, so that the corner location is ill defined at high resolution. Another source of error is distortions in the sections such as compression or folds, even tears. The map assumes a rigid area that is mapped by a coordinate system, and these irregularities introduce errors in locating structures within the section.
In practice, the mapping error can be as large as 50 microns or more. If the final imaging resolution is 5 nm per pixel, 50 microns corresponds to an error of 10,000 pixels, and the number of pixels imaged must be significantly larger than this to obtain any useful data by automated collection. There are three kinds of solutions to this problem: 1) user assisted automatic imaging, 2) imaging of a field of view much larger than the mapping error, and 3) a local coordinate system based on image alignment.
User assisted automated imaging:
For this approach, the program moves the stage to the approximate position of the structure of interest, and the user adjusts the stage position. The coordinates are either saved for a later automated imaging run, or the user immediately acquires a data image. The second alternative can require the user to spend long stretches of time at the microscope.
Large field of view:
In this approach, the automated imaging run is programmed with a field of view that is compatible with the map error. For instance, to accommodate a relatively small mapping error of 20 microns, one would want a ~200 micron field of view. If the desired resolution is 5 nm per pixel, this requires a 40,000 × 40,000 pixel image. The SEM cannot acquire a single image of this size but creates a tile of smaller images to cover this field of view. For instance, 16,000 × 16,000 pixel images can be tiled 3 × 3 to cover this field of view. The drawback of this approach is that acquisition of large fields of view comes at the expense of much longer imaging times.
Local coordinate system:
Instead of using the corner of the section as the origin of the coordinate system, the origin is centered on a feature of interest in the specimen (Fig. 7). The origin changes if one decides to image another feature of interest on the sections. This approach was taken in Kasthuri et al (2015) by using Wafer Mapper, a Matlab program written in Jeff Lichtman’s lab (2014). As mentioned above, this program is not yet compatible with later versions of Atlas 5 AT software.
Figure 7. Two different strategies for mapping sections.
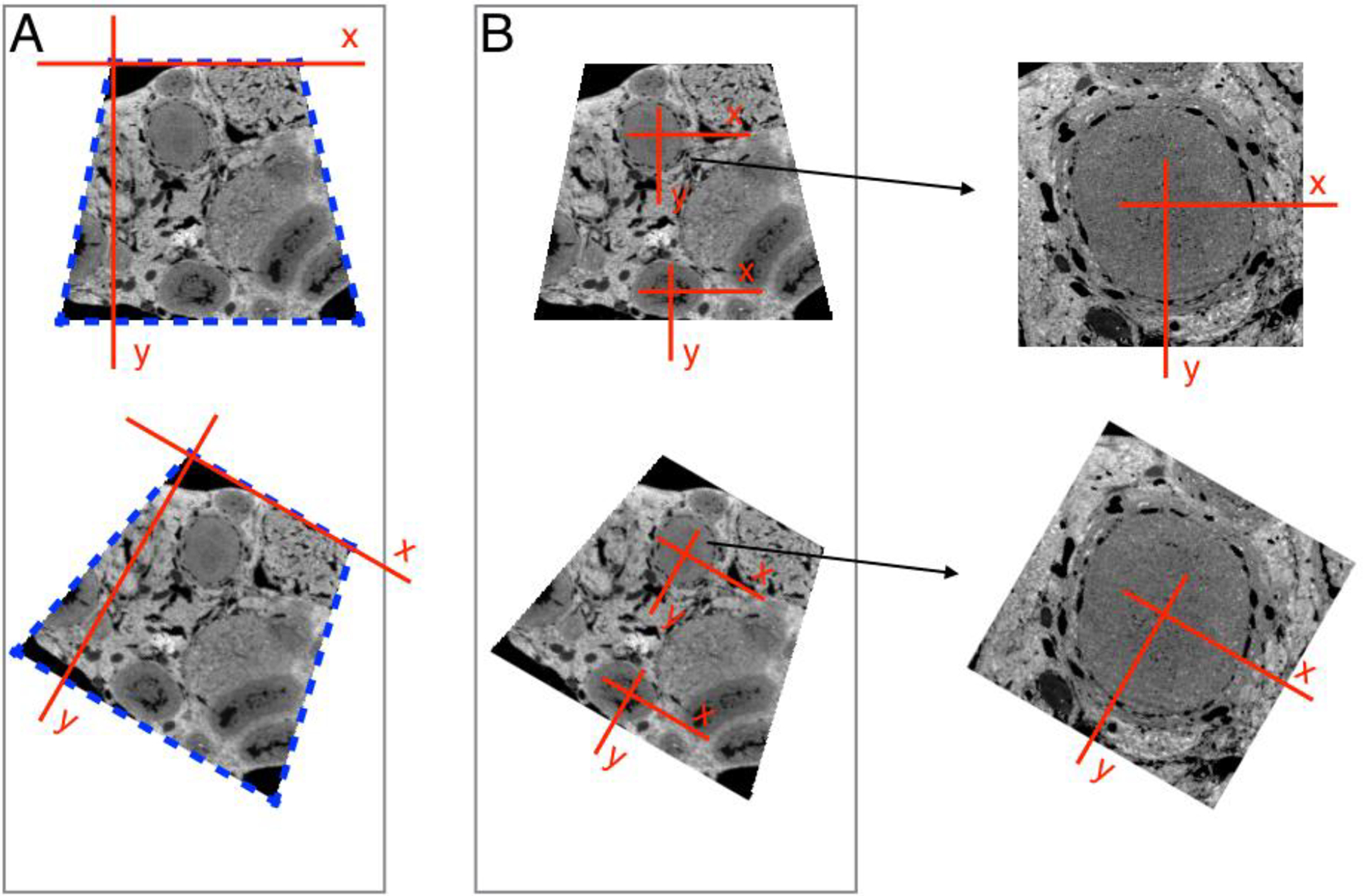
(A) With the Atlas 5 AT software, a template for the section outline is made of the first section (blue dashed lines), and this is aligned with each of the following sections. The origin of the coordinate system for each section is at one corner of the section. (B) In the alternative method, the coordinate system is defined relative to a biological structure in the specimen. An alignment algorithm operates on the image, and shifts the origin from section to section. The text explains in detail why this is advantageous. Briefly, this procedure locates the coordinate system origin with higher accuracy. In addition, a local coordinate system reduces mapping errors due to wrinkles / compression in the sections.
Another approach uses FIJI / Image J alignment algorithms in conjunction with the Atlas 4 or 5 software (Terasaki et al., 2018; Baena and Terasaki, 2019). The following is a brief description of the workflow: The initial wafer photograph is opened in Image J, and by clicking on the approximate centers of each section, their relative locations are recorded. To translate these relative locations to microscope stage coordinates, the EM grids (Figure 6) are used as fiducials. The center of the EM grids locations on the stage are matched with the locations in the image, and a Python script calculates the stage coordinates for an initial map. The Atlas 5 AT program stores the map information in an xml file, and Python scripts are used to modify them. The initial map is used for a mapping imaging run at low resolution (typically 2048 pixels, 1000 nm per pixel). The images are aligned using the FIJI plugin Register Virtual Stack Slices. This plugin produces the alignment parameters of rotation and displacement. A Python script uses these parameters to make a more accurate map. A second imaging run at intermediate resolution (2048 × 2048 pixels, 100–300 nm per pixel) is done and aligned in the same way. After this, the map is often accurate enough for a data imaging run, though a third mapping imaging run is not uncommon. A Python script can be used to change the center point (origin) if the user wants to image another structure in the section. Other scripts help with the auto-focus, remove damaged sections from the queue, and reorient the wafer if it is re-imaged at another time.
Although the workflow is well-defined, it may not go smoothly for various reasons, such as problems with the alignment algorithms. The convenience of automated imaging sometimes requires solving a long series of small and occasionally not so small problems. Setting up the imaging runs with the right parameters for autofocus is crucial. The autofocus procedure starts at an initial focus value, and the success rate is significantly better if this initial value is close to the true focus level. To get closer to the true focus level, we use autofocus in all (excluding the first) mapping runs. With each mapping run, we get an approximate focus value for each section. These values are then used as the initial focus level for the final imaging run.
Image alignment
Post-acquisition alignment is required because the sequence of acquired images is not well aligned, if only due to the stage positioning error. Images are aligned with reasonable accuracy and speed by FIJI/Image J alignment plug-ins. With the “Linear Stack Alignment with SIFT” plug-in, the entire series of images are loaded into RAM and the aligned images are also stored in RAM. This algorithm is impractical when the size of the stack is comparable to the RAM size. The “Register Virtual Stack Slices “ plug-in holds only two images at a time in RAM as it aligns, so it is less limited to RAM size. In addition, this plug-in has a “save transforms” option to save the alignment parameters of rotation and displacement for each section, which can be used in other manipulations.
The alignment may fail for several reasons. One is the lack of features to align. This can come about from a damaged section, by low image contrast, or by dust contamination. The user can try to remedy this by retaking the image, deleting a section, enhancing contrast or deleting regions of an image that cause problems, etc.
The alignment may take an inordinate amount of time. One can align reduced versions of the original images and then scale the alignment parameters to modify the original images.
The alignment may fail at a more basic level because the FIJI/Image J plug-ins are not powerful enough to align complex patterns of compression / distortion in the image. This occurs particularly with large images. Supercomputer alignment methods have been used in this case (Morgan et al., 2016; Hildebrand et al., 2017).
The most satisfying part of the ATUM workflow is to finally view a well-aligned data set. We take leave now as the investigator settles down to analyze the 3 dimensional data for biological content.
III. INSTRUMENTS AND MATERIALS
A. Ultramicrotome and ATUM (ATUMtome)
ATUMtome (RMC Boeckeler, Tucson, AZ). Ultramicrotome and tape collector sold together.
B. Diamond knives and trimming tools
Ultra diamond knife, 35° or 45° angle, 3–4 mm edge size, wet (Ultra, Diatome-US, Hatfield, PA) for cutting thin sections (≤60nm).
Ultra diamond knife with a large boat, 35° or 45° angle, 3–4 mm edge size (Ultra Jumbo, Diatome, Hatfield, PA) for cutting thin sections (≤60nm).
Histo diamond knife, 4 mm edge size (Histo, Diatome-US, Hatfield, PA) for cutting thick sections (60–1000 nm).
Trimming diamond knife, 45° angle (Trimtool, Diatome-US, Hatfield, PA) for trimming the block face and mesa.
Trimming diamond knife, 90° angle (Trimtool, Diatome-US, Hatfield, PA) for trimming the block face and mesa.
Single-edged razor blades (55411–055, VWR, Radnor, PA) for rough trimming of the block face.
C. Tools for ultramicrotomy
Eyelash with handle, No.1 superfine eyelash (113, Ted Pella, Inc., Redding CA) for manipulating sections in the knife boat and cleaning the knife edge.
Disposable syringe with needle, 1 ml syringe with 25 G needle (14-826-88, Fisher Scientific, Hampton, NH) for finely adjusting the water level in the knife boat.
Disposable syringe, 10 ml (14-826-88, Fisher Scientific, Hampton, NH) to add water to the knife boat.
Syringe filter, 25 mm (05-713-401 Fisher Scientific, Hampton, NH) to be used with 10 ml syringe above for adding water to the knife boat.
Ionizer (Static Line Ionizer, Diatome-US, Hatfield, PA) for neutralizing electrostatic charge on the block face.
D. Wafers, tapes, and tools for wafer fabrication
Silicon wafers, 100 mm diameter (see other specifics on section II.G) (3246, University Wafer, South Boston, MA).
Kapton tape, 8 mm wide (item number not available currently).
Carbon conductive tape, 8 mm wide, double coated (16073, Ted Pella, Inc., Redding CA).
Sharp scalpel (21909–654 disposable #10 scalpel, VWR International, Radnor, PA) for cutting Kapton and double-sided carbon tape.
Cutting board (63308, Electron Microscopy Sciences, Hatfield, PA) to use as a surface for cutting tape.
Serrated forceps (5441, Ted Pella, Inc., Redding, CA)
TEM grids with center mark (G150, Ted Pella, Inc., Redding, CA) used as fiducials for wafer mapping.
Dust blower (81710, Ted Pella, Inc., Redding, CA) for removing dust from the surface of the sections.
Plastic reels for reeling the tape (RMC Boeckeler, Tucson, AZ).
Seam roller (62395T69, McMaster-Carr, Robbinsville, NJ) to press the Kapton tape flat against the double-sided carbon adhesive tape.
E. Glow discharge
Glow discharge system, PELCO easiGlow glow discharge unit (91000, Ted Pella, Inc., Redding, CA).
Rotary pump for glow discharge unit (92081, RVP 200–7, Ted Pella, Inc., Redding, CA). Although the Glow discharge system has a suggested pump, we found that it was not powerful enough for the custom-built chamber used for glow discharge of tape.
Dynamic O-Ring Shaft Seal (2) (FMH-25A, ¼” aluminum, Kurt J. Lesker Co., Jefferson Hills, PA). For rotating the tape reels.
Plexi Glass (2) 12 × 6, (2) 3.5 × 6, (2) 3.5 × 11. (Chemcast GP Acrylic Sheet, 0.5” clear, Tap Plastics, San Leandro, CA).
Plexiglas adhesive (Weld-On 4 Cement, 4oz, Tap Plastics, San Leandro, CA).
Plexiglas adhesive applicator (SY20–65, Syringe Hypodermic Applicator, 18 gauge, Tap Plastics, San Leandro, CA).
Motor for automatic turning (ROB-09238, Stepper motor with cable, Sparkfun, Niwot, CO).
Stepper motor driver (ROB-12779, EasiDriver, Sparkfun, Niwot, CO).
Arduino board for motor (DEV-11021, Arduino Uno – R3, Sparkfun, Niwot, CO).
F. Carbon coating
Carbon evaporator (DV-502B High Vacuum Evaporator, Denton Vacuum, Moorestown, NJ).
Carbon rods pointed and presharpened (70220–01, Electron Microscopy Sciences, Hatfield, PA).
G. Supplies for post-staining wafers
Uranyl acetate (22400, Electron Microscopy Sciences, Hatfield, PA).
Lead citrate (17800, Electron Microscopy Sciences, Hatfield, PA).
Petri dishes 150 mm diameter Falcon (351058, VWR, Radnor, PA)
Syringe disposable 10 ml (72510, Electron Microscopy Sciences, Hatfield, PA).
Syringe filters 0.2 um (67002–25, 25 mm, Electron Microscopy Sciences, Hatfield, PA).
ACKNOWLEDGEMENTS
We would like to thank Martina Schifferer for useful suggestions on the manuscript.
REFERENCES
- Ai-Awami AK, Beyer J, Haehn D, Kasthuri N, Lichtman JW, Pfister H, & Hadwiger M (2016). NeuroBlocks--Visual Tracking of Segmentation and Proofreading for Large Connectomics Projects. IEEE Transactions on Visualization and Computer Graphics, 22(1), 738–746. 10.1109/TVCG.2015.2467441 [DOI] [PubMed] [Google Scholar]
- Baena V, & Terasaki M (2019). Three-dimensional organization of transzonal projections and other cytoplasmic extensions in the mouse ovarian follicle. Scientific Reports, 9(1), 1262. 10.1038/s41598-018-37766-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger DR, Seung HS, & Lichtman JW (2018). VAST (Volume Annotation and Segmentation Tool): Efficient Manual and Semi-Automatic Labeling of Large 3D Image Stacks. Frontiers in Neural Circuits, 12. 10.3389/fncir.2018.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzola JJ, & Russell LD (1999). Electron Microscopy: Principles and Techniques for Biologists. Jones & Bartlett Learning. [Google Scholar]
- Eberle AL, Mikula S, Schalek R, Lichtman J, Tate MLK, & Zeidler D (2015). High-resolution, high-throughput imaging with a multibeam scanning electron microscope. Journal of Microscopy, 259(2), 114–120. 10.1111/jmi.12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang T, Lu X, Berger D, Gmeiner C, Cho J, Schalek R, et al. (2018). Nanobody immunostaining for correlated light and electron microscopy with preservation of ultrastructure. Nature Methods, 15(12), 1029. 10.1038/s41592-018-0177-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler HK (2007). Ultramicrotomy for biological electron microscopy. Methods in Molecular Biology (Clifton, N.J.), 369, 67–96. 10.1007/978-1-59745-294-6_5 [DOI] [PubMed] [Google Scholar]
- Hayworth KJ, Morgan JL, Schalek R, Berger DR, Hildebrand DGC, & Lichtman JW (2014). Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Frontiers in Neural Circuits, 8. 10.3389/fncir.2014.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, et al. (2017). Whole-brain serial-section electron microscopy in larval zebrafish. Nature, 545(7654), 345–349. 10.1038/nature22356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszewski M, Kornfeld J, Li PH, Pope A, Blakely T, Lindsey L, et al. (2018). High-precision automated reconstruction of neurons with flood-filling networks. Nature Methods, 15(8), 605. 10.1038/s41592-018-0049-4 [DOI] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, et al. (2015). Saturated Reconstruction of a Volume of Neocortex. Cell, 162(3), 648–661. 10.1016/j.cell.2015.06.054 [DOI] [PubMed] [Google Scholar]
- Kubota Y, Sohn J, Hatada S, Schurr M, Straehle J, Gour A, et al. (2018). A carbon nanotube tape for serial-section electron microscopy of brain ultrastructure. Nature Communications, 9(1), 437. 10.1038/s41467-017-02768-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunsbach AB, & Afzelius BA (1998). Biomedical Electron Microscopy: Illustrated Methods and Interpretations. Elsevier. [Google Scholar]
- McDonald K, Schwarz H, Müller-Reichert T, Webb R, Buser C, & Morphew M (2010). “Tips and tricks” for high-pressure freezing of model systems. Methods in Cell Biology, 96, 671–693. 10.1016/S0091-679X(10)96028-7 [DOI] [PubMed] [Google Scholar]
- Morgan JL, Berger DR, Wetzel AW, & Lichtman JW (2016). The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus. Cell, 165(1), 192–206. 10.1016/j.cell.2016.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris RP, Baena V, & Terasaki M (2017). Localization of phosphorylated connexin 43 using serial section immunogold electron microscopy. Journal of Cell Science, 130(7), 1333–1340. 10.1242/jcs.198408 [DOI] [PubMed] [Google Scholar]
- Tapia JC, Kasthuri N, Hayworth KJ, Schalek R, Lichtman JW, Smith SJ, & Buchanan J (2012). High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nature Protocols, 7(2), 193–206. 10.1038/nprot.2011.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M (2018). Axonal endoplasmic reticulum is very narrow. Journal of Cell Science, 131(4). 10.1242/jcs.210450 [DOI] [PubMed] [Google Scholar]


