Abstract
SARS-CoV-2, the causative virus for COVID-19 has now super-mutated into the Omicron (Om) variant. On its spike (S) glycoprotein alone, more than 30 substitutions have been characterized with 15 within the receptor binding domain (RBD); It therefore calls to question the transmissibility and antibody escapability of Omicron. This study was setup to investigate the Omicron RBD's interaction with ACE2 (host receptor) and a SARS-CoV-2 neutralizing monoclonal antibody (mAb). In-silico mutagenesis was used to generate the Om-RBD in complex with ACE2 or mAb from the wildtype. HDOCK server was used to redock and score the mAbs in Om-RBD bound state relative to the wildtype. Stability of interaction between all complexes were investigated using all-atom molecular dynamics (MD). Analyses of trajectories showed that Om-RBD has evolved into an efficient ACE2 binder, via pi-pi (Om-RBD-Y501/ACE2-Y41) and salt-bridge (Om-RBD-K493/ACE2-Y41) interactions. Conversely, in binding mAb, it has become less efficient (Center of mass distance of RBD from mAb complex, wildtype ≈ 30 Å, Omicron ≈ 41 Å). Disruption of Om-RBD/mAb complex resulted from loose interaction between Om-RBD and the light chain complementarity-determining region residues. Omicron is expected to be better transmissible and less efficiently interacting with neutralizing convalescent mAbs with consequences on transmissibility provided other mutations within the S protein similarly promote cell fusion and viral entry.
Keywords: COVID-19, Omicron variant, Spike glycoprotein, Receptor binding domain (RBD), Monoclonal antibody (mAbs)
1. Introduction
COVID-19 (coronavirus disease 2019) is caused by the novel coronavirus severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) (Zhou et al., 2020) and despite the best efforts of WHO, COVID has remained a threat to humanity, causing more than 5.2 million deaths globally whilst infecting more than 265 million. SARS-CoV-2 tropism is initiated when its spike (S) glycoprotein binds to the host angiotensin-converting enzyme 2 (ACE2) and its partner transmembrane serine protease 2 (TMPRSS2) [1] serving as door-way to cellular entry. RBD/ACE2/TMPRSS2 interaction event ultimately initiates the life cycle of SARS-CoV-2, as intracellular injection of the viral RNA (genome) initiates the proteolytic processing [2] of structural and non-structural proteins [3] from the polyproteins pp1a and pp1ab [4]; being translational products of ORF1a and ORF1ab translation respectively followed by viron assembly, endoplasmic reticulum budding and viral particle release via endocytosis [5].
In addition to its role as the cellular receptor, the S protein has continued to draw attention for two key reasons, first, as veritable drug/vaccine target [6] and secondly, as a hotbed for clinically-relevant mutations [7].
Vaccine and antibody-based prophylactic/therapeutic treatment against the COVID-19 virus [8] currently available target the S (mRNA-1273 (Moderna), BNT162b2 (Pfizer-BioNTech), Ad26.COV2·S (Johnson&Johnson), AZD1222 (Oxford/AstraZeneca), Ad5-nCoV (Convidecia)) or its RBD (BNT162b1, ZF2001, and ARcoVax) [9]. Furthermore, many characterized convalescent monoclonal antibodies (mAbs) also target the RBD [[10], [11], [12]]; thus, making mutations around the spike glycoprotein and indeed RBD very concerning.
Indeed, the SARS-CoV-2 B.1.617 lineage (Delta variant; subtypes: B1.617.1, B.1.617.2 and B.1.617.3) first identified in India and the UK, contains mutations within the N-terminal domain (NTD) and RBD which result in immune evasion, and faster transmission [13]. The newest in the long list of variants of concerns is the B.1.1.529 lineage (also termed the Omicron variant) [14]. In Omicron, at least thirty (30) amino acid substitutions on the S protein and intriguingly, about 11 (K417 N, N440K, G446S, S477 N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H) of the substitutions occur at the ACE2-binding site of the RBD [15], thus, raising concerns about the transmissibility and antibody escapability.
In order to answer the question whether SARS-COV-2 Omicron is more transmissible, this study focused on the evaluating S protein RBD/ACE2 interaction in Omicron in comparison with the wildtype using in silico mutagenesis and all-atom MD simulation in explicit water. Antibody escape potency was similarly investigated in RBD/mAB complex of the wildtype and Omicron RBD mutations following similar protocols.
2. Methodology
2.1. Starting structures
2.1.1. RBD-ACE2 complex
Wildtype RBD in ACE2 bound state previously resolved (PDB ID: 7KMB) [16] was retrieved. All broken chains and incomplete residues were reconstructed in protein preparation module of Schrodinger Suite. In order to generate the Omicron RBD, the following substitutions (N440K, G446S, S477 N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H) were made using PyMol Mutagenesis plugin.
2.1.2. RBD-mAB complex
Different neutralizing mABs bound to unique regions of the RBD previously deposited: 7DEO/7DEU/7DET [11], 7CJF [12], 7B3O [10] were retrieved. Prior to antibody re-docking experiment, all broken chains and incomplete residues were reconstructed in protein preparation module of Schrodinger Suite. Omicron RBD was generated using PyMol Mutagenesis plugin.
2.1.3. Antibody docking
The HDOCK server [17] for integrated protein–protein docking was used to reproduce crystallographic poses and scoring of the RBD-mAB poses for both wildtype and Omicron.
2.1.4. Biosystem generation for atomistic simulation
To generate RBD-ACE2 (PDB ID: 7KMB: ACE2 (resid: 19–614), RBD (resid: 335–526)) or RBD-mAB (PDB ID: 7B3O: mAB-H (resid: 1–219)/mAB-L (resid: 1–215), RBD (resid: 335–517)) biosystems for Omicron and Wildtype for simulation, CHARMM-GUI webserver (www.charmm-gui.org) [18] was used. All protein were parameterized in CHARMM36 all-atom additive protein force field [19] while glycan parameterization was performed using ParamChem service (https://cgenff.paramchem.org) as implemented on CHARMM-GUI webserver interface. Each biosystem was solvated in TIP3P explicit water model [20] and neutralized with Na+/CL−. Wildtype RBD-ACE2 biosystem (107,690 atoms) was simulated in 11.4 × 10.7 × 9.3 nm box containing 31,638 molecules of water, and 44 ions while the Omicron-RBD-ACE2 biosystem (107,688 atoms) was simulated in the same box dimension but containing 31,650 molecules of water, and 50 ions. Both wildtype (181,608 atoms) and Omicron RBD-mAB (181,463 atoms) biosystems were simulated in 12.4 × 12.4 × 12.4 nm box.
2.1.5. Molecular dynamics (MD) simulation
All molecular dynamics simulation was run on NAMD molecular dynamics software [21] in three stages of minimization, equilibration and production. During equilibration, the biosystems were under constant pressure and temperature (NPT; 298K, 1 bar) conditions using Berendsen temperature and pressure coupling algorithms. All van der Waals interactions were estimated at 10 Å, while electrostatic interactions were estimated using particle mesh Ewald (PME) summation equation and equation of atomic motion was integrated using the leap-frog algorithm at 2 fs time step for a total time of 30 ns with positional restraints imposed on the heavy atoms in all directions.
In order to generate the two independent states for production stage MD simulation, equilibration stage trajectories were loaded into VMD [22] and two structures with the largest rmsd were retrieved and simulated as discussed for equilibration above for 50 ns with the removal of restraints. All trajectories were checked for convergence simulations. All calculations were performed on SuperMicro workstations (32-E2600 Intel Xeon CPUs, 2 M 6000 GPUs Accelerator PCI-E x16 Card/node) housed at the S.E. Bogoro Center, Afe Babalola University, Ado-Ekiti, Nigeria.
2.1.6. Post-MD simulation analyses and data presentation
Dynamical networks for RBD-ACE2 interaction for both wildtype and Omicron systems were calculated as described [23], we have previously described the use of Carma (ver. 1.4), gncommunities and subopt scripts for generating files for network analysis [24]. Network tools implemented in VMD was used to visualize the source-sink pairs. A pair of nodes was connected by an edge if the corresponding residues were resident within 4.5 Å distance for at least 80% of the frames analyzed while the edge size is weighted.
Unless otherwise stated, all inter-group, inter-residue or inter-atomic distances were calculated using PLUMED plugin for molecular dynamics [25].
All line graphs, bar charts, or population counts were plotted as mean from 2 independent runs using GraphPad prism (ver. 9.0), all 3D representations were done using VMD or PyMol.
3. Results and discussion
3.1. Comparative binding dynamics of Omicron and Wildtype RBDs to ACE2
The S protein is also the key targets for several antibodies currently in use as treatment options for COVID-19; especially the receptor binding domain (RBD) [26]. Notably, the antibodies generated by the Pfizer-BioNTech mRNA-BNT162b2, Moderna mRNA-1273 ultimately targets SARS-CoV2 spike glycoprotein (Fig. 1 a, b-ii) at the receptor-binding domain (RBD, Fig. 1b, i-ii)) while Astra-Zeneca-ChAdOx1-S and Janssen-Ad26.COV2·S were primarily designed to express SARS-CoV-2 spike protein as immunogen [27]. Curiously, many convalescent monoclonal antibodies (mAbs) bind at the ACE2 site on the RBD [10]. It is therefore not surprising that as SARS-CoV-2 variants with mutations in the RBD begin to emerge [28], so is concern over transmissibility and antibody escape [27]. Variants whose RBD mutations eventually resulted in worse clinical outcomes include: B.1.1.7 (N501Y), B.1.351/P1 (K417T/N, E484K, N501Y) [29], and B.1.617.2 (Delta variant; L452R, T478K); then the most recent (B.1.1.529), also termed the Omicron variant [14]. Intriguingly most S protein substitutions (N440K, G446S, S477 N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, Fig. 1c) occur at the ACE2-binding site of the RBD (Fig. 1d).
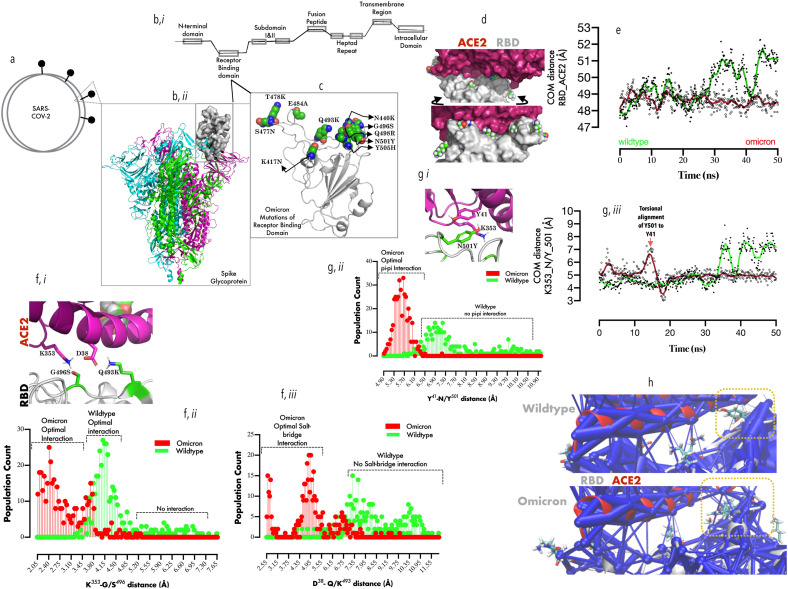
Fig. 1.0.
Comparative binding dynamics of Omicron and Wildtype RBDs to ACE2:1a: Representation of SARS-CoV-2 and S protein, (b, i) Representation of the different constituent regions in a typical monomeric S protein. (b, ii) Cartoon representation of the trimeric S protein, showing one of the three RBDs in up configuration (surface representation). (c). Cartoon representation of the RBD, showing the cluster of substitutions (represented in VMD spheres) that define the Omicron variant. (d, upper and lower plane) Surface representation of ACE2/RBD complex showing the spatial distribution of the Omicron substitutions around the RBD. (e) Smoothened line graph showing the mean center of mass distance between RBD and ACE2 with time. (f, i-iii) Spatial projection of ACE2-K353/D38 proximal to the RBD-G966S/Q493K (i), and population count distributions of side-chain atom distance between K353/G966S (ii) and D38/Q493K (iii). (g, i-iii) Spatial projection of ACE2-K353/Y41 proximal to the RBD-N501Y (i), and population count distributions of side-chain atom distance between Y41/Y501 (ii) and time-evolved smoothened mean distance between K353 and Y501 (iii). (h) The network data showing weighted interaction between ACE2/RBD in wildtype (upper plane) and Omicron (lower plane). Yellow rectangles indicate the substitution cluster and their effect on the weight of RBD-ACE2 interaction.
First, in order to provide insight into how the substitutions affect ACE2 binding, all-atom MD simulation in explicit water was set up (Methods) and checked for important biological events every 10 ns. Surprisingly, Wildtype- but not Omicron RBD exhibit intermittent dissociation from ACE2. The largest amplitude of dissociation occurs at 30 ns, (Om-vs Wt-: ≈ 48.5 Å vs 51.5 Å) and 45 ns (Om-vs Wt-: ≈ 48.0 Å vs 52.5 Å, Fig. 1e), indicating that Omicron, but not the wildtype RBD has better ACE2 binding capacity.
Whilst it is to be noted that previous reports identified T478K, and N501Y substitutions are associated with increased ACE2 binding [30], in Omicron RBD, S496 resides within hydrogen-bond distance (≈3.5 Å) from ε-amino group of ACE2-K353 (Fig. 1f, i), which is absent in Wildtype (G496, distance >4.5 Å, Fig. 1f, ii). Omicron K493 also evolved salt-bridge interaction with ACE2 D38 (distance ≈ 2.5–4.5 Å) as opposed to Q493 which fails to form hydrogen bond (distance >6.0 Å, Fig. 1f, ii). Further investigation showed that G496 (wildtype) allowed ACE2-K353/D38 salt-bridge interaction rather than engaging RBD (data not shown), thus, further weakening ACE2 binding in the wildtype.
We further elucidate that the stabilizing effect of N501Y mutation on ACE2 occurs through ACE2-K353 interaction (Fig. 1g, i). Here, the measurement of the inter-atomic distance between the phenolic side chains of Y501 (RBD) and Y41 (ACE2) indicated a possible pi-pi interaction (distance <6.0 Å, Fig. 1g, ii); a feature non-existent in wildtype (N501, distance >6.9 Å, Fig. 1g, ii). Y501/41 stacking spatially locks Y501 in place allowing cation-pi interaction (Fig. 1g, ii) with K353 (ACE2). A representation of the difference in the strengths of ACE2 interaction offered by wildtype (Fig. 1h, upper plane) and Omicron (Fig. 1h, lower plane) RBDs were also projected using a weighted network representation, with Omicron RBD residues displaying stronger network interaction within the substitution clusters. Without a doubt, Omicron S protein RBD exhibits super-binder ability with ACE2 with resulting higher transmissibility potentials.
3.2. Comparative binding dynamics of Omicron and Wildtype RBDs to convalescent mAb
Next, we sought to understand how the Omicron RBD substitutions affect mAb binding, protein-protein docking and scoring with several available antibodies was initially performed. A cursory look suggests that Omicron binding is associated with improved binding scores (data no shown) but when one of the complexes [10] was subjected to simulation, the events were different. First, the mean COM distance separating RBD from the mAb complex (heavy & light chains, Fig. 2 a, i) over the entire trajectories showed that the Omicron was more loosely bound to the mAb in comparison with wildtype (Om-vs Wt-: ≈ 41.0 Å vs 30.0 Å, Fig. 2a, ii), and we showed that the antigen-binding fragments (Fab, residues 1–108 (heavy chain), residues 1–106 (light chain), Fig. 2a, iii), for the first 30 ns was ≈3 Å less compact in binding Omicron RDB in comparison with the wildtype (Fig. 2a, iv), This result suggest that local events at the RBD-Fab interface is responsible for the difference in mAb binding. Therefore, we further investigated the roles of each chain as most of the mutations cluster at the VLCDR binding interface (Fig. 2b, i). A population plot of the COM distance between the mAb-light chain and RBD strongly suggest that Omicron RBD binds beyond sub-optimal distance (distance >50 Å, Fig. 2b, ii) in comparison with the wildtype (distance <47 Å) and surprisingly a similar pattern was observed in the heavy chain (Wildtype ≈ 37.5 Å vs. Omicron ≈38.5 Å, Fig. 2b, iii).
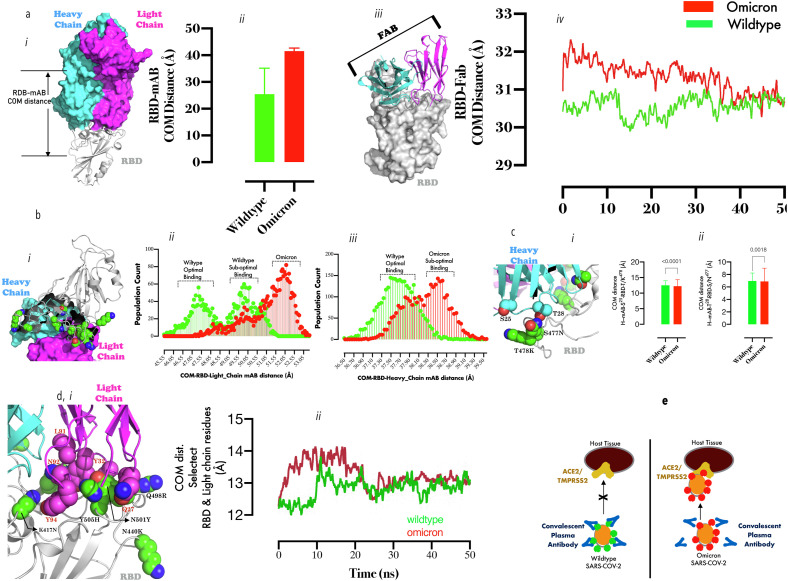
Fig. 2.0.
Comparative binding dynamics of Omicron and Wildtype RBDs to convalescent mAb: 2a (i) Representation of convalescent mAb (blue surface = heavy chain, pink surface = light chain) in RBD bound (gray cartoon) state. (a, ii) The bar chart plot of the COM distance between RBD and mAb during MD simulation. (a, iii) Representation of MAb-Fab (cartoon)/RBD (surface) complex. (2, iv) Smoothened line graph of showing the mean center of mass distance between RBD and Fab with time. (2b, i) A spatial representation of Omicron RBD substitutions relative to the light and heavy chains of mAb. (2b,ii) population count distributions of COM distance between the light chain/RBD and heavy-chain/RBD (iii). (2c, i) Zoomed representation of heavy chain CDR loop residues (S25/T28) proximal to some substituted RBD residues (T478K, S477 N). (2c, ii) Bar graph plots of the inter-residue distance between S25 and T/K478(ii) and T28/S/N477 (T-test comparison were made at p < 0.05 using two-tailed paired sampling and parametric test). (2d, i) Zoomed representation of light chain CDR loop residues (Q27, Y32, L91, N92, and Y94) proximal to substituted RBD residue cluster (K417 N, N440K, Q498R, Y505H). Smoothened line graph showing the mean center of mass distance between the clustered RBD substitutions and the light chain CDR loop residues with time. Fig. 2e: A mechanistic projection showing a tightly bound convalescent mAbs to Wildtype SARS-Cov-2-RBD thus, protecting ACE2/TMPRSS2-experessing cells from infection; while loosely bound convalescent mAbs to Omicron SARS-Cov-2-RB coupled with high-potency interaction between ACE2 and Omicron-RBD promotes higher infectivity in ACE2/TMPRSS2-experessing cells.
Finally, specific mAb residues which account for the sub-optimal binding were identified. Loss of interaction between S25/T28 (heavy chain CDRs) and T478K and S477 N respectively (Fig. 2c, i, right and left panels) partially explain the loose binding with Omicron RBD and time-evolved dynamics setup for to monitor the interaction between clustered RBD substitutions (K417 N, N440K, Q493R, G496S, Q498R, N501Y, and Y505H) and the VLCDR loop residues (Fig. 2d, i). The 2 Å separation of VLCDR loop residues from Omicron RBD for the first 20 ns of simulation (reconverged afterwards) is consistent with a previous studies where N501Y and K417 N were associated with detached RBD from mAb light chain CDR1 loop [31]. These results suggest strongly that Omicron RBD is ACE2 super binder but damped convalescent mAb binder (Fig. 2e).
4. Conclusion
Data from this study have provided the mechanistic basis for the increased ACE2 binding by Omicron RBD, which may have serious consequences on SARS-CoV-2 transmission and disease severity; it is however worthy of note that RBD/ACE2 interaction is just single step in the array of events that must occur in order to establish cellular entry [32]. The other factors worth considering is the effect of the 30 S protein mutations on (i) its overall structure, (ii) the conformational transitions required to load “up” conformation for RBD in readiness for ACE2 binding and (iii) how the mutations affect proteolytic (cathepsin L furin, and TMPRSS2) activation of the S protein [32,33]. The mAb tested in this study has slightly reduced affinity to Omicron RBD and therefore may only have reduced potency.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was partly supported by ABUAD Research Foundation to S.E. Bogoro Center, Afe Babalola University (Dr. Omotuyi, I.O: grant no: SEBC/IDRD/01/2021/0004) and in part by H3-Africa Bio-informatics Network: (to Prof. Nash Oyekanmi) (NIH Common Fund Award/NHGRI Grant no: U41HG006941).
References
- 1.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak J.J., Gray H.B., Garza-Lopez R.A. Structural stability of the SARS-CoV-2 main protease: can metal ions affect function? J. Inorg. Biochem. 2020;211:111179. doi: 10.1016/j.jinorgbio.2020.111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Xu Z., Lei Q., Lai D.Y., Hou H., Jiang H.W., Zheng Y.X., Wang X.N., Wu J., Ma M.L., Zhang B., Chen H., Yu C., Xue J.B., Zhang H.N., Qi H., Guo S.J., Zhang Y., Lin X., Yao Z., Sheng H., Sun Z., Wang F., Fan X., Tao S.C. Antibody landscape against SARS-CoV-2 reveals significant differences between non-structural/accessory and structural proteins. Cell Rep. 2021;36:109391. doi: 10.1016/j.celrep.2021.109391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardenas-Conejo Y., Linan-Rico A., Garcia-Rodriguez D.A., Centeno-Leija S., Serrano-Posada H. An exclusive 42 amino acid signature in pp1ab protein provides insights into the evolutive history of the 2019 novel human-pathogenic coronavirus (SARS-CoV-2) J. Med. Virol. 2020;92:688–692. doi: 10.1002/jmv.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju X., Zhu Y., Wang Y., Li J., Zhang J., Gong M., Ren W., Li S., Zhong J., Zhang L., Zhang Q.C., Zhang R., Ding Q. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durmaz B., Abdulmajed O., Durmaz R. Mutations observed in the SARS-CoV-2 spike glycoprotein and their effects in the interaction of virus with ACE-2 receptor. Medeni Med J. 2020;35:253–260. doi: 10.5222/MMJ.2020.98048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayat M., Asemani Y., Mohammadi M.R., Sanaei M., Namvarpour M., Eftekhari R. An overview of some potential immunotherapeutic options against COVID-19. Int. Immunopharm. 2021;95:107516. doi: 10.1016/j.intimp.2021.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Q., Mao Q., Zhang J., Bian L., Gao F., Wang J., Xu M., Liang Z. COVID-19 vaccines: current understanding on immunogenicity, safety, and further considerations. Front. Immunol. 2021;12:669339. doi: 10.3389/fimmu.2021.669339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertoglio F., Fuhner V., Ruschig M., Heine P.A., Abassi L., Klunemann T., Rand U., Meier D., Langreder N., Steinke S., Ballmann R., Schneider K.T., Roth K.D.R., Kuhn P., Riese P., Schackermann D., Korn J., Koch A., Chaudhry M.Z., Eschke K., Kim Y., Zock-Emmenthal S., Becker M., Scholz M., Moreira G., Wenzel E.V., Russo G., Garritsen H.S.P., Casu S., Gerstner A., Roth G., Adler J., Trimpert J., Hermann A., Schirrmann T., Dubel S., Frenzel A., Van den Heuvel J., Cicin-Sain L., Schubert M., Hust M. A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations. Cell Rep. 2021;36:109433. doi: 10.1016/j.celrep.2021.109433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu D., Zhang G., Wang Y., Zhang Z., Hu H., Shen S., Wu J., Li B., Li X., Fang Y., Liu J., Wang Q., Zhou Y., Wang W., Li Y., Lu Z., Wang X., Nie C., Tian Y., Chen D., Wang Y., Zhou X., Wang Q., Yu F., Zhang C., Deng C., Zhou L., Guan G., Shao N., Lou Z., Deng F., Zhang H., Chen X., Wang M., Liu L., Rao Z., Guo Y. Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y., Huang L., Zhang G., Yao Y., Zhou H., Shen S., Shen B., Li B., Li X., Zhang Q., Chen M., Chen D., Wu J., Fu D., Zeng X., Feng M., Pi C., Wang Y., Zhou X., Lu M., Li Y., Fang Y., Lu Y.Y., Hu X., Wang S., Zhang W., Gao G., Adrian F., Wang Q., Yu F., Peng Y., Gabibov A.G., Min J., Wang Y., Huang H., Stepanov A., Zhang W., Cai Y., Liu J., Yuan Z., Zhang C., Lou Z., Deng F., Zhang H., Shan C., Schweizer L., Sun K., Rao Z. A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes. Nat. Commun. 2021;12:2623. doi: 10.1038/s41467-021-22926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Pere H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Loriere E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 14.Gao S.J., Guo H., Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert. J. Med. Virol. 2021 doi: 10.1002/jmv.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaughan A. Omicron emerges, New Sci. 2021;252:7. doi: 10.1016/S0262-4079(21)02140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schon A., Bimela J., Boyington J.C., Nazzari A., Olia A.S., Shi W., Sastry M., Stephens T., Stuckey J., Teng I.T., Wang P., Wang S., Zhang B., Friesner R.A., Ho D.D., Mascola J.R., Shapiro L., Kwong P.D. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879. doi: 10.1016/j.chom.2020.11.004. e865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Y., Tao H., He J., Huang S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 18.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 19.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Florova P., Sklenovsky P., Banas P., Otyepka M. Explicit water models affect the specific solvation and dynamics of unfolded peptides while the conformational behavior and flexibility of folded peptides remain intact. J. Chem. Theor. Comput. 2010;6:3569–3579. doi: 10.1021/ct1003687. [DOI] [PubMed] [Google Scholar]
- 21.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 23.Sethi A., Eargle J., Black A.A., Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omotuyi O.I., Nagai J., Ueda H. Lys39-Lysophosphatidate carbonyl oxygen interaction locks LPA1 N-terminal cap to the orthosteric site and partners Arg 124 during receptor activation. Sci. Rep. 2015;5:13343. doi: 10.1038/srep13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bussi G., Tribello G.A. Analyzing and biasing simulations with PLUMED. Methods Mol. Biol. 2019;2022:529–578. doi: 10.1007/978-1-4939-9608-7_21. [DOI] [PubMed] [Google Scholar]
- 26.Zhou M., Zhang X., Qu J. Coronavirus disease 2019 (COVID-19): a clinical update. Front. Med. 2020;14:126–135. doi: 10.1007/s11684-020-0767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascellino M.T., Di Timoteo F., De Angelis M., Oliva A. Overview of the main anti-SARS-CoV-2 vaccines: mechanism of action, efficacy and safety. Infect. Drug Resist. 2021;14:3459–3476. doi: 10.2147/IDR.S315727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omotuyi I.O., Nash O., Ajiboye O.B., Iwegbulam C.G., Oyinloye E.B., Oyedeji O.A., Kashim Z.A., Okaiyeto K. Atomistic simulation reveals structural mechanisms underlying D614G spike glycoprotein-enhanced fitness in SARS-COV-2. J. Comput. Chem. 2020;41:2158–2161. doi: 10.1002/jcc.26383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Queiros-Reis L., Gomes da Silva P., Goncalves J., Brancale A., Bassetto M., Mesquita J.R. SARS-CoV-2 virus-host interaction: currently available structures and implications of variant emergence on infectivity and immune response. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S., Liu Y., Lei Z., Dicker J., Cao Y., Zhang X.F., Im W. bioRxiv; 2021. Differential Interactions between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Lopez-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Costa Clemens S.A., Naveca F.G., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Levin R., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Paterson N.G., Williams M.A., Hall D.R., Hulswit R.J.G., Bowden T.A., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954 e2939. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2021 doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]