Highlights
-
•
Both DCLK1 long (DCLK1-iso1) and short (DCLK1-iso4) isoforms promote epithelial-mesenchymal transition in pancreatic cancer cells.
-
•
Immunosuppressive components are highly enriched in “mesenchymal” pancreatic cancer.
-
•
The infiltration of CD4 and CD8+ T cells decrease while the M2 macrophages increase in DCLK1-overexpressing tumors.
-
•
DCLK1 inhibitor could reverse EMT program and restore T-cells activity.
Keywords: DCLK1, EMT, Tumor immune microenvironment
Abstract
Immunotherapy has recently become a promising cancer therapy with extensive applications of immune checkpoint inhibitors (ICIs). However, pancreatic ductal adenocarcinoma (PDAC) appears to be unresponsive to immunotherapy due to the immunosuppressive microenvironment. Recent studies showed that cancer stem cell marker DCLK1 promoted the initiation and development of PDAC. Nevertheless, the mechanism driving this process remains unclear. Here, by performing gain-of-function investigations in PDAC cell lines, we demonstrate that both DCLK1 long (DCLK1-iso1, DCLK1-AS) and short (DCLK1-iso4, DCLK1-BL) isoforms can efficiently activate EMT leading to tumor migration and invasion. Consistent with experiments in vitro, bioinformatic analysis demonstrates that DCLK1 may act as a driver of EMT activation in PDAC. Further analysis showed that EMT was associated with an immunosuppressive microenvironment, which includes more immunosuppressive cells and chemokines, and patients with a higher EMT score were less sensitive to immune checkpoint inhibitors according to the TIDE (Tumor Immune Dysfunction and Exclusion) algorithm. Multiplexed immunofluorescence results demonstrated the close correlation between DCLK1, EMT and immunosuppression in PDAC patients. The findings were further confirmed in vivo reflected by decreased CD4+, CD8+ T cells and increased M2 macrophages as well as E-cad loss in DCLK1-overexpressing subcutaneous tumors. Importantly, the highly-specific DCLK1 inhibitor (DCLK1-IN-1) was able to effectively block EMT process and restore T-cell activity. Altogether, our data demonstrate that DCLK1 is strongly associated with tumor immune escape in PDAC and inhibiting DCLK1 kinase activity may be a promising therapeutic modality.
Abbreviations
- DCLK1
(doublecortin-like kinase 1)
- ICIs
(immune checkpoint inhibitors)
- PDAC
(pancreatic ductal adenocarcinoma)
- EMT
(epithelial–mesenchymal transition)
- TIDE
(tumor immune dysfunction and exclusion)
- TME
(tumor microenvironment)
- PanIN
(pancreatic intraepithelial neoplasia)
- CSC
(cancer stem cell)
- qPCR
(quantitative real-time PCR)
- PBMC
(peripheral blood mononuclear cell)
- TCGA
(the cancer genome atlas)
- MsigDB
(molecular signatures database)
- TIMER
(tumor immune estimation resource)
- mIF
(multiplexed immunofluorescence)
- EMT-TFs
(EMT- transcription factors)
- MDSCs
(myeloid-derived suppressive cells)
- TAMs
(tumor-associated macrophages)
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal diseases with 5-year survival rates less than 10% [1]. Poor outcomes in PDAC are mainly due to a tumor-promoting microenvironment, which causes the resistance to current therapy regimens [2]. A distinctive characteristic of the PDAC tumor microenvironment (TME) is an abundance of non-tumor cell components including both immunosuppressive molecular signals and cell types [3]. Recent reports demonstrate that more mesenchymal-like tumors generally induce a more immunosuppressive TME compared to more epithelial-like tumors in several cancers like breast cancer and non-small cell lung cancer [4], [5], [6]. EMT is an important program through which epithelial cells lose their cell–cell adhesion and apico-basal polarity, resulting in increased migration and invasion abilities. Importantly, EMT is a dynamic process that may render tumor cells non-responsive to ICIs [7,8] and drive the recruitment of immunosuppressive cells via the regulation of key chemokines [4,9]. Current knowledge suggests that EMT activation drives multiple parallel pathways to alter the landscape of immune cell infiltrates [10]. For instance, EMT promotes the infiltration of activated B cells and regulatory T-cells as well as the exclusion of CD4/CD8+ Tcells in NSCLC [6]. Moreover, higher EMT-related gene expression is correlated with lower ICI response rates and worse prognosis in patients with urothelial cancer [11]. Similarly, breast cancer with more epithelial characteristics has higher infiltration of CD8+ T cells and M1 macrophages as well as lower PD-L1 expression compared to more mesenchymal breast cancer [12]. In PDAC, it has recently been demonstrated that EMT occurred in pancreatic intraepithelial neoplasia (PanIN) lesions before tumor formation, and that pancreatic carcinogenesis was accompanied by immunosuppression [13]. Notably, whether EMT is associated with immune cell infiltration in PDAC has not been investigated.
Doublecortin-like kinase 1 (DCLK1) was initially identified as a crucial regulator of neurogenesis and neuronal migration [14]. More recently, it has emerged as a cancer stem cell (CSC) marker in intestine and pancreas [15], [16], [17], [18], [19]. DCLK1 is also overexpressed in many cancers, including colorectal cancer [20], PDAC [21], hepatocellular carcinoma [22], and renal clear cell carcinoma [23]. Nakanishi et al. firstly revealed that DCLK1 marked intestinal CSCs specifically using lineage-tracing experiments. Further research by Westphalen et al. demonstrated that DCLK1-positive cells were long-lived and could function as tumor-initiating cells in colorectal cancer [24]. Likewise, DCLK1-expressing epithelial cells were identified as progenitors in conditions of pancreatic injury and tumor-initiating cells in PDAC [25]. However, it is unclear whether DCLK1-promoted tumorigenesis in PDAC is related to the formation of immunosuppressive microenvironment. The DCLK1 gene has two distinct promoter regions. The upstream 5′(α)-promoter encodes a full-length DCLK1 (∼82 kDa long forms DCLK1/2), and the β-promoter encodes a C-terminus kinase-containing region (termed as DCLK1-S or DCLK3/4, 45–50 kDa,) of the full-length DCLK1 [26]. Whether the difference in structures lead to different functions are still unknown. Here, the potential tumor-promoting role of these isoforms is a subject of continued study.
In this study, by performing multiple bioinformatics as well as in vitro and in vivo experiments, we found that DCLK1 played an important role in EMT activation which may be involved in the formation of immunosuppressive microenvironment characterized by the enrichment of M2 macrophages as well as decreased CD4+ and CD8+ T cells. Finally, targeting DCLK1 might be an effective therapeutic target to improve the efficacy of immunotherapies.
Materials and methods
Cell culture and drug administration
Human PDAC cells including AsPC-1, PANC-1 and BxPC-3 and mouse cell line Pan02 were purchased from the Beijing Union Cell Resource Center (Beijing, China). AsPC-1 and BxPC-3 were propagated in RPMI-1640 medium (SIGMA) and PANC-1, Pan02 were cultured in DMEM-high glucose (SIGMA). All cells were cultured with 10% fetal bovine serum (FBS) (Ausbian) at 37 °C and 5% CO2. DCLK1 inhibitor (DCLK1-IN-1) was dissolved in DMSO and three different working concentrations (0 μM, 5 μM, 10 μM) were put in the cell culture medium of PDAC cells and incubated at 37 °C for 48 h.
Construction of DCLK1 overexpressing cell lines
DCLK1 isoforms sequences were cloned into pCDH-MSCV-MCS-EF1α-puro vector for construction of overexpressing cell lines. Then the constructed plasmids and packaging plasmids including pMD2G and psPAX2 were co-transfected into 293T packaging cells by lipofectamine 3000 (Invitrogen, USA) for lentiviral amplification, and the lentivirus was obtained at 48 h and 72 h post-transfection. Next, the harvested viruses were filtered by 0.45 μm filters and co-cultivated with PDAC cell lines (AsPC-1 and PANC-1, Pan02). After 3 days of co-cultivation, those cells stably overexpressing DCLK1 were picked out with 2 μg/ml puromycin to construct stable cell lines.
Western blotting
The whole-cell protein was extracted by RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) and the protein concentration was detected using a BCA protein assay kit (Thermo Fisher Scientic). Extracted proteins were denatured at 95 °C for 10 min and separated on a 10% SDS polyacrylamide gel. After electrophoresis, these proteins were transferred onto a PVDF membrane using semi-dry transfer at 200 mA for 90 min (EMD Millipore). All membranes were blocked for one hour using 5% nonfat milk and probed overnight with specific primary antibodies at 4 °C. After that, membranes were washed with TBST and incubated with secondary antibodies (ZSGB-BIO, China) at room temperature for one hour. Signals were detected using chemiluminescence kit (Millipore) with the Bio-Rad imaging system. The antibodies used above were anti-DCLK1 (1:1000, ab31704, Abcam, 62257S, Cell Signaling Technology), anti-ZO-1(1:1000, 8193T, Cell Signaling Technology), anti-Snail (1:1000, 3879T, Cell Signaling Technology), anti-Vimentin (1:1000, 5741T, Cell Signaling Technology), anti-ZEB1(1:1000, 3396T, Cell Signaling Technology), anti-N-cad (1:1000, 13116T, Cell Signaling Technology), anti-E-cad (1:1000, 3195T, Cell Signaling Technology), anti-GAPDH (1:1000, abs830030, Univ).
Quantitative real-time PCR
Total cellular RNA from PDAC cells was extracted using Trizol reagent (Yeasen, China). Then 1ug RNA was reversely transcribed into cDNA by applying Hifair III 1st Strand cDNA Synthesis SuperMix (11141ES10, Yeasen, China). Real-time PCR analysis was carried out to detect DCLK1-isoforms mRNA expression levels on the 7500 Sequence Detection System (Applied Biosystems, China). During this process, the SYBR Green Premix (Invitrogen, USA) was used with GAPDH as an internal control. The sequences of primers were as follows:
GAPDH-F: 5′-AATCCCATCACCATCTTCCA-3′
GAPDH-R: 5′-TGGACTCCACGACGTACTCA-3′
DCLK1-l-F: 5′-GGAGTGGTGAAACGCCTGTAC-3′
DCLK1-l-R: 5′-GGTTCCATTAACTGAGCTGG-3′
DCLK1-S-F: 5′-ACACTAAGACTGTGTCCATGTTAGAACTC-3′
DCLK1-S-R: 5′-AAGCCTTCCTCCGACACTTCT-3′
The relative mRNA expression levels of DCLK1-isoforms were calculated by 2−ΔΔCT methods.
Wound healing assay
The empty control, DCLK1iso1 OE, and DCLK1iso4 OE cells of PANC-1 and AsPC-1 (2 × 105) were respectively seeded into 6-well culture plates and permitted to grow to approximately 60% confluence. The confluent cell layers were then scratched by a sterile 200 μl pipette tip, thereby generating a linear wound. Next, the wound areas were viewed and detected at 0 h, 6 h, 12 h and 24 h. Finally, the ImageJ was used to quantify the unmigrated area.
Transwell migration and invasion assay
Transwell assays were performed to evaluate the migration and invasion potential of cancer cells. The empty control, DCLK1iso1 OE, and DCLK1iso4 OE cells of PANC-1 and AsPC-1 (10 × 104 in 0.2 mL serum-free medium) were seeded into the upper chamber of 24-well plate (Corning, USA) and 0.6 ml DMEM comprising 10% FBS was added to the lower chamber. After incubating for 24 h at 37 °C, the migrated or invaded cells were fixed with paraformaldehyde for 10 min and dyed with crystal violet for 5 min. Three random fields of view were counted and the average was calculated. For invasion assay, matrigel (BD Bioscience, USA) was needed in the upper chamber.
PBMC separation and co-culture experiments
Peripheral blood mononuclear cell (PBMC) was isolated from healthy donors by ficoll density gradient centrifugation (GE Healthcare, USA). CD4+ and CD8+ T cells were depurated from the collected PBMC using a Human CD4+ and CD8+ T cell kit (Stem cell, Canada). CFSE was used to tag CD4+ and CD8+ T cells by the manufacturer's instructions (1:1000, Life Technologies, C34554) and CFSE-labeled human CD4+ and CD8+ T cells were co-cultured with DCLK1 iso1-overexpressing cells which were treated with DCLK1-IN-1 for 48 h before co-culture. Anti-CD3 (eBioscience, USA) and Anti-CD28 (eBioscience, USA) was used to stimulate T cell. After three days of co-culture, the proliferation of T cells was detected via measuring CFSE fluorescence using flow cytometry on a Fortessa (BD) Flow Cytometer.
The cancer genome atlas (TCGA) dataset analysis
mRNA expression data of TCGA-PDAC dataset (n = 148) were downloaded from UCSC Xena (https://xenabrowser.net/datapages/). EMT score was calculated by the mean of expression values of 200 EMT-related genes, which were acquired from Molecular Signatures Database (MsigDB, HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION) [27]. Tumor samples were then grouped by EMT score as either EMT-low (lowest 1/3 of EMT scores) or EMT-high (highest 1/3 of EMT scores). We defined the EMT-low group as “epithelial” PDAC while the EMT-high group was defined as “mesenchymal” PDAC. Likewise, we divided tumor samples into DCLK1-high expression group (highest 1/3 of DCLK1 expression levels) and DCLK1-low expression group (lowest 1/3 of DCLK1 expression levels). The methods to classify TCGA-PDAC patients into established molecular subtypes were obtained from Collisson et al. [28] and Bailey et al. [29].
We applied CIBERSORT deconvolution method to evaluate the differential abundance of tumor-infiltrating immune cells in “epithelial” and “mesenchymal” groups [30]. Additionally, the distribution of 10 key immunosuppressive cytokines/chemokines (CCL2, CCL5, CSF1, CXCL1, CXCL2, CXCL12, CXCL8, IL10, IL6, TGFB1) and 6 immune checkpoint molecules (CD274, CTLA-4, TIM3, BTLA, LAG3, VSIR) were compared between “epithelial” and “mesenchymal” groups. Moreover, the Tumor Immune Dysfunction and Exclusion (TIDE, http://tide.dfci.harvard.edu./) online tool was utilized to analyze the effect of EMT status on ICIs efficacy, and the T cell dysfunction and T cell exclusion scores were also calculated [31]. T-cell subpopulations refers to cytotoxic T lymphocytes (CTL).
Multiplexed immunofluorescence (mIF)
Two consecutive PDAC tissue microarrays containing 28 primary PDAC tumor and matching para-cancerous tissues and 2 normal pancreatic tissues were used for multiplexed Immunofluorescence staining with Opal™ 4 Multiplex reagents (Perkinelmer, USA) by the manufacturer's instructions, as in the previous studies described [32], [33], [34]. These six markers were stained on two consecutive tissue microarrays rather than one microarray mainly to avoid the cross-color. Staining was conducted as two interdependent panels with DAPI as a nuclear stain: Panel 1: DCLK1 (ab31704,1:500) opal 690, CD163(ab189915, 1:500) opal 570, E-cad (ab40772,1:500) opal 520, panel 2: DCLK1 (ab31704,1:500) opal 690, ZEB1(ab203829, 1:500) opal 570, CD8 (ab93278, 1:500) opal 520. Multiplexed fluorophore-stained slides were scanned by Shanghai Outdo Biotechnology. For all multicolor fluorescence images, each channel was obtained separately to further evaluate the gene expression levels according to mean fluorescence intensity of the whole slide by ImageJ software. When the background color or the cross color cannot be eliminated, we randomly selected three slide fields (× 100) and evaluated the mean fluorescence intensity. These scores from ImageJ were used for statistical analysis. Human tissue microarrays of PDAC (HPanA060CS03-M-111, HPanA060CS03-M-110) were purchased from Shanghai Outdo Biotechnology.
In vivo experiments
Adult femal C57BL/C (6 to 8 weeks) mice were purchased from Charles River Laboratories (Beijing, China). 1 × 106 cells diluted in 100 μl PBS were transplanted subcutaneously into the abdomen of mice (n = 6 per group) and tumors size were measured every 3–5 days after 3 days of injection. Tumor volume was calculated by the following methods: Volume = (Length × Width2)/2. After 16 days, the subcutaneous tumors were harvested and photographed.
Immunohistochemistry (IHC)
The harvested subcutaneous tumors were fixed with 4% paraformaldehyde (Solarbio, Beijing, China) and embedded in paraffin, finally sliced into 6 µm-thick sections for IHC analysis. Briefly, these tissues sections were deparaffinized in xylene for 30mins and hydrated in a descending alcohol series (100% alcohol, 95% alcohol, 80% alcohol) for 5 min respectively. After antigen retrieval with EDTA (ZSGB-BIO, China) and blocking with goat serum for one hour at room temperature, the sections were incubated with the corresponding antibody at 4 °C overnight. Followed by incubation with enhanced enzyme-labeled goat anti-mouse/rabbit IgG polymer (ZSGB-BIO, China), IHC signal was detected by 3,3′-diaminobenzidine (DAB, ZSGB-BIO, China). The antibodies used above were: anti-CD4 (1:500, ab183685, Abcam), anti-CD8 (1:500, ab217344, Abcam), anti-CD163(1:500, ab182422, Abcam), anti-E-cadherin (1:500, 3195T, Cell Signaling Technology), anti-IFN-γ (1:100, AF-585-NA, R&D), anti-DCLK1 (1:500, ab31704, Abcam)
Statistical analyses
The student's t-test was performed by Graph Pad Prism 7 and nonparametric data were analyzed by the Mann-Whitney U test and Wilcoxon using R software (version 3.6.3). P values < 0.05 were considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). The Spearman's correlations between DCLK1 expressions and EMT-TFs expressions as well as EMT score in TCGA dataset were analyzed. All experiments were independently performed for at least three times.
Results
DCLK1 may be a key contributor to EMT program activation in PDAC patients
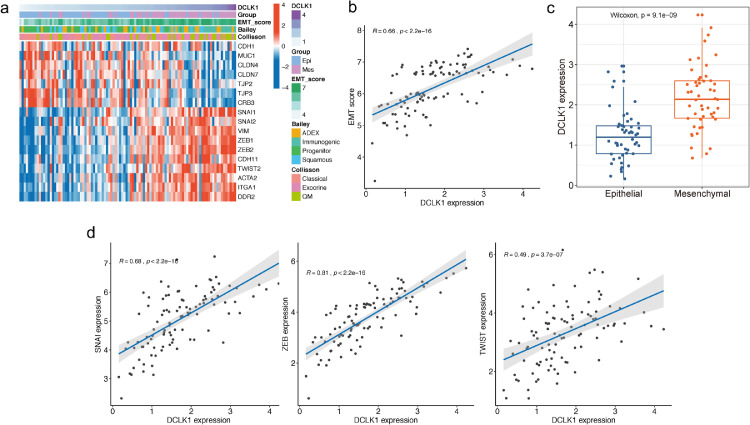
EMT is a critical factor for cancer initiation and metastasis in PDAC and considered as a hallmark of CSCs [35]. Based on the TCGA-PDAC dataset, we found that CSC marker DCLK1 was closely associated with EMT in PDAC patients as evidenced by increased mesenchymal marker (SNAIL1, Vimentin, ZEB1, ZEB2, CDH11, TWIST2, ACAT2, ITGA1, DDR2) and decreased epithelial marker (CDH1, MUC1, CLDN4, CLDN7, TJP3, TJP2, CRB3) expression (Fig. 1a). According to established classifications in PDAC, the Quasi-mesenchymal (QM- PDA) subtype characterized by high expression of mesenchyme-associated genes which provided by Collisson et al. [28] and the immunogenic subtype involved upregulated acquired immune suppression pathways which provided by Bailey et al. [29] were enriched by high DCLK1 expression (Fig. 1a). DCLK1 expressions were also positively and significantly correlated with EMT scores (Spearman's test, R = 0.66, p < 0.001, Fig. 1b). Moreover, DCLK1 labeled EMT-activated subtypes of PDAC (Fig. 1c). We next analyzed the association between DCLK1 and EMT transcription factors (EMT-TFs), the main drivers of EMT program activation. The results showed that DCLK1 was positively associated with all three families of EMT-TFs including SNAI (mean expression of Snail and Slug), ZEB (mean expression of ZEB1 and ZEB2), and Twist (mean expression of Twist1 and Twist 2) (Fig. 1d).
Fig. 1.
DCLK1 is closely accociated with EMT program according to TCGA database analysis. (a) Heatmap analysis demonstrating the correlation between DCLK1 expression levels and EMT-related genes as well as established classifications by Collisson et al., Bailey et al. (b) DCLK1 expression was positively correlated with EMT score (R = 0.66, p<0.0001). (c) DCLK1 expression was significantly higher in “mesenchymal” group than “epithelial” group. (d) Spearman correlation analysis showing the positive association between DCLK1 expression and EMT-TFs.
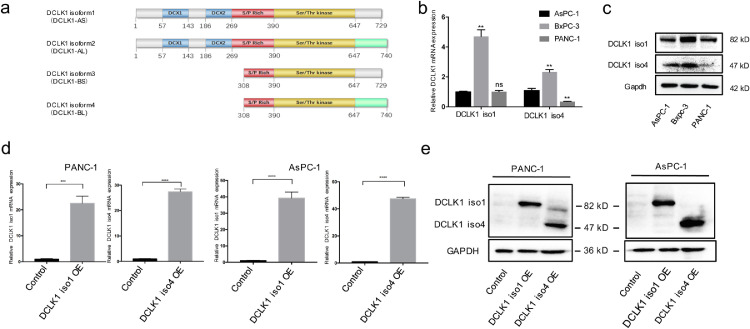
DCLK1 isoform 1 (DCLK1-AS) and isoform 4 (DCLK1-BL) activate EMT and enhance migration and invasion in PDAC cells
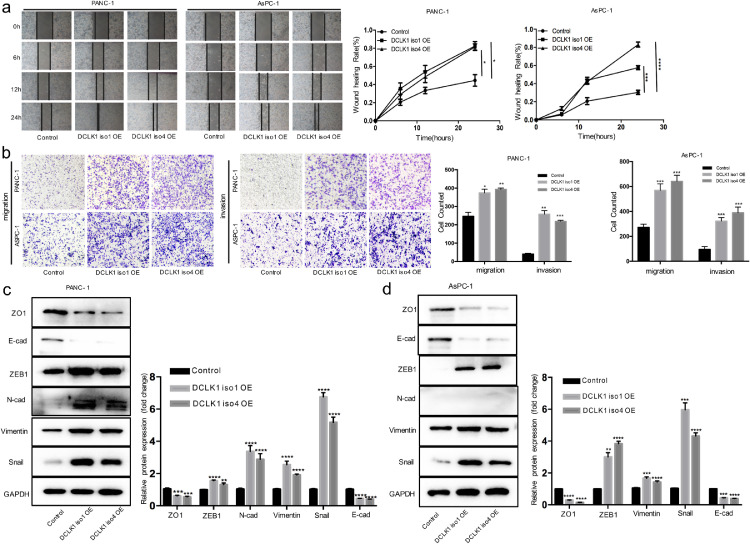
Although the existence of alternative splice variants leading to a lengthened C-terminus from the DCLK1 gene is known (Fig. 2a), most prior studies have not distinguished their individual functions. To obtain more precise knowledge of the role of DCLK1, we performed gain-of-function studies with an alpha-promoter driven (DCLK1 iso1, DCLK1-AS) and beta-promoter driven (DCLK1 iso4, DCLK1-BL) isoforms. Baseline expression levels of DCLK1 isoforms by qPCR and Western blotting indicated relative lower DCLK1-iso1 and DCLK1-iso4 expression levels in AsPC-1 and PANC-1 cells compared to BxPC-3 cells (Fig. 2b-c). Therefore, AsPC-1 and PANC-1 cell lines were selected to overexpress DCLK1 iso1 and DCLK1 iso4 for subsequent study. Overexpression of DCLK1 isoforms in AsPC-1 and PANC-1 cells were confirmed by qPCR and Western blotting analyses (Fig. 2d-e). Wound healing and transwell assays revealed that both DCLK1-iso1 and DCLK1-iso4 significantly promoted cell migratory and invasive abilities of PDAC cells (Fig. 3a-b). Moreover, EMT was activated in DCLK1-overexpressing cells as demonstrated by increased expression of mesenchymal markers and EMT-TFs including Snail, ZEB1, N—Cadherin and Vimentin concurrent with decreased expression of epithelial marker ZO1, E-cad, although N—Cadherin was not detected in AsPC-1 cells. (Fig. 3c-d). Taken together, these results reveal that both the alpha and beta-promoter driven DCLK1 isoforms are closely associated with EMT in PDAC cells.
Fig. 2.
Establishment of stable cell lines with DCLK1 isoforms overexpression in PDAC cells. (a) Schematic demonstrating differences in protein domains of DCLK1 isoforms. (b-c) Relative mRNA and protein expressions of DCLK1 isoform1 and 4 in three PDAC cell lines (AsPC-1, Bxpc-3 and PANC-1) were detected by Real-time PCR (b) and Western blotting (c). (d-e) Validation of the upregulation of DCLK1 mRNA (d) and protein (e) expressions in DCLK1 iso1 and iso4-overexpressing (DCLK1 iso1 OE, DCLK1 iso4 OE) PANC-1 and AsPC-1 cells.
Fig. 3.
Overexpression of DCLK1 isoform1 and isoform4 activates EMT in PDAC cells. (A-B) Wound-healing (a) and transwell assays (b) showing both DCLK1 isoform1 and isoform4 significantly increased the migratory and invasive abilities of PDAC cells. (c-d) Western blotting analysis of the effects of DCLK1 isoform1 and isoform 4 on EMT marker proteins in PANC-1 (c) and AsPC-1 (d) cells.
Immunosuppressive components are highly enriched in “mesenchymal” PDAC and may result in low response rates to ICIs
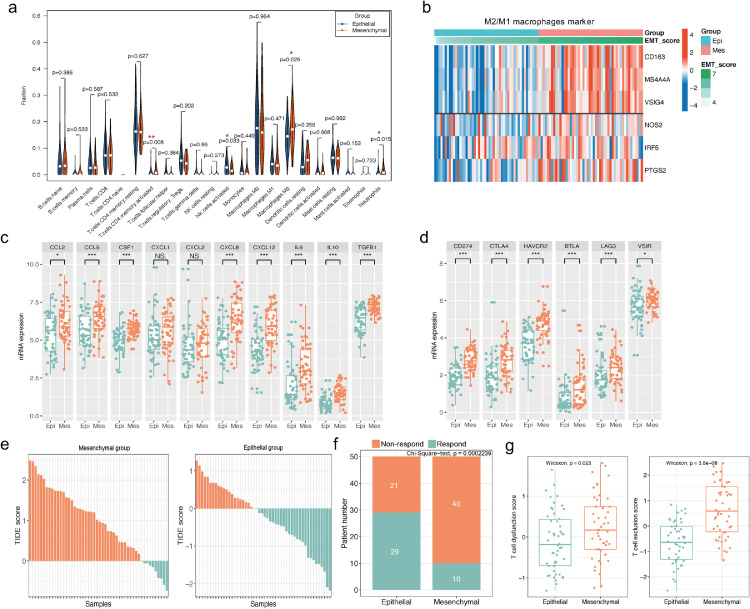
Recent reports pointed out that EMT played a pivotal role in immunosuppression in many cancers such as NSCLC, triple-negative breast cancer, and urothelial cancer [5,[9], [10], [11],36]. We firstly analyzed the relationship between EMT and immune cell infiltration by quantified the percentages of 22 types of immune cells in the PDAC TME via CIBERSORT. The results revealed that M2-macrophages were the most abundant immunosuppressive cells in the PDAC TME and represented a higher percentage in the “mesenchymal” group than in the “epithelial” group, while there was no significant difference was observed in the proportion of M1 macrophages (Fig. 4a). The next heatmap analysis also revealed a positive correlation between EMT score and M2-macrophage markers, while there was no clear correlation between M1-macrophage markers and EMT score (Fig. 4b). M2 macrophages could directly promote immunosuppression by secreted cytokines and indirectly by inducing tumor-cell–associated immune checkpoint molecules to block spontaneous antitumor immune responses [37]. Afterwards, we compared differences in macrophage-secreted immunosuppressive cytokines/chemokines and immune checkpoint molecules between “mesenchymal” and “epithelial” groups. Our results showed that the distribution of ten chemokines (CCL2, CCL5, CSF1, CXCL1, CXCL2, CXCL12, CXCL8, IL10, IL6, TGFB1) associated with macrophages recruitments and M2-macrophages polarization were increased in “mesenchymal” PDAC, although the difference was not statistically significant in CXCL1 and CXCL2. (Wilcoxon test, all p <0.01, Fig. 4c). Moreover, six immune checkpoint markers (CD274, CTLA-4, TIM3, BTLA, LAG3, VSIR) were also significantly higher in “mesenchymal” group (Wilcoxon test, all p <0.001, Fig. 4d). These data demonstrated a strong association between EMT and the formation of a tumor immunosuppressive microenvironment. Subsequently, TIDE algorithm was used to evaluate the effects of EMT status on T cell function and ICIs efficacy [31]. As shown in Fig. 4e-f, the “epithelial” group PDAC patients (58%, 29/50) may be more likely to respond to ICIs (anti-PD-1 and anti-CTLA4) compared with the “mesenchymal” group (20%, 10/50) (Chi-Square test, P <0.001). Furthermore, as shown in Fig. 4g, T cell dysfunction and T cell exclusion scores were both significantly higher in “mesenchymal” group, which might account for the limited ICIs response in “mesenchymal” PDAC patients according to TIDE algorithm (Fig. 4g).
Fig. 4.
EMT is closely associated with immune suppression in PDAC patients and might affect ICIs efficacy. (a) Wilcoxon analysis of 22 immune cell proportions estimated via CIBERSORT method between “epithelial” and “mesenchymal” groups of TCGA-PDAC patients. (b) Heatmap visualizing the correlations of EMT score with M1/2 macrophages markers, and EMT score was positively associated with M2 macrophage markers (CD163, MS4A4A, VSIG4) rather than M1 macrophage markers (NOS2, PTGS2, IPF5). (c-d) Wilcoxon analysis of the expressions of ten immunosuppressive chemo/cytokines (c) and six immune checkpoint markers (d) between “mesenchymal” and “epithelial” groups. (e-f) The “mesenchymal” group (20%) was predicted to be less likely to respond to ICIs compared to the “epithelial” group (58%) in the TCGA-PDAC dataset according to TIDE algorithm. (g) T cell dysfunction score and T cell exclusion score (calculated via TIDE algorithm) were higher in “mesenchymal” group than “epithelial” group according to TIDE.
High DCLK1 expression are associated with EMT activation and immunosuppression in PDAC patients
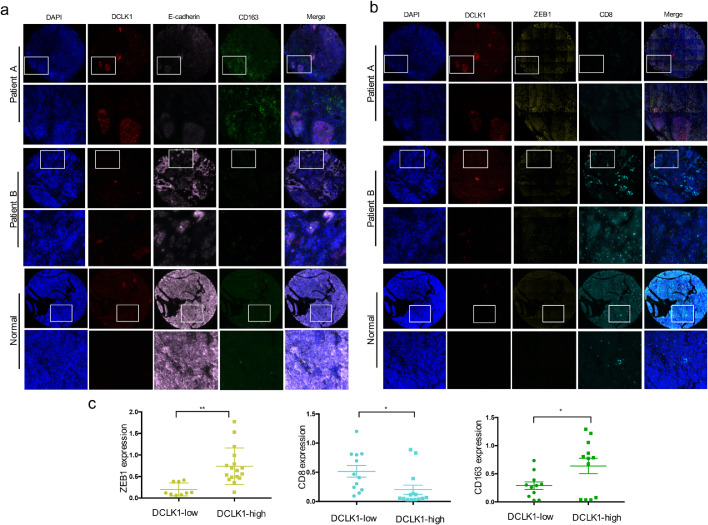
To explore the association among DCLK1, EMT program and tumor immune infiltrations, we preformed multiplexed immunofluorescence analysis of tissue microarrays including 30 early-stage PDAC samples. The representative images of multiplexed immunofluorescence stained for DCLK1, CD163, ZEB1, CD8, E-cad from two consecutive microarrays were presented in Fig. 5. The results revealed that DCLK1 was directly related to EMT program activation, with DCLK1 expression increased, the expression of E-cad was decreased and the expression of ZEB1 was increased. Besides, more M2 macrophages and less CD8+ T-cells infiltrations could also be found in patients’ samples with higher DCLK1 expression. The roughly converse phenomenon (low ZEB1 expression, high E-cad expression, low density of M2 macrophages and abundant CD8+ T cell) were presented in patients with low DCLK1 expression and normal pancreatic tissues (Fig. 5a-b). Applying the median DCLK1-expression level as the cutoff value, we classified the tumors into high- and low-DCLK1 groups. We observed that the expression scores of CD163 and ZEB1 in the high DCLK1 group were significantly higher than that in the low DCLK1 group. Conversely, the expression scores of E-cad and CD8 were significantly lower in the high DCLK1 group than that in the low DCLK1 group (Fig. 5c). It's worth noting that almost all patients’ samples used in our tissue microarrays were in the early-stage, which suggested that the tumor- promoting role of EMT may occurred in the formation of tumor, which was firstly proved by Ben Z. Stanger in 2012 [38]. And the cross-talk between EMT and immunosuppression has been described comprehensively in recent reviews [8,39]. To some extent, our multiplexed immunofluorescence results verified the close association between EMT and immunosuppressive environment.
Fig. 5.
Multiplexed immunofluorescence displays the close association among DCLK, EMT activation and immunosuppression. (a-b). Multiplexed immunofluorescence analysis showing that high DCLK1 expression is often accompanied by increased CD163 and ZEB1 expression levels as well as decreased CD8 and E-cad expression levels. (c) Immunofluorescence quantitative analysis results showing ZEB1 and CD163 expression levels are higher in DCLK1-high group, while CD8 expression level was lower in DCLK1-high group than DCLK1-low group.
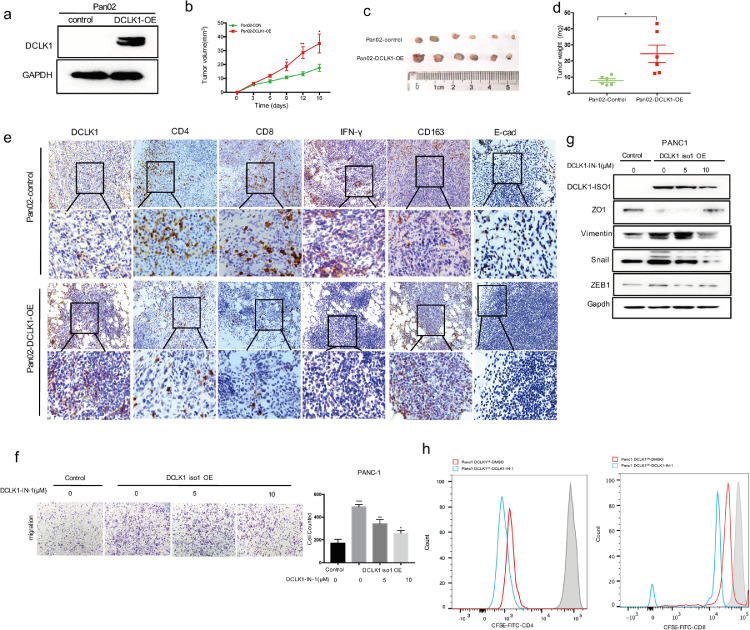
DCLK1 promotes tumor growth and reduces T cells infiltration in vivo
According to our previous findings, we supposed that DCLK1 might also affect tumor-infiltrating immune cells. So, we constructed the mouse DCLK1 gene overexpression vector which was then stably transferred into Pan02 cells (Fig. 6a). DCLK1-overexpressing Pan02 cells and the corresponding control cells were subcutaneously injected into the abdomen of mice (n = 6). 16 days after inoculation, the tumor growth curve achieved by regularly measuring tumor size showed that DCLK1 significantly promoted the tumor growth when compared to the control group (Fig. 6b). And the average weight of tumor tissues from DCLK1-overexpressing group was also higher (Fig. 6c-d). Next, the tumor-infiltrating immune cells were detected by IHC using the obtained subcutaneous tumors. The results demonstrated that the tumors with high DCLK1 expression were often accompanied with E-cad loss and decreases in CD4+, CD8+ T cells, especially CD8+ cells, as well as the decrease of IFN-γ secretion, an important effector cytokine of CD8+ cells (Fig. 6e). At the same time, M2 macrophages were also increased in DCLK1-overexpressing tumors (Fig. 6e). In vivo results further revealed the importance of DCLK1 in EMT activation and formation of immunosuppressive environment. TCGA-PDAC dataset analysis also supported that DCLK1 expression had strongly positive correlations with expression of M2 macrophages marker (Supplementary Fig.1a). Immune checkpoint molecules and immunosuppressive chemokines/cytokines were also elevated in high DCLK1 expressing patients (Supplementary Fig. 1b-c). The TIDE analysis revealed that T cell dysfunction score and T cell exclusion score were significantly higher in patients with high DCLK1 expression (Supplementary Fig.1d).
Fig. 6.
DCLK1 kinase inhibition can block EMT program and restore T-cells activity.(a) Western blotting verifying the overexpression of DCLK1 in Pan02 cells. (b) Tumor growth curve showed that the tumors derived from DCLK1-overexpressing group grew much more quickly than those from the control group (n = 6). (c-d) 16 days after subcutaneous injections, tumors were excised to weighed (c) and photographed (d). Results showed that DCLK1 overexpression led to increased tumor weights. (e) Immunohistological staining of harvested tumor tissues and results demonstrated that the infiltration of CD4+, CD8+ T cells, IFN-γ secretion and E-cad expression levels were decreased while M2 macrophages were increased in DCLK1-overexpressing group. (f-g) Transwell assay (f) and western blot (g) analysis demonstrated that DCLK1 inhibitor DCLK1-IN-1 could attenuate DCLK1-induced EMT in PDAC cells. (h) DCLK1-IN-1 restored CD4+ and CD8+ T-cell activity as measured by CFSE-based T-cell proliferation assays.
DCLK1 kinase inhibition blocks EMT program and restores T-cell activity
These findings hinted that DCLK1 might be an effective target to improve the efficacy of immunotherapies in PDAC. We used DCLK1 inhibitor (DCLK1-IN-1) on DCLK1-overexpressing PDAC cells. The results demonstrated that EMT could be blocked by DCLK1 inhibitor, as evidenced by decreased migratory and invasive abilities (Fig. 6f) as well as decreased expression levels of EMT-TFs (ZEB1, Snail) and mesenchymal marker Vimentin (Fig. 6g). Finally, in order to assess the effect of DCLK1 inhibitor on T-cell activity, we conducted an experiment of co-culture of CD4+ and CD8+ T cells with tumor cells. The results revealed that DCLK1-IN-1 was able to effectively restore T cell activity (Fig. 6h).
Discussion
Numerous clinical trials have intended to find effective immunotherapy targets in PDAC, but strategies including ICIs and adoptive cellular therapies have largely failed due to a highly immunosuppressive microenvironment. Immunosuppressive cells including myeloid-derived suppressive cells (MDSCs), tumor-associated fibroblasts, tumor-associated macrophages (TAMs) appear and function in early PanIN lesions [40]. Therefore, a better understanding of critical factors driving immunosuppression in PDAC could help in identifying more effective therapies.
CSCs are key drivers of tumorigenesis and have gained attention for modulating the tumor immune microenvironment. Recent studies have demonstrated that CSCs promoted the recruitment of TAMs and MDSCs [41], [42], [43], which may in turn inhibit the anti-tumor immune response by inducing an immunosuppressive microenvironment. As a specific CSC marker in PDAC, the role of DCLK1 in interactions between tumor cells and TME components requires further investigation. According to our bioinformatics analysis, DCLK1 is closely associated with EMT activation in PDAC, which concurs with previous findings [44], [45], [46], [47]. To verify these results experimentally, we overexpressed both DCLK1-long (DCLK1-iso1, ∼80–82 KDa) and DCLK1-short (DCLK1-iso4, ∼45–50 KDa) isoforms in PDAC cell lines and detected changes in EMT-associated cell functions and molecular markers. We found that both DCLK1 long and short isoforms effectively activated EMT in PDAC cells as demonstrated by the enhanced migration/invasion abilities as well as the acquisition of mesenchymal molecular characteristics. These data suggest that DCLK1 acts as both a marker and driver of EMT activation.
A recent study reported that EMT occurred during early stages of precancerous lesions before tumor formation in PDAC [38]. However, whether EMT activation relates to the immunosuppressive nature of PDAC, as it does in other cancers, remains unknown. Lou et al. found that EMT was closely associated with multiple immune checkpoint molecules and could be identified as a predictive biomarker for the efficacy of ICIs in NSCLC, and this finding was confirmed by a retrospective study analyzing the transcriptional profiles of 52 advanced NSCLC patients treated with anti-PD1/PD-L1 therapy [5,48]. In melanoma, Snail, one of the most important EMT-TFs, promotes immunotherapy resistance by induction of Treg and DC [49]. It has also been shown that EMT-related gene expression is associated with T cell infiltration and the efficacy of ICIs in urothelial cancer [11]. Similar results have been reported in other cancers including ovarian cancer, head and neck squamous cell carcinoma, and esophageal squamous cell carcinoma [50], [51], [52]. To investigate the role of EMT in PDAC immunosuppression, we divided PDAC patients into epithelial and mesenchymal groups according to the expression of EMT-related genes obtained from The Molecular Signatures Database. The CIBERSORT algorithm was then used to evaluate the landscape of TME immune cells in both groups. The relative proportion of M2 macrophages, which could inhibit anti-tumor immunity by inducing T-cell suppression, were significantly higher in “mesenchymal” group. In addition, the expressions of immunosuppressive cytokines and immune checkpoint molecules were also significantly higher in “mesenchymal” group. These results strongly suggest that an immunosuppressive microenvironment induced by EMT might be a potential limitation to immunotherapy. To further explore this concept, we applied TIDE online algorithm for the prediction of immunotherapy responses in TCGA-PDAC patients and found that PDAC patients in the “mesenchymal” group were less likely to respond to immunotherapy compared to the “epithelial” group.
The Pan02 syngeneic mouse model further verified the above results in vivo as demonstrated by E-cad loss accompanied by an immunosuppressive environment in DCLK1-overexpressing tumors. EMT-induced immunosuppression promotes tumor progression yet also brings a new therapeutic direction of treatment. DCLK1 is a critical driver in the EMT program, and we hypothesized that EMT program could be blocked by DCLK1 kinase inhibition in patients with high DCLK1 expression. Our analysis showed that DCLK1 was positively correlated with immune cells, especially tumor associated macrophages, and that high DCLK1 expression can inactivate T cells. Compounds that can inhibit DCLK1’s kinase activity have been studied in several cancers [53], [54], [55]. Among them, DCLK1-IN-1 is the first highly selective inhibitor of this target. Using this inhibitor, we found that EMT program was blocked and T-cell activity was restored in DCLK1-overexpressing PDAC cells. These data suggest that DCLK1 is a promising target to inhibit EMT-associated immunosuppression. To verify the association among DCLK1, EMT and immune infiltrate in patients, we applied multiplexed immunofluorescence analysis of DCLK1, ZEB1, E-cad, CD8, CD163 with PDAC's tissue microarrays. The results showed that patients with high DCLK1 expressions were often accompanied by EMT program activation and increasing immunosuppressive cells infiltrations. However, we couldn't yet capture these events in an autochthonous model of PDAC due to the limitations of our experimental conditions.
In conclusion, our results suggest that DCLK1 is a critical activator in the EMT program, which may result in an immunosuppressive microenvironment and render cancer cells non-responsive to immunotherapies. DCLK1 inhibitors might be of great utility for application in conjunction with immunotherapies to elevate their clinical efficacy.
Availability of data and materials
The datasets supporting the findings of this study are available in the The Cancer Genome Atlas (TCGA) (https://gdc.xenahubs.net). And the original data presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
Funding
This work was supported by Beijing Natural Science Foundation (7202051 toYang Ge), and National Natural Science Foundation of China (81802738 to Yang Ge).
CRediT authorship contribution statement
Yang Ge: Writing – review & editing, Formal analysis. Heshu Liu: Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Methodology, Validation. Yuanyuan Zhang: Writing – original draft, Methodology, Validation. Jian Liu: Writing – review & editing, Formal analysis. Rui Yan: Writing – review & editing, Data curation, Validation. Zeru Xiao: Writing – review & editing, Data curation, Validation. Xiaona Fan: Writing – review & editing, Data curation, Validation. Xuying Huang: Writing – review & editing, Data curation, Validation. Guangyu An: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101317.
Contributor Information
Yang Ge, Email: interna-1@163.com.
Guangyu An, Email: agybjcy@163.com.
Appendix. Supplementary materials
References
- 1.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., et al. Pancreatic cancer. Nat. Rev. Dis. Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. Epub 2016/05/10.PubMed PMID: 27158978. [DOI] [PubMed] [Google Scholar]
- 2.Wolfgang C.L., Herman J.M., Laheru D.A., Klein A.P., Erdek M.A., Fishman E.K., et al. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013;63(5):318–348. doi: 10.3322/caac.21190. Epub 2013/07/17PubMed PMID: 23856911; PubMed Central PMCID: PMCPMC3769458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho W.J., Jaffee E.M., Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 2020;17(9):527–540. doi: 10.1038/s41571-020-0363-5. Epub 2020/05/14PubMed PMID: 32398706; PubMed Central PMCID: PMCPMC7442729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu D.S., Wang H.J., Tai S.K., Chou C.H., Hsieh C.H., Chiu P.H., et al. Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell. 2014;26(4):534–548. doi: 10.1016/j.ccell.2014.09.002. Epub 2014/10/15PubMed PMID: 25314079. [DOI] [PubMed] [Google Scholar]
- 5.Lou Y., Diao L., Cuentas E.R., Denning W.L., Chen L., Fan Y.H., et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016;22(14):3630–3642. doi: 10.1158/1078-0432.ccr-15-1434. Epub 2016/02/07PubMed PMID: 26851185; PubMed Central PMCID: PMCPMC4947453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae Y.K., Chang S., Ko T., Anker J., Agte S., Iams W., et al. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC) Sci. Rep. 2018;8(1):2918. doi: 10.1038/s41598-018-21061-1. Epub 2018/02/15PubMed PMID: 29440769; PubMed Central PMCID: PMCPMC5811447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chouaib S., Janji B., Tittarelli A., Eggermont A., Thiery J.P. Tumor plasticity interferes with anti-tumor immunity. Crit. Rev. Immunol. 2014;34(2):91–102. doi: 10.1615/critrevimmunol.2014010183. Epub 2014/06/19PubMed PMID: 24940910. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y., Zhan H. Communication between EMT and PD-L1 signaling: new insights into tumor immune evasion. Cancer Lett. 2020;468:72–81. doi: 10.1016/j.canlet.2019.10.013. Epub 2019/10/13PubMed PMID: 31605776. [DOI] [PubMed] [Google Scholar]
- 9.Low-Marchelli J.M., Ardi V.C., Vizcarra E.A., van Rooijen N., Quigley J.P., Yang J. Twist1 induces CCL2 and recruits macrophages to promote angiogenesis. Cancer Res. 2013;73(2):662–671. doi: 10.1158/0008-5472.can-12-0653. Epub 2013/01/19PubMed PMID: 23329645; PubMed Central PMCID: PMCPMC3566985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soundararajan R., Fradette J.J., Konen J.M., Moulder S., Zhang X., Gibbons D.L., et al. Targeting the interplay between epithelial-to-mesenchymal-transition and the immune system for effective immunotherapy. Cancers. 2019;11(5) doi: 10.3390/cancers11050714. (Basel)Epub 2019/05/30PubMed PMID: 31137625; PubMed Central PMCID: PMCPMC6562947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Saci A., Szabo P.M., Chasalow S.D., Castillo-Martin M., Domingo-Domenech J., et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat. Commun. 2018;9(1):3503. doi: 10.1038/s41467-018-05992-x. Epub 2018/08/31PubMed PMID: 30158554; PubMed Central PMCID: PMCPMC6115401 Bristol-Myers Squibb. A.S., P.M.S., S.D.C., and A.A. are employees of Bristol-Myers Squibb. The remaining authors declare no competing interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dongre A., Rashidian M., Reinhardt F., Bagnato A., Keckesova Z., Ploegh H.L., et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77(15):3982–3989. doi: 10.1158/0008-5472.can-16-3292. Epub 2017/04/22PubMed PMID: 28428275; PubMed Central PMCID: PMCPMC5541771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark C.E., Hingorani S.R., Mick R., Combs C., Tuveson D.A., Vonderheide R.H. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67(19):9518–9527. doi: 10.1158/0008-5472.can-07-0175. Epub 2007/10/03PubMed PMID: 17909062. [DOI] [PubMed] [Google Scholar]
- 14.Lin P.T., Gleeson J.G., Corbo J.C., Flanagan L., Walsh C.A. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J. Neurosci. 2000;20(24):9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. PubMed PMID: 11124993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi Y., Seno H., Fukuoka A., Ueo T., Yamaga Y., Maruno T., et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 2013;45(1):98–103. doi: 10.1038/ng.2481. PubMed PMID: 23202126. [DOI] [PubMed] [Google Scholar]
- 16.Bailey J.M., Alsina J., Rasheed Z.A., McAllister F.M., Fu Y.Y., Plentz R., et al. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146(1):245–256. doi: 10.1053/j.gastro.2013.09.050. Epub 2013/10/08PubMed PMID: 24096005; PubMed Central PMCID: PMCPMC3910427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sureban S.M., May R., Qu D., Weygant N., Chandrakesan P., Ali N., et al. DCLK1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms in pancreatic cancer. PLoS ONE. 2013;8(9):e73940. doi: 10.1371/journal.pone.0073940. PubMed PMID: 24040120; PubMed Central PMCID: PMCPMC3767662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Bellows C.F. Doublecortin-like kinase 1 exhibits cancer stem cell-like characteristics in a human colon cancer cell line. Chin. J. Cancer Res. 2013;25(2):134–142. doi: 10.3978/j.issn.1000-9604.2013.03.02. PubMed PMID: 23592893; PubMed Central PMCID: PMC3626979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrakesan P., Yao J., Qu D., May R., Weygant N., Ge Y., et al. Dclk1, a tumor stem cell marker, regulates pro-survival signaling and self-renewal of intestinal tumor cells. Mol. Cancer. 2017;16(1):30. doi: 10.1186/s12943-017-0594-y. PubMed PMID: 28148261; PubMed Central PMCID: PMC5286867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sureban S.M., May R., Mondalek F.G., Qu D., Ponnurangam S., Pantazis P., et al. Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a notch-1 dependent mechanism. J. Nanobiotechnol. 2011;9:40. doi: 10.1186/1477-3155-9-40. PubMed PMID: 21929751; PubMed Central PMCID: PMCPMC3200989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sureban S.M., May R., Lightfoot S.A., Hoskins A.B., Lerner M., Brackett D.J., et al. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 2011;71(6):2328–2338. doi: 10.1158/0008-5472.CAN-10-2738. PubMed PMID: 21285251; PubMed Central PMCID: PMCPMC3072762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sureban S.M., Madhoun M.F., May R., Qu D., Ali N., Fazili J., et al. Plasma DCLK1 is a marker of hepatocellular carcinoma (HCC): targeting DCLK1 prevents HCC tumor xenograft growth via a microRNA-dependent mechanism. Oncotarget. 2015;6(35):37200–37215. doi: 10.18632/oncotarget.5808. PubMed PMID: 26468984; PubMed Central PMCID: PMC4741924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weygant N., Qu D., May R., Tierney R.M., Berry W.L., Zhao L., et al. DCLK1 is a broadly dysregulated target against epithelial-mesenchymal transition, focal adhesion, and stemness in clear cell renal carcinoma. Oncotarget. 2015;6(4):2193–2205. doi: 10.18632/oncotarget.3059. PubMed PMID: 25605241; PubMed Central PMCID: PMCPMC4385845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westphalen C.B., Asfaha S., Hayakawa Y., Takemoto Y., Lukin D.J., Nuber A.H., et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest. 2014;124(3):1283–1295. doi: 10.1172/jci73434. Epub 2014/02/04PubMed PMID: 24487592; PubMed Central PMCID: PMCPMC3934168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westphalen C.B., Takemoto Y., Tanaka T., Macchini M., Jiang Z., Renz B.W., et al. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell. 2016;18(4):441–455. doi: 10.1016/j.stem.2016.03.016. Epub 2016/04/09PubMed PMID: 27058937; PubMed Central PMCID: PMCPMC4826481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen C.B., Houchen C.W., Ali N. APSA awardee submission: tumor/cancer stem cell marker doublecortin-like kinase 1 in liver diseases. Exp. Biol. Med. 2017;242(3):242–249. doi: 10.1177/1535370216672746. (Maywood)PubMed PMID: 27694285; PubMed Central PMCID: PMCPMC5384496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. Epub 2016/01/16PubMed PMID: 26771021; PubMed Central PMCID: PMCPMC4707969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. Epub 2011/04/05PubMed PMID: 21460848; PubMed Central PMCID: PMCPMC3755490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. Epub 2016/02/26PubMed PMID: 26909576. [DOI] [PubMed] [Google Scholar]
- 30.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. Epub 2015/03/31PubMed PMID: 25822800; PubMed Central PMCID: PMCPMC4739640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. Epub 2018/08/22PubMed PMID: 30127393; PubMed Central PMCID: PMCPMC6487502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim J.C.T., Yeong J.P.S., Lim C.J., Ong C.C.H., Wong S.C., Chew V.S.P., et al. An automated staining protocol for seven-colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Pathology. 2018;50(3):333–341. doi: 10.1016/j.pathol.2017.11.087. Epub 2018/02/13PubMed PMID: 29429740. [DOI] [PubMed] [Google Scholar]
- 33.Stack E.C., Wang C., Roman K.A., Hoyt C.C. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. doi: 10.1016/j.ymeth.2014.08.016. (San Diego, Calif)Epub 2014/09/23PubMed PMID: 25242720. [DOI] [PubMed] [Google Scholar]
- 34.Yeong J., Lim J.C.T., Lee B., Li H., Ong C.C.H., Thike A.A., et al. Prognostic value of CD8 + PD-1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J. Immunother. Cancer. 2019;7(1):34. doi: 10.1186/s40425-019-0499-y. Epub 2019/02/08PubMed PMID: 30728081; PubMed Central PMCID: PMCPMC6366051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibue T., Weinberg R.A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. Epub 2017/04/12PubMed PMID: 28397828; PubMed Central PMCID: PMCPMC5720366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.H., Byers L.A., et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. Epub 2014/10/29PubMed PMID: 25348003; PubMed Central PMCID: PMCPMC4212319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Velez-Delgado A., Mathew E., Li D., Mendez F.M., Flannagan K., et al. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut. 2017;66(1):124–136. doi: 10.1136/gutjnl-2016-312078. Epub 2016/07/13PubMed PMID: 27402485; PubMed Central PMCID: PMCPMC5256390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. Epub 2012/01/24PubMed PMID: 22265420; PubMed Central PMCID: PMCPMC3266542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taki M., Abiko K., Ukita M., Murakami R., Yamanoi K., Yamaguchi K., et al. Tumor immune microenvironment during epithelial-mesenchymal transition. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021 doi: 10.1158/1078-0432.ccr-20-4459. Epub 2021/04/09PubMed PMID: 33827891. [DOI] [PubMed] [Google Scholar]
- 40.Liou G.Y., Bastea L., Fleming A., Döppler H., Edenfield B.H., Dawson D.W., et al. The presence of interleukin-13 at pancreatic ADM/PanIN lesions alters macrophage populations and mediates pancreatic tumorigenesis. Cell Rep. 2017;19(7):1322–1333. doi: 10.1016/j.celrep.2017.04.052. Epub 2017/05/18PubMed PMID: 28514653; PubMed Central PMCID: PMCPMC5510483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maccalli C., Volontè A., Cimminiello C., Parmiani G. Immunology of cancer stem cells in solid tumours. A review. Eur. J. Cancer. 2014;50(3):649–655. doi: 10.1016/j.ejca.2013.11.014. (Oxford, England: 1990)Epub 2013/12/18PubMed PMID: 24333096. [DOI] [PubMed] [Google Scholar]
- 42.Volonté A., Di Tomaso T., Spinelli M., Todaro M., Sanvito F., Albarello L., et al. Cancer-initiating cells from colorectal cancer patients escape from T cell-mediated immunosurveillance in vitro through membrane-bound IL-4. J. Immunol. 2014;192(1):523–532. doi: 10.4049/jimmunol.1301342. (Baltimore, Md: 1950)Epub 2013/11/28PubMed PMID: 24277698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maccalli C., Parmiani G., Ferrone S. Immunomodulating and immunoresistance properties of cancer-initiating cells: implications for the clinical success of immunotherapy. Immunol. Invest. 2017;46(3):221–238. doi: 10.1080/08820139.2017.1280051. Epub 2017/03/14PubMed PMID: 28287848. [DOI] [PubMed] [Google Scholar]
- 44.Liu H., Wen T., Zhou Y., Fan X., Du T., Gao T., et al. DCLK1 plays a metastatic-promoting role in human breast cancer cells. Biomed Res. Int. 2019;2019 doi: 10.1155/2019/1061979. Epub 2019/06/22PubMed PMID: 31223610; PubMed Central PMCID: PMCPMC6541964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makino S., Takahashi H., Okuzaki D., Miyoshi N., Haraguchi N., Hata T., et al. DCLK1 integrates induction of TRIB3, EMT, drug resistance and poor prognosis in colorectal cancer. Carcinogenesis. 2020;41(3):303–312. doi: 10.1093/carcin/bgz157. Epub 2019/09/29PubMed PMID: 31562741. [DOI] [PubMed] [Google Scholar]
- 46.Liu W., Wang S., Sun Q., Yang Z., Liu M., Tang H. DCLK1 promotes epithelial-mesenchymal transition via the PI3K/Akt/NF-κB pathway in colorectal cancer. Int. J. Cancer. 2018;142(10):2068–2079. doi: 10.1002/ijc.31232. Epub 2017/12/27PubMed PMID: 29277893. [DOI] [PubMed] [Google Scholar]
- 47.Ikezono Y., Koga H., Akiba J., Abe M., Yoshida T., Wada F., et al. Pancreatic neuroendocrine tumors and emt behavior are driven by the CSC marker DCLK1. Mol. Cancer Res. MCR. 2017;15(6):744–752. doi: 10.1158/1541-7786.mcr-16-0285. Epub 2017/02/10PubMed PMID: 28179411. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J.C., Hwang W.T., Davis C., Deshpande C., Jeffries S., Rajpurohit Y., et al. Gene signatures of tumor inflammation and epithelial-to-mesenchymal transition (EMT) predict responses to immune checkpoint blockade in lung cancer with high accuracy. Lung Cancer. 2020;139:1–8. doi: 10.1016/j.lungcan.2019.10.012. Epub 2019/11/05PubMed PMID: 31683225; PubMed Central PMCID: PMCPMC7176049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kudo-Saito C., Shirako H., Takeuchi T., Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195–206. doi: 10.1016/j.ccr.2009.01.023. Epub 2009/03/03PubMed PMID: 19249678. [DOI] [PubMed] [Google Scholar]
- 50.Taki M., Abiko K., Baba T., Hamanishi J., Yamaguchi K., Murakami R., et al. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat. Commun. 2018;9(1):1685. doi: 10.1038/s41467-018-03966-7. Epub 2018/04/29PubMed PMID: 29703902; PubMed Central PMCID: PMCPMC5923228 of DSK project, Medical Innovation Center, Kyoto University sponsored by Sumitomo Dainippon Pharma Co Ltd. The remaining authors declare no competing interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ock C.Y., Kim S., Keam B., Kim M., Kim T.M., Kim J.H., et al. PD-L1 expression is associated with epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Oncotarget. 2016;7(13):15901–15914. doi: 10.18632/oncotarget.7431. Epub 2016/02/20PubMed PMID: 26893364; PubMed Central PMCID: PMCPMC4941285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsutsumi S., Saeki H., Nakashima Y., Ito S., Oki E., Morita M., et al. Programmed death-ligand 1 expression at tumor invasive front is associated with epithelial-mesenchymal transition and poor prognosis in esophageal squamous cell carcinoma. Cancer Sci. 2017;108(6):1119–1127. doi: 10.1111/cas.13237. Epub 2017/03/16PubMed PMID: 28294486; PubMed Central PMCID: PMCPMC5480087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weygant N., Qu D., Berry W.L., May R., Chandrakesan P., Owen D.B., et al. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer. 2014;13:103. doi: 10.1186/1476-4598-13-103. Epub 2014/06/03PubMed PMID: 24885928; PubMed Central PMCID: PMCPMC4030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sureban S.M., May R., Weygant N., Qu D., Chandrakesan P., Bannerman-Menson E., et al. XMD8-92 inhibits pancreatic tumor xenograft growth via a DCLK1-dependent mechanism. Cancer Lett. 2014;351(1):151–161. doi: 10.1016/j.canlet.2014.05.011. Epub 2014/06/01PubMed PMID: 24880079. [DOI] [PubMed] [Google Scholar]
- 55.Ferguson F.M., Nabet B., Raghavan S., Liu Y., Leggett A.L., Kuljanin M., et al. Discovery of a selective inhibitor of doublecortin like kinase 1. Nat. Chem. Biol. 2020;16(6):635–643. doi: 10.1038/s41589-020-0506-0. Epub 2020/04/07PubMed PMID: 32251410; PubMed Central PMCID: PMCPMC7246176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the findings of this study are available in the The Cancer Genome Atlas (TCGA) (https://gdc.xenahubs.net). And the original data presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.