Abstract
Major depressive disorder (MDD) is a complex and multifactorial psychiatric disorder that causes serious health, social, and economic concerns worldwide. The main treatment of the symptoms is through antidepressant (AD) drugs. However, not all patients respond properly to these drugs. Omic sciences are widely used to analyze not only biomarkers for the AD response but also their molecular mechanism. In this review, we aimed to focus on omics data to better understand the molecular mechanisms involving AD effects on MDD. We consistently found, from preclinical to clinical data, that glutamatergic transmission, immune/inflammatory processes, energy metabolism, oxidative stress, and lipid metabolism were associated with traditional and potential new ADs. Despite efforts of studies investigating biomarkers of response to ADs, which could contribute to personalized treatment, there is no biomarker panel available for clinical application. From clinical genomic studies, we found that the main findings contribute to the development of pharmacogenomic tests for AD efficacy for each patient. Several studies pointed at DRD2, PXDNL, CACNA1E, and CACNA2D1 genes as potential targets for MDD treatment and the efficacy and rapid-antidepressant effect of ketamine. Finally, more in-depth studies of the molecular targets pointed here are needed to determine the clinical relevance and provide further evidence for precision MDD treatment.
Keywords: Proteomics, Transcriptomics, Genomics, Metabolomics, Antidepressant
Introduction
As one of the most common and disabling psychiatric disorders, major depressive disorder (MDD) affects >320 million people worldwide and has a lifetime prevalence of 10.8–16.2% [1, 2]. MDD symptoms include depressed mood, loss of pleasure, appetite and sleep disturbance, feelings of extreme sadness or guilt, low self-esteem or self-worth, feelings of tiredness, and poor concentration, which can negatively affect the subject's social and family life, besides reducing work productivity, causing great rates of unemployment [3, 4]. Antidepressants (ADs) are the main pharmacologic strategy to reverse depressive symptoms. However, up to 30% of patients are refractory to currently available ADs [5].
These drugs act mainly as selective serotonin reuptake inhibitors (SSRIs), enhancing the synaptic levels of 5-HT and decreasing the firing activity of norepinephrine and dopamine neurons [6]. On the other hand, not all molecular mechanisms associated with MDD are known and are therefore not targeted by ADs. Furthermore, choosing the optimal AD for a patient is based on hit-and-miss attempts, where several unsuccessful attempts can end up burdening them and lead to a late treatment-resistant depression (TRD) diagnostic [7, 8]. Thus, it remains necessary to develop more precise treatment approaches, designed selectively for biologically defined subtypes of this disorder, as well as to improve the knowledge about molecular mechanisms of ADs and new molecules [9]. As MDD is a multifactorial disorder with heterogeneous symptoms and complex pathophysiology, omic approaches are strategies that can provide a better comprehension of this disorder.
Omic studies can potentially provide biomarkers for patient response to ADs and be helpful to identify new therapeutic targets [10, 11, 12]. Taking that into account, we searched, in the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed), for studies using omic techniques to shed light on MDD treatments from 2014 to 2020. We used the search terms major depressive disorder* combined with drug treatment* AND proteomics* OR transcriptomics* OR genomics* OR metabolomics*. Also, antidepressant drug* AND depression* combined with the “omics” terms already mentioned. An initial selection of articles was conducted by 2 independent authors. Reviews and book chapters were excluded. Only studies based on omic sciences related to MDD were selected, which resulted in 61 articles.
Animal Models
Proteomic Studies
Animal models have been widely used to comprehend the effects of ADs. Among the pharmacologic treatments investigated, several articles studied the effects of chronic use of fluoxetine (FLX), an SSRI AD [13, 14]. For instance, a study analyzed the hippocampus (HPC) of chronically socially isolated rats after FLX treatment [13]. In terms of the proteome, the cytosolic and nonsynaptic mitochondrial subproteome involved in the tricarboxylic acid cycle and oxidative phosphorylation was upregulated after treatment [13]. Still, in the cytosolic and nonsynaptic mitochondrial hippocampal proteomes of FLX treated mice versus controls, another study revealed that the most relevant differentially expressed proteins in the HPCs cytoplasm were related to the chaperone system, cytoskeletal components, and calcium signaling [14], while the most relevant in nonsynaptic mitochondria were catalytic enzymes involved in the pyruvate metabolism, citric acid cycle, oxidative phosphorylation, synthesis and transduction of ATP, and glutamate metabolism [14]. Collectively, these results suggest a relationship between FLX mechanism of action and energy metabolism, as well as a complex effect on neuronal signaling mechanisms potentially involved in restoring plasticity in the adult brain.
Studies have also compared the effects of FLX with other ADs and molecules. Comparing FLX and donepezil, an acetylcholinesterase inhibitor, after a chronic social defeat depression model in mice, a study [15] showed that FLX reversed anhedonia-like behavior and improved recognition memory and inhibitory learning in a stressor-related context, whereas donepezil only improved fear of extinction [15]. Proteomic analysis of the HPC revealed that altered proteins in the social defeat model were mainly related to reduced glutamatergic neurotransmission, increased glutamate clearance, impaired Ca2+ homeostasis, and learning processes. These alterations were reversed by FLX but not by donepezil [15]. Collectively, these results showed a possible association between the FLX effects with glutamatergic AMPA receptor transmission and Ca2+ responsive intracellular signaling, potentially influencing synaptic plasticity and therefore learning and memory [15].
FLX effects were also compared to imipramine (IMP), a tricyclic AD in an animal model of depression based on the prenatal stress procedure [16, 17]. Despite their different mechanisms of action, mitochondrial activity was modulated by both ADs, normalizing the depression-like behavior in rats [16, 17]. Moreover, IMP modulated proteins involved in synaptic function, learning and memory processes, and respiration, whereas FLX modulated proteins involved in the attenuation of neurodegenerative processes, mitochondrial biogenesis, and oxidative defense, as well as in the improvement of learning and memory processes [16, 17]. IMP also modulated proteins involved in oxidative stress and immune responses, phagocytosis, and cell migration on a rat primary mixed glial culture [18]. It upregulated proteins related to glycolysis but negatively influenced proteins which determine the mitochondrial membrane potential [18].
Other studies measured enzyme activities on male rats' HPC treated with desipramine (tricyclic AD) and FLX [19, 20]. They showed that both treatments led to alterations in mitochondrial bioenergetics, especially related to energy requirements of pre and postsynaptic compartments [19]. Further, results from the catalytic activities of regulatory enzymes from mitochondrial energy-yielding metabolic pathways highlighted that these ADs modified the activities of selected enzymes of mitochondria from different cell types, leading to metabolic modifications in the energy metabolism of the brain cortex [20].
Regarding potential new pharmacologic approaches, the fatty amide oleamide had AD-like effects in the chronic mild stress rat model [21]. After treatment, proteomic analysis of the HPC showed altered proteins involved in processes such as energy metabolism, oxidative stress, and cell communication of stressed oleamide-treated rats [21]. Based on these findings, the altered proteins reveal potential candidates as therapeutic targets. In this line, ketamine (KET) has recently been approved as a medication for TRD [22]. An animal study that evaluated the effect of low doses of KET in the HPC found a correlation between the FST floating time and the ATP/ADP metabolite ratio, which decreased after a single injection of the drug [23], suggesting that KET AD effect may be mediated by energy metabolism and the antioxidant defense system. A study comparing the amygdala and HPC in a chronic unpredictable mild stress (CUMS) rat model showed a proteomic signature of the acute esketamine effect in the amygdala with actin nucleation and glutamate signaling pathways as the most significant biologic processes affected [24]. Acute KET treatment also led to significant changes in signaling kinases and proteases in the HPC, synaptic and energy pathways in the prefrontal cortex, and peripheral inflammation and growth factor dysregulation in serum analysis of a corticosterone mice model of depression [25]. It is interesting to note that the fibroblast growth factor 9 has not been reported previously and modulates the expression of myelin-related proteins and multiple fibroblast growth factor receptors in developing oligodendrocytes [25]. Impairments in these cells have been reported in MDD patients, and it might be an important key in treatment and pathophysiology of the disorder. Taken together, these findings indicate that KET-induced effects are involved in synaptic plasticity, inflammation, growth factor signaling, and in the balance between excitatory and inhibitory signals.
Nimesulide, a classical nonsteroidal anti-inflammatory, had an AD-like effect when administered in a CUMS rat model [26]. Proteins modulated in the animal's HPC were mainly involved in the formation of cellular components, such as cytoplasm and extracellular exosome, and the regulation of cellular response to oxidative stress. Differentially expressed proteins are also mainly involved in metabolic and endocytosis pathways [26]. Overall, these results show that nimesulide might mediate its AD effect through the inhibition of oxidative stress and inflammatory response.
Last, proteomic analysis of the hippocampal tissue from neutral amino acid transporter SLC6A15 knock-out mice together with several in vitro assays in primary hippocampal neurons showed that the differentially expressed proteins identified were involved in pathways of metabolic, mitochondrial, and structural functions [27]. Besides that, a reduced release probability at glutamatergic synapses, increased mitochondrial function, higher glutathione/glutathione disulfide redox ratio, and an improved neurite outgrowth in primary neurons lacking SLC6A15 was also reported [27]. Taken together, these findings provide indications that a pharmacologic or genetic reduction of SLC6A15 activity may be a promising approach for AD therapy.
Other Omics
A genomic study showed that berberine, a natural alkaloid, was associated with gastrointestinal flora alterations in a chronic unpredictable stress rat model [28]. The treatment with the inulin-type prebiotic fructo-oligosaccharides purified from Morinda officinalis in a CUMS rat model resulted in AD-like effects and repair of intestinal epithelial damages, reinstating the dysbiosis in depressive rats' gut [29]. Moreover, inulin-type prebiotic fructo-oligosaccharides promoted the abundance of the bacterial phylum Cyanobacteria and decreased corticosterone levels [29]. Together, these analyses implied that AD-like efficacy was strongly associated with the modulation of the microbiota-gut-brain axis.
Regarding treatment response, an increase of methylation in the P11 promoter was related to a good response in the chronic mild stress model after electroconvulsive therapy (ECT) in a genomic landscape [30]. These data can help diagnose and properly treat patients avoiding repeated trial and error treatment attempts.
The hippocampal transcriptional profile in postmortem samples was also evaluated to identify common signatures in the MDD rat model and humans [31]. Data allowed the identification of a hippocampal transcriptional signature of resilient or susceptible responses in rat MDD models which overlapped with gene expression alterations observed in patients with depression. Also through a transcriptomic approach, biomarkers predictive of FLX response, candidate trait biomarkers, and candidate state biomarkers were also found in the CUMS animal model, followed by validation in humans [32]. Regarding KET effects, transcriptomics analyses showed an abnormal expression of the 2 circular RNAs in the HPC of rats after treatment [33]. Besides that, several biologic processes were regulated by the target genes of the 2 circular RNAs [33].
Other possible treatments were investigated by metabolomics. For instance, the chronic treatment with the minocycline antibiotic had an AD-like effect, inhibited neuroinflammation in the HPC, and altered species abundance and metabolites of gut microbiota in CUMS mice [34]. It also ameliorated intestinal barrier disruption and reduced the bacteriological indexes, such as diamine oxidase, C-reactive protein, and endotoxin in peripheral blood of CUMS mice. Therefore, the AD-like mechanism of chronic minocycline treatment is maybe due to the combined action of neuroinflammation and gut microbiota modulation [34]. Regarding herbal extracts, the significant AD-like activity of the alkaloidal fraction of M. cordifolium (A. cordifolia) root extract, might be attributed in part to its considerable content of mesembrane alkaloid 5, which is postulated to have potent inhibitory activity in serotonin transporter, as suggested by in silico virtual screening [35].
Proteomic analysis combined with metabolomics also indicated that chronic paroxetine (SSRI) treatment affects purine, pyrimidine, glutamatergic, and ubiquitin-proteasome system, having a crucial role in the AD treatment response in both mice and human [36, 37]. Moreover, transcriptomics combined with metabolomics also showed that the treatment with paroxetine has an influence not only on the composition of gut microbes in mice but also on the levels of animal bile acids, which in turn may be linked with behavior alterations [38]. Additionally, the microbiota alpha diversity between paroxetine- and vehicle-treated mice, suggest that microbiota functions are altered by the drug [38].
Clinical Studies
Proteomic Studies
Biomarker studies allow distinguishing between responder and nonresponder patients to AD treatments. Investigations of peripheral blood mononuclear cells collected from AD responder and nonresponder MDD patients identified alterations in GTPase activity pathway, platelet activation, antigen processing, and presentation pathways [39]. Members of the integrin signaling pathway, RAS-related proteins, and ubiquitin-proteasome system were differentially expressed between blood mononuclear cells of responders and nonresponders patients as well [40]. Turck et al. [41] showed that differences in the levels of 29 proteins at baseline were found in the group with a favorable treatment response, where most of these proteins were components of metabolism or immune response pathways as well as multiple components of the coagulation cascade. After the treatment period, 2 proteins that were altered in responders and 9 in nonresponders overlapped with those proteins that had been identified in the baseline group [41]. Of this, the fibrinogen alpha chain was 1 of the 9 altered proteins in nonresponders as shown by our group [42].
The search for biomarkers of response to venlafaxine (a serotonin-norepinephrine reuptake inhibitor), IMP, and mixed ADs was also accessed by Chan et al. [43]. The identified biomarkers in the patient's serum were predominantly involved in immune/inflammatory processes and blood coagulation followed by endocrine and growth factor signaling [43]. Apolipoprotein A-IV was the most reproducible biomarker of treatment response and also 1 validated through LC-MS, being significantly associated with response to treatment with venlafaxine and mixed ADs [43].
In this line, an exploratory study analyzed plasma protein signatures associated with the response to escitalopram (SSRI), and the authors found that 37 proteins were significantly associated with response/time [44]. In addition, gene ontology annotation showed that response to the stimulus was the biologic process that had more altered proteins during 10 weeks of treatment [44].
Further, Lee et al. [45] showed a serum biomarker panel consisting of 6 proteins which results suggest that the modulation of the immune and inflammatory systems and lipid metabolism are involved in the pathophysiology of MDD [45]. Many of the markers to treatment response point in interesting directions which could lead to a better comprehension not only about the treatment of MDD but also in the pathophysiology of this disorder, which in turn shed light on the investigations of potential new targets.
Other Omics
A genomic study using DNA extracted from saliva to derive polygenic risk scores for MDD reported that after multiple testing corrections, polygenic risk scores in the drug metabolic pathway significantly predicted recurrent depression [46]. Among genes in this pathway, CYP2C19 and CBR1 appear to contribute the most to MDD liability [46]. These results seem to be potentially relevant to pharmacokinetics since CYP2C19 protein is involved in the metabolism of ADs. Another study investigated the role of genetic variants in predicting rapid-AD response to KET and scopolamine in subjects diagnosed with MDD or bipolar disorder [47]. This study pointed out that SEC11A, KRASP1, and FAM83B genes seem to be associated with the rapid-AD effect of KET. On the other hand, Li et al. [48] pointed out the IRAK3 and NME7 genes as ones associated with the efficacy of esketamine treatment in MDD patients. Moreover, pathways relevant to neuronal and synaptic function, immune signaling, and glucocorticoid receptor/stress response showed enrichment among the suggestive GWAS signals [48].
Comparing Chinese patients in remission and nonremission states, high-throughput single nucleotide polymorphism (SNP) genotyping showed an association among venlafaxine (serotonin-norepinephrine reuptake inhibitor) effects, NR3C2 gene polymorphisms, and the neuroendocrine thyroid-stimulating hormone, in which the concentration significantly increased after venlafaxine treatment in the remission group [49]. The mineralocorticoid receptor NR3C2 has been pointed out in the pathophysiology of MDD, and this study showed a novel NR3C2 gene polymorphism involved in the MDD treatment response. However, more studies are demanded to validate these results. Specific to SSRIs, a functional SNP in the serotonin receptor 7 (HTR7) gene was associated with response to citalopram, paroxetine, FLX, and sertraline in both bipolar disorder and MDD providing novel pharmacogenomic evidence to support the role of HTR7 in association with AD response [50].
Single nucleotide variants were also evaluated in modulating AD response, demonstrating that the SNV burden had a significant impact on the AD drug response group but was limited in the repetitive transcranial magnetic stimulation group [51]. Accordingly, 7 SNPs at 5 loci, namely, IL1A, GNA15, PPP2CB, PLA2G4C, and GBA, were identified as affecting the AD response [51]. These results showed some promising evidence on genetic variants that could be used as individualized therapeutic guides for MDD patients. Regarding TRD, Maffioletti et al. [52] reported that the alleles associated with lower serum vascular endothelial growth factor concentrations in MDD patients were associated with nonresponse to ECT.
Unlike traditional treatments, a group investigated the effects of changes in the bacteria Firmicutes phylum, related to a protective function in the intestinal barrier, in the feces of MDD patients and healthy controls [53]. They found that the variety of alpha bacteria, especially Firmicutes sp, were considerably lower in MDD patients [53], which point out to possible alteration promoted by ADs in the gut microbiota.
A study conducted with a subset of patients with MDD who did not respond to at least 1 prior medication trial and were taking medication with predicted gene-drug interactions was compared with patients under the treatment selected as usual [54]. The authors found that, by analyzing the gene-drug interaction, clinical improvements can be reached. Despite the study contributing to better AD prescribing, it did not show the gene targets.
Regarding metabolomic studies, plasma samples were analyzed before and after ECT in TRD patients [55]. The treatment with ECT was associated with a significant decrease in the plasma levels of tryptophan, kynurenine (KYN), and quinolinic acid, but no significant alterations were found in the levels of kynurenic acid. On the other hand, only the kynurenic acid was detected in overlapped biomarkers between MDD diagnosis and treatment response to escitalopram; MDD patients with lower levels of kynurenic acid showed a better therapeutic response to escitalopram [56]. Together, these data confirm an imbalance at the KYN pathway in MDD and overlapping biomarkers that facilitate diagnosis and prediction of the treatment response may help improve disorder classification and reduce the exposure of patients to fewer effective treatments in MDD.
In a large case-control treatment study of major depressive episodes before and after AD treatment, results from the metabolomic analysis showed that depressed patients had a lower nitric oxide synthase (NOS) activity than healthy controls at baseline and lower NOS activity at baseline predicted a higher response rate [57]. In addition, NOS activity in depressed patients increased after AD monotherapy treatment.
Some omic studies have been done through the analyses of databanks and published data. One of them, through the investigation of possible drug targets and repurposing opportunities outside the major histocompatibility complex polygenic risk scores region, found 153 protein-coding genes which are associated with MDD, 5 potentially up- or downregulated in the brain and 24 of which are druggable [58]. Another conducted a systematic review in the search for genes and pathways which could be frequently associated with MDD and AD response, where they found 2 sets of genes, associated with SSRIs AD response and MDD [59]. They highlighted the Cushing syndrome, axon guidance, cAMP signaling pathway, insulin secretion, and glutamatergic synapse pathways [59]. To predict medication blood levels in MDD patients, a combinatorial pharmacogenomic algorithm through the analysis of Genomics Used to Improve Depression Decisions databank incorporated the CYP2C19, CYP2D6, and CYP3A4 genes and showed these 3 genes are a superior predictor of citalopram and escitalopram blood levels when compared to individual genes commonly used [60]. Further, a repurposing drug study found molecules, such as modulators of inflammation, renin-angiotensin system, proliferator-activated receptor agonists, glycogen synthase kinase 3 beta inhibitors, the rho associated kinase inhibitor fasudil, and also nutraceuticals, as potential treatments [61], which in turn can be valuable for future studies that may lead to new treatments for TRD.
Discussion and Conclusion
From the established scope reviewed here, we found 25 studies on proteomics, 16 on genomics, 3 on transcriptomics, 11 on metabolomics, and 6 on multi-omics (see online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000518098; Fig. 1). There is a potential comprehension of MDD combining different omic landscapes; however, the lack of integration of omics data jeopardizes the discussion and conclusion. Despite some successful integration of omic approaches on animal models of stress processes [62], no preclinical or clinical studies integrating all omics on the treatment of MDD have been reported. Nevertheless, omic studies reviewed here can individually contribute to providing pathways and molecular targets associated with new potential treatments for MDD patients, which do not respond to current ADs [5].
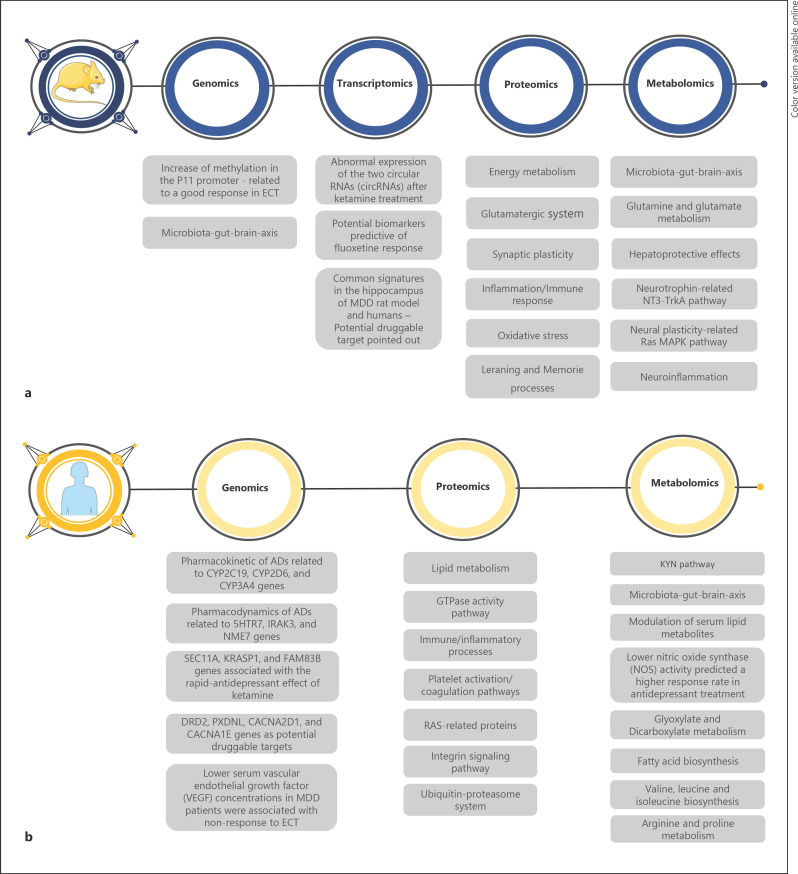
Fig. 1.
Summary of main findings associated with MDD treatment reported by animal (a) and human (b) studies. MDD, major depressive disorder; circRNAs, circular RNAs; AD, antidepressant; VEGF, vascular endothelial growth factor; NOS, nitric oxide synthase.
Animal studies are useful to comprehend the molecular mechanism of ADs on brain regions associated with the pathophysiology of depression since human samples present challenges to be studied. Despite that, modeling complex human disorders like MDD in animals is a challenge represented not only by the model per se but also by the way the tests are selected, conducted, and interpreted. Herewith, understanding pharmacologic effects and looking for new drugs to treat this disorder has limitations that should be taken into account when animal studies are carried out in this field. Apart from that, the studies described here show that ADs can attenuate or reverse depressive-like behaviors, and the authors associated these effects with alterations mainly on energy metabolism with the involvement of several mitochondrial proteins, glutamatergic transmission, and neuroplasticity [14, 15, 16, 17, 25, 27, 63]. These studies reinforce the effects on those pathways but do not add new advances for the field. However, more studies using different animal models are encouraged, as well as other ADs classes should be investigated.
Studying complex disorders in humans is limited, despite them being the bearers of the disorders, many aspects involving the study cannot be controlled. For one, the person's background prior to the study, what they are exposed to during the treatment, and even if the treatment was done correctly, in case it requires taking the medication regularly. Moreover, many sample acquisition procedures are invasive, making them limited to chance. Nevertheless, humans are the most complete system to understanding the efficacy of medications, and this kind of study is extremely important to cross with molecular data and better comprehend drugs' mechanisms of action. In reference to that, while some proteomic and omic studies point to the pathways and genes affected by ADs, others analyze differences between responder and nonresponder patients. The omic data revealed biomarkers that have a key impact on ADs response prediction [30, 32, 39, 40, 41, 42, 43, 44, 52, 56, 57], which can be investigated in terms of new druggable targets. However, the different biomarker panels resulted from these studies, and no validation of those potential biomarkers are the main limitations for clinical application. Therefore, integrating omic studies on MDD is demanded to provide a useful panel of markers that could be investigated as new treatment targets.
Here, we observed that both animal and clinical studies pointed to alterations in inflammatory pathways [18, 25, 34, 43, 45, 48, 61, 64]. Whereas, oxidative stress [18, 21, 23, 26, 27] and energy metabolism [13, 14, 15, 16, 17, 18, 19, 20, 21, 23, 65] were reported more in animal studies. To note, a drug repositioning study reported several potential modulators, and among them, drugs acting on neuroplasticity and inflammation, which has been described for several animal and clinical studies here, are the most promising targets [61]. It is also interesting to note other targets pointed out, including the nutraceuticals.
Efforts have been made to find new molecules as a potential treatment of MDD symptoms, such as studies using herbal formulations [28, 35, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74] or isolated active components [29] from traditional Chinese medicine. Antibiotic [34] and anti-inflammatory compounds [26, 61] were pointed out as well. These findings can contribute to understanding other potential molecular mechanisms for the treatment of MDD. However, these nontraditional treatments do not show clinical relevance for MDD treatment. Therefore, studies are demanded to corroborate the molecular findings and make these data more robust and valuable at the systems level.
Studies have gone beyond brain and blood samples showing the importance of the microbiota-gut-brain axis for the treatment of MDD [28, 29, 34, 38, 53, 66, 67]. Furthermore, pathways that have not been widely described to be associated with AD effects, such as KYN [55, 56], and lipid metabolism [45, 64, 70, 73] seem to be promising since their role on neuroplasticity, signaling transducing in the brain, and myelination.
Regarding new treatments, KET and esketamine seem to be the most valuable on MDD [23, 24, 25, 48]. The AD effect of KET and esketamine seems to be involved in synaptic plasticity, inflammation, growth factor signaling, energy metabolism, calcium channels, and the balance between excitatory and inhibitory signals [23, 24, 25, 48] and with the genes IRAK3 and NME7 [48]. Moreover, the rapid onset of KET ADs effects was associated with SEC11A, KRASP1, and FAM83B genes [47]. These data should be deeply investigated to understand and discover rapid-acting AD mechanisms.
From genomic studies, an important contribution to personalized treatment has been noted. The pharmacogenomic tests including genes related to pharmacokinetic of ADs (CYP2C19, CYP2D6, and CYP3A4) until rapid-acting effect (SEC11A, KRASP1, and FAM83B) and pharmacodynamics (5HTR7, IRAK3, and NME7) will be valuable to increase the current treatment efficacy. Despite not being our focus, we highlighted that an important drug repositioning study showed potential druggable targets [58] that were also reported in studies described here, such as DRD2, PXDNL, CACNA2D1, and CACNA1E genes [31, 45, 59]. These 4 druggable targets discussed may aid in the development or searching of drug repurposing to the treatment of MDD.
In conclusion, we highlight that the omic data described here contributed to understanding the molecular mechanisms of ADs and possible new molecules on MDD therapy (Fig. 2). However, efforts are demanded to apply these findings to the development of new drugs or repurposing strategies for others. We also noticed the lack of studies using cell culture models, such as techniques using induced pluripotent stem cells from MDD patients that can be useful not only to comprehend the neurobiology of MDD but also molecular mechanisms of ADs. Finally, overall omics data are in the way to contribute with personalized therapy for MDD and other subtypes of depression.
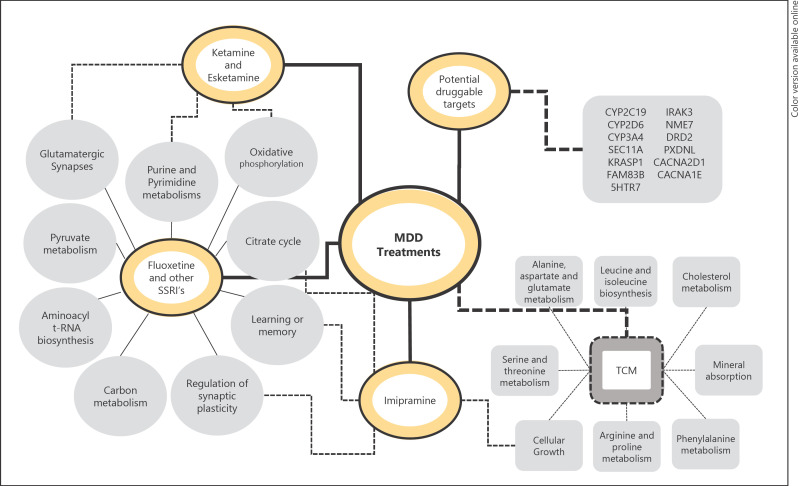
Fig. 2.
Representative pathways, biologic processes, and genes related to alterations promoted by the most explored drugs in recent years. The dashed lines represent pathways that may be in common between treatments. TCM, traditional Chinese medicine.
Conflict of Interest Statement
The authors declare no conflict of interest.
Funding Sources
This work was supported by FAPESP (São Paulo Research Foundation, grants 2017/18242-1, 2017/25588-1, 2018/03422-7, 2018/03673-0 and 2018/25818-0), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, grant 123887/2019-4), and SAE (Student Support Service of Unicamp, grant 01.P.96/2020).
Author Contributions
V.A. and D.M.S. supervised, edited, and revised the manuscript. L.R.S. and P.C.T. selected the manuscripts here and considered, wrote and revised the manuscript. L.R.S. and L.C.S.-C. analyzed the data and built the figures. All the authors agreed with the final version.
Supplementary Material
Supplementary data
Acknowledgements
The authors thank Prof. Brett Vern Carlson (Technological Institute of Aeronautics [ITA]) for typing assistance with the manuscript. We dedicate our work to patients with depression.
References
- 1.Patten SB, Kennedy SH, Lam RW, O'Donovan C, Filteau MJ, Parikh SV, et al. Canadian network for mood and anxiety treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults. I. Classification, burden and principles of management. J Affect Disord. 2009;117(Suppl 1):S5–14. doi: 10.1016/j.jad.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Depression and other common mental disorders: global health estimates. Geneva: World Health Organization; 2017. WHO/MSD/MER/2017.2. Available from: https://apps.who.int/iris/handle/10665/254610 Accessed 2020 Jan 31. [Google Scholar]
- 3.Adler DA, McLaughlin TJ, Rogers WH, Chang H, Lapitsky L, Lerner D. Job performance deficits due to depression. Am J Psychiatry. 2006;163:1569–76. doi: 10.1176/appi.ajp.163.9.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DSM-5 diagnostic classification. Diagnostic and statistical manual of mental disorders. Forthcoming. 2013 [Google Scholar]
- 5.Al-Harbi KS. Treatment-resistant depression: therapeutic trends, challenges, and future directions. Patient Prefer Adherence. 2012;6:369. doi: 10.2147/PPA.S29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blier P. Neurobiology of depression and mechanism of action of depression treatments. J Clin Psychiatry. 2016;77((3)):e319. doi: 10.4088/JCP.13097tx3c. [DOI] [PubMed] [Google Scholar]
- 7.Kraus C, Kadriu B, Lanzenberger R, Zarate CA, Kasper S. Prognosis and improved outcomes in major depression: a review. Focus. 2020;18((2)):220–35. doi: 10.1176/appi.focus.18205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thase ME, Schwartz TL. Choosing medications for treatment-resistant depression based on mechanism of action. J Clin Psychiatry. 2015;76((6)):720–7. doi: 10.4088/JCP.14052ah2c. [DOI] [PubMed] [Google Scholar]
- 9.Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. Erratum: resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23:264–38. doi: 10.1038/nm0217-264d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarazona S, Balzano-Nogueira L, Conesa A. Multiomics data integration in time series experiments. Compr Anal Chem. 2018;82:505–32. [Google Scholar]
- 11.Zarayeneh N, Ko E, Oh JH, Suh S, Liu C, Gao J, et al. Integration of multi-omics data for integrative gene regulatory network inference. Int J Data Min Bioinform. 2017;18:223–39. doi: 10.1504/IJDMB.2017.10008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennington K, Cotter D, Dunn MJ. The role of proteomics in investigating psychiatric disorders. Br J Psychiatry. 2005;187:4–6. doi: 10.1192/bjp.187.1.4. [DOI] [PubMed] [Google Scholar]
- 13.Perić I, Costina V, Stanisavljević A, Findeisen P, Filipović D. Proteomic characterization of hippocampus of chronically socially isolated rats treated with fluoxetine: depression-like behaviour and fluoxetine mechanism of action. Neuropharmacology. 2018;135:268–83. doi: 10.1016/j.neuropharm.2018.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Filipović D, Costina V, Perić I, Stanisavljević A, Findeisen P. Chronic fluoxetine treatment directs energy metabolism towards the citric acid cycle and oxidative phosphorylation in rat hippocampal nonsynaptic mitochondria. Brain Res. 2017;1659:41–54. doi: 10.1016/j.brainres.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalk MG, Mortas P, Haman M, Ozcan S, Biemans B, Bahn S. Fluoxetine, not donepezil, reverses anhedonia, cognitive dysfunctions and hippocampal proteome changes during repeated social defeat exposure. Eur Neuropsychopharmacol. 2018;28:195–210. doi: 10.1016/j.euroneuro.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Głombik K, Stachowicz A, Trojan E, Olszanecki R, Ślusarczyk J, Suski M, et al. Evaluation of the effectiveness of chronic antidepressant drug treatments in the hippocampal mitochondria − a proteomic study in an animal model of depression. Prog Neuropsychopharmacol Biol Psychiatry. 2017;78:51–60. doi: 10.1016/j.pnpbp.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Głombik K, Stachowicz A, Trojan E, Ślusarczyk J, Suski M, Chamera K, et al. Mitochondrial proteomics investigation of frontal cortex in an animal model of depression: Focus on chronic antidepressant drugs treatment. Pharmacol Rep. 2018;70((2)):322–30. doi: 10.1016/j.pharep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Kedracka-Krok S, Swiderska B, Bielecka-Wajdman AM, Prus G, Skupien-Rabian B, Jankowska U, et al. Impact of imipramine on proteome of rat primary glial cells. J Neuroimmunol. 2018;320:25–37. doi: 10.1016/j.jneuroim.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Villa RF, Ferrari F, Bagini L, Gorini A, Brunello N, Tascedda F. Mitochondrial energy metabolism of rat hippocampus after treatment with the antidepressants desipramine and fluoxetine. Neuropharmacology. 2017;121:30–8. doi: 10.1016/j.neuropharm.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 20.Villa RF, Ferrari F, Gorini A, Brunello N, Tascedda F. Effect of desipramine and fluoxetine on energy metabolism of cerebral mitochondria. Neuroscience. 2016;330:326–34. doi: 10.1016/j.neuroscience.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 21.Ge L, Zhu MM, Yang JY, Wang F, Zhang R, Zhang JH, et al. Differential proteomic analysis of the anti-depressive effects of oleamide in a rat chronic mild stress model of depression. Pharmacol Biochem Behav. 2015;131:77–86. doi: 10.1016/j.pbb.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Kernel Networks Inc FDA approves new nasal spray medication for treatment-resistant depression; available only at a certified doctor's office or clinic. Case Med Res. 2019 [Google Scholar]
- 23.Weckmann K, Deery MJ, Howard JA, Feret R, Asara JM, Dethloff F, et al. Ketamine's antidepressant effect is mediated by energy metabolism and antioxidant defense system. Sci Rep. 2017;7:15788. doi: 10.1038/s41598-017-16183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al Shweiki R, Oeckl P, Steinacker P, Barschke P, Pryce C, Dorner-Ciossek C, et al. S-ketamine induces acute changes in the proteome of the mouse amygdala. J Proteomics. 2020;216:103679. doi: 10.1016/j.jprot.2020.103679. [DOI] [PubMed] [Google Scholar]
- 25.Wesseling H, Rahmoune H, Tricklebank M, Guest PC, Bahn S. A targeted multiplexed proteomic investigation identifies ketamine-induced changes in immune markers in rat serum and expression changes in protein kinases/phosphatases in rat brain. J Proteome Res. 2015;14:411–21. doi: 10.1021/pr5009493. [DOI] [PubMed] [Google Scholar]
- 26.Luo W, Luo Y, Yang J. Proteomics-based screening of the target proteins associated with antidepressant-like effect and mechanism of nimesulide. Sci Rep. 2020;10:11052. doi: 10.1038/s41598-020-66420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schraut KG, Kalnytska O, Lamp D, Jastroch M, Eder M, Hausch F, et al. Loss of the psychiatric risk factor SLC6A15 is associated with increased metabolic functions in primary hippocampal neurons. Eur J Neurosci. 2021;53((2)):390–401. doi: 10.1111/ejn.14990. [DOI] [PubMed] [Google Scholar]
- 28.Zhu X, Sun Y, Zhang C, Liu H. Effects of berberine on a rat model of chronic stress and depression via gastrointestinal tract pathology and gastrointestinal flora profile assays. Mol Med Rep. 2017;15:3161–71. doi: 10.3892/mmr.2017.6353. [DOI] [PubMed] [Google Scholar]
- 29.Chi L, Khan I, Lin Z, Zhang J, Lee MYS, Leong W, et al. Fructo-oligosaccharides from Morinda officinalis remodeled gut microbiota and alleviated depression features in a stress rat model. Phytomedicine. 2020;67:153157. doi: 10.1016/j.phymed.2019.153157. [DOI] [PubMed] [Google Scholar]
- 30.Neyazi A, Theilmann W, Brandt C, Rantamäki T, Matsui N, Rhein M, et al. P11 promoter methylation predicts the antidepressant effect of electroconvulsive therapy. Transl Psychiatry. 2018;8:25. doi: 10.1038/s41398-017-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carboni L, Marchetti L, Lauria M, Gass P, Vollmayr B, Redfern A, et al. Cross-species evidence from human and rat brain transcriptome for growth factor signaling pathway dysregulation in major depression. Neuropsychopharmacology. 2018;43:2134–45. doi: 10.1038/s41386-018-0117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hervé M, Bergon A, Le Guisquet AM, Leman S, Consoloni JL, Fernandez-Nunez N, et al. Translational identification of transcriptional signatures of major depression and antidepressant response. Front Mol Neurosci. 2017;10:248. doi: 10.3389/fnmol.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao J, Li T, Fan D, Zhou H, Feng J, Liu L, et al. Abnormal expression of rno_circRNA_014900 and rno_circRNA_005442 induced by ketamine in the rat hippocampus. BMC Psychiatry. 2020;20:1. doi: 10.1186/s12888-019-2374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Q, Luo L, Sun T, Yang L, Cheng LF, Wang Y, et al. Chronic minocycline treatment exerts antidepressant effect, inhibits neuroinflammation, and modulates gut microbiota in mice. Psychopharmacology. 2020;237:3201–13. doi: 10.1007/s00213-020-05604-x. [DOI] [PubMed] [Google Scholar]
- 35.Said AAE, Ali TFS, Attia EZ, Ahmed AF, Shehata AH, Abdelmohsen UR, et al. Antidepressant potential of Mesembryanthemum cordifolium roots assisted by metabolomic analysis and virtual screening. Nat Prod Res. 2020:1–5. doi: 10.1080/14786419.2020.1788019. [DOI] [PubMed] [Google Scholar]
- 36.Park DI, Dournes C, Sillaber I, Uhr M, Asara JM, Gassen NC, et al. Purine and pyrimidine metabolism: convergent evidence on chronic antidepressant treatment response in mice and humans. Sci Rep. 2016;6:35317. doi: 10.1038/srep35317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park DI, Dournes C, Sillaber I, Ising M, Asara JM, Webhofer C, et al. Delineation of molecular pathway activities of the chronic antidepressant treatment response suggests important roles for glutamatergic and ubiquitin-proteasome systems. Transl Psychiatry. 2017;7:e1078. doi: 10.1038/tp.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dethloff F, Vargas F, Elijah E, Quinn R, Park DI, Herzog DP, et al. Paroxetine administration affects microbiota and bile acid levels in mice. Front Psychiatry. 2020;11:518. doi: 10.3389/fpsyt.2020.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park DI, Štambuk J, Razdorov G, Pučić-Baković M, Martins-de-Souza D, Lauc G, et al. Blood plasma/IgG N-glycome biosignatures associated with major depressive disorder symptom severity and the antidepressant response. Sci Rep. 2018;8:179. doi: 10.1038/s41598-017-17500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins-de-Souza D, Maccarrone G, Ising M, Kloiber S, Lucae S, Holsboer F, et al. Blood mononuclear cell proteome suggests integrin and Ras signaling as critical pathways for antidepressant treatment response. Biol Psychiatry. 2014;76:e15–7. doi: 10.1016/j.biopsych.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Turck CW, Guest PC, Maccarrone G, Ising M, Kloiber S, Lucae S, et al. Proteomic differences in blood plasma associated with antidepressant treatment response. Front Mol Neurosci. 2017;10:272. doi: 10.3389/fnmol.2017.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martins-de-Souza D, Maccarrone G, Ising M, Kloiber S, Lucae S, Holsboer F, et al. Plasma fibrinogen: now also an antidepressant response marker? Transl Psychiatry. 2014;4:e352. doi: 10.1038/tp.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan MK, Cooper JD, Bot M, Birkenhager TK, Bergink V, Drexhage HA, et al. Blood-based immune-endocrine biomarkers of treatment response in depression. J Psychiatr Res. 2016;83:249–59. doi: 10.1016/j.jpsychires.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Kim EY, Ahn HS, Lee MY, Yu J, Yeom J, Jeong H, et al. An exploratory pilot study with plasma protein signatures associated with response of patients with depression to antidepressant treatment for 10 weeks. Biomedicines. 2020 Oct 28;8((11)):455. doi: 10.3390/biomedicines8110455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MY, Kim EY, Kim SH, Cho KC, Ha K, Kim KP, et al. Discovery of serum protein biomarkers in drug-free patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2016;69:60–8. doi: 10.1016/j.pnpbp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Docherty AR, Moscati A, Bigdeli TB, Edwards AC, Peterson RE, Adkins DE, et al. Pathway-based polygene risk for severe depression implicates drug metabolism in CONVERGE. Psychol Med. 2020;50((5)):793–8. doi: 10.1017/S0033291719000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo W, Machado-Vieira R, Mathew S, Murrough JW, Charney DS, Grunebaum M, et al. Exploratory genome-wide association analysis of response to ketamine and a polygenic analysis of response to scopolamine in depression. Transl Psychiatry. 2018;8:280. doi: 10.1038/s41398-018-0311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li QS, Wajs E, Ochs-Ross R, Singh J, Drevets WC. Genome-wide association study and polygenic risk score analysis of esketamine treatment response. Sci Rep. 2020;10:12649. doi: 10.1038/s41598-020-69291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan F, Yuan R, Ren D, Bi Y, Niu W, Guo Z, et al. A novel NR3C2 polymorphism and the increased thyroid-stimulating hormone concentration are associated with venlafaxine treatment outcome in Chinese Han MDD patients. Psychiatry Res. 2020;284:112690. doi: 10.1016/j.psychres.2019.112690. [DOI] [PubMed] [Google Scholar]
- 50.Wei YB, McCarthy M, Ren H, Carrillo-Roa T, Shekhtman T, DeModena A, et al. A functional variant in the serotonin receptor 7 gene (HTR7), rs7905446, is associated with good response to SSRIs in bipolar and unipolar depression. Mol Psychiatry. 2020;25:1312–22. doi: 10.1038/s41380-019-0397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Z, Xie C, Xia L, Yuan Y, Zhu H, Huang X, et al. Targeted exome sequencing identifies five novel loci at genome-wide significance for modulating antidepressant response in patients with major depressive disorder. Transl Psychiatry. 2020;10:30. doi: 10.1038/s41398-020-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maffioletti E, Gennarelli M, Magri C, Bocchio-Chiavetto L, Bortolomasi M, Bonvicini C, et al. Genetic determinants of circulating VEGF levels in major depressive disorder and electroconvulsive therapy response. Drug Dev Res. 2020;81:593–9. doi: 10.1002/ddr.21658. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, Shi X, Li Z, Shen Y, Shi X, Wang L, et al. Possible association of Firmicutes in the gut microbiota of patients with major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:3329–37. doi: 10.2147/NDT.S188340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thase ME, Parikh SV, Rothschild AJ, Dunlop BW, DeBattista C, Conway CR, et al. Impact of pharmacogenomics on clinical outcomes for patients taking medications with gene-drug interactions in a randomized controlled trial. J Clin Psychiatry. 2019;80((6)):19m12910. doi: 10.4088/JCP.19m12910. [DOI] [PubMed] [Google Scholar]
- 55.Schwieler L, Samuelsson M, Frye MA, Bhat M, Schuppe-Koistinen I, Jungholm O, et al. Electroconvulsive therapy suppresses the neurotoxic branch of the kynurenine pathway in treatment-resistant depressed patients. J Neuroinflammation. 2016;13:51. doi: 10.1186/s12974-016-0517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erabi H, Okada G, Shibasaki C, Setoyama D, Kang D, Takamura M, et al. Kynurenic acid is a potential overlapped biomarker between diagnosis and treatment response for depression from metabolome analysis. Sci Rep. 2020;10:16822. doi: 10.1038/s41598-020-73918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loeb E, El Asmar K, Trabado S, Gressier F, Colle R, Rigal A, et al. Nitric oxide synthase activity in major depressive episodes before and after antidepressant treatment: results of a large case-control treatment study. Psychol Med. 2020 Jun 11;:1–10. doi: 10.1017/S0033291720001749. [DOI] [PubMed] [Google Scholar]
- 58.Gaspar HA, Gerring Z, Hübel C, au, Middeldorp CM, Derks EM, et al. Using genetic drug-target networks to develop new drug hypotheses for major depressive disorder. Transl Psychiatry. 2019;9:117. doi: 10.1038/s41398-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srivastava A, Singh P, Gupta H, Kaur H, Kanojia N, Guin D, et al. Systems approach to identify common genes and pathways associated with response to selective serotonin reuptake inhibitors and major depression risk. Int J Mol Sci. 2019 Apr 23;20((8)):1993. doi: 10.3390/ijms20081993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shelton RC, Parikh SV, Law RA, Rothschild AJ, Thase ME, Dunlop BW, et al. Combinatorial pharmacogenomic algorithm is predictive of citalopram and escitalopram metabolism in patients with major depressive disorder. Psychiatry Res. 2020;290:113017. doi: 10.1016/j.psychres.2020.113017. [DOI] [PubMed] [Google Scholar]
- 61.Fabbri C, Kasper S, Zohar J, Souery D, Montgomery S, Albani D, et al. Drug repositioning for treatment-resistant depression: hypotheses from a pharmacogenomic study. Prog Neuropsychopharmacol Biol Psychiatry. 2021;104:110050. doi: 10.1016/j.pnpbp.2020.110050. [DOI] [PubMed] [Google Scholar]
- 62.Floriou-Servou A, von Ziegler L, Stalder L, Sturman O, Privitera M, Rassi A, et al. Distinct proteomic, transcriptomic, and epigenetic stress responses in dorsal and ventral hippocampus. Biol Psychiatry. 2018;84:531–41. doi: 10.1016/j.biopsych.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 63.Wang T, Bai S, Wang W, Chen Z, Chen J, Liang Z, et al. Diterpene ginkgolides exert an antidepressant effect through the NT3-TrkA and Ras-MAPK pathways. Drug Des Devel Ther. 2020;14:1279–94. doi: 10.2147/DDDT.S229145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C, Hu Y, Dong XZ, Zhou XJ, Mu LH, Liu P. Proteomic analysis of the antidepressant effects of Shen-Zhi-Ling in depressed patients: identification of proteins associated with platelet activation and lipid metabolism. Cell Mol Neurobiol. 2018;38:1123–35. doi: 10.1007/s10571-018-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C, Yin Q, Tian J, Gao X, Qin X, Du G, et al. Studies on the potential link between antidepressant effect of Xiaoyao San and its pharmacological activity of hepatoprotection based on multi-platform metabolomics. J Ethnopharmacol. 2020;249:112432. doi: 10.1016/j.jep.2019.112432. [DOI] [PubMed] [Google Scholar]
- 66.Yu M, Jia HM, Zhang T, Shang H, Zhang HW, Ma LY, et al. Gut microbiota is the key to the antidepressant effect of Chaihu-Shu-Gan-San. Metabolites. 2020 Feb 10;10((2)):63. doi: 10.3390/metabo10020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng Y, Gao X, Meng M, Xue H, Qin X. Multi-omics reveals the mechanisms of antidepressant-like effects of the low polarity fraction of Bupleuri Radix. J Ethnopharmacol. 2020;256:112806. doi: 10.1016/j.jep.2020.112806. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Zhang S, Hu M, Chen Y, Wang W, Zhang K, et al. An integrative metabolomics and network pharmacology method for exploring the effect and mechanism of Radix Bupleuri and Radix Paeoniae Alba on anti-depression. J Pharm Biomed Anal. 2020;189:113435. doi: 10.1016/j.jpba.2020.113435. [DOI] [PubMed] [Google Scholar]
- 69.Liu T, Zhou N, Xu R, Cao Y, Zhang Y, Liu Z, et al. A metabolomic study on the anti-depressive effects of two active components from Chrysanthemum morifolium. Artif Cells Nanomed Biotechnol. 2020;48:718–27. doi: 10.1080/21691401.2020.1774597. [DOI] [PubMed] [Google Scholar]
- 70.Zhou X, Wang J, Lu Y, Chen C, Hu Y, Liu P, et al. Anti-depressive effects of Kai-Xin-San on lipid metabolism in depressed patients and CUMS rats using metabolomic analysis. J Ethnopharmacol. 2020;252:112615. doi: 10.1016/j.jep.2020.112615. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Liu C, Tian J, Gao X, Li K, Du G, et al. Plasma metabolomics of depressed patients and treatment with Xiaoyaosan based on mass spectrometry technique. J Ethnopharmacol. 2020;246:112219. doi: 10.1016/j.jep.2019.112219. [DOI] [PubMed] [Google Scholar]
- 72.Zhang K, He M, Su D, Pan X, Li Y, Zhang H, et al. Quantitative proteomics reveal antidepressant potential protein targets of xiaochaihutang in corticosterone induced model of depression. J Ethnopharmacol. 2019;231:438–45. doi: 10.1016/j.jep.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 73.Du H, Wang K, Su L, Zhao H, Gao S, Lin Q, et al. Metabonomic identification of the effects of the Zhimu-Baihe saponins on a chronic unpredictable mild stress-induced rat model of depression. J Pharm Biomed Anal. 2016;128:469–79. doi: 10.1016/j.jpba.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 74.Wang B, Lu S, Zhang C, Zhu L, Li Y, Bai M, et al. Quantitative proteomic analysis of the liver reveals antidepressant potential protein targets of Sinisan in a mouse CUMS model of depression. Biomed Pharmacother. 2020;130:110565. doi: 10.1016/j.biopha.2020.110565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data