Abstract
Methicillin-resistant staphylococci (MRS) are one of the most common causes of nosocomial infections and bacteremia. Standard bacterial identification and susceptibility testing frequently require as long as 72 h to report results, and there may be difficulty in rapidly and accurately identifying methicillin resistance. The use of the PCR is a rapid and simple process for the amplification of target DNA sequences, which can be used to identify and test bacteria for antimicrobial resistance. However, many sample preparation methods are unsuitable for PCR utilization in the clinical laboratory because they either are not cost-effective, take too long to perform, or do not provide a satisfactory DNA template for PCR. Our goal was to provide same-day results to facilitate rapid diagnosis and therapy. In this report, we describe a rapid method for extraction of bacterial DNA directly from blood culture bottles that gave quality DNA for PCR in as little as 20 min. We compared this extraction method to the standard QIAGEN method for turnaround time (TAT), cost, purity, and use of template in PCR. Specific identification of MRS was determined using intragenic primer sets for bacterial and Staphylococcus 16S rRNA and mecA gene sequences. The PCR primer sets were validated with 416 isolates of staphylococci, including methicillin-resistant Staphylococcus aureus (n = 106), methicillin-sensitive S. aureus (n = 134), and coagulase-negative Staphylococcus (n = 176). The total supply cost of our extraction method and PCR was $2.15 per sample with a result TAT of less than 4 h. The methods described herein represent a rapid and accurate DNA extraction and PCR-based identification system, which makes the system an ideal candidate for use under austere field conditions and one that may have utility in the clinical laboratory.
Since the introduction of methicillin, methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci (MRCoNS) have spread worldwide and have become an important cause of nosocomial infections (10, 11, 21). Nationally, 5 to 10% of all hospitalized patients in a given facility will be colonized or infected with MRSA (24). There has been a steady increase in the prevalence of MRSA isolated in U.S. hospitals over the years, and now approximately 25% of all S. aureus nosocomial isolates are methicillin resistant (8). Also, as many as 60 to 90% of all clinical CoNS isolates are resistant to methicillin (2, 42). These increased rates of methicillin resistance worldwide make it crucial for clinical laboratories to have rapid, accurate, and simple methods for the identification and confirmation of MRSA and MRCoNS.
MRSA produce a novel penicillin binding protein (PBP) (PBP 2a or PBP 2′) in addition to the usual PBPs (18, 46). This is the primary mechanism of staphylococcal methicillin resistance and is referred to as intrinsic resistance (38). PBP 2a has a low affinity for β-lactam antibiotics and is thought to function in their presence to confer resistance to the bacteria. Resistance can also be heterogeneous, because factors other than PBP 2a influence the degree to which it is expressed (6, 17, 28). In addition, bacterial strains with low-level resistance to methicillin may produce large amounts of β-lactamase and therefore not exhibit intrinsic resistance. MRCoNS also become resistant by acquisition of the PBP 2a-encoding gene, mecA (9, 44). mecA is a chromosomally derived gene that has been cloned and sequenced (5, 25, 39). It has a very high level of homology in MRSA and MRCoNS and is absent from methicillin-susceptible staphylococcal isolates (32, 40). Additionally, the mecA gene is virtually identical in all staphylococcal strains and thus is a useful molecular marker of methicillin resistance (3, 48).
Automated systems have excellent specificity but often lack sensitivity in detecting methicillin-resistant staphylococci (MRS), particularly coagulase-negative strains (8). Currently, methods for identification of methicillin resistance in isolates from clinical laboratories include agar dilution, disk diffusion, and broth dilution. These methods detect phenotypic expression rather than the presence of the mecA gene, and their results depend on numerous variables, specially requiring isolated colonies from an overnight subculture on solid agar from the positive blood culture, sputum, or urine sample (8). For CoNS, some laboratories have reported a false-susceptibility rate of 16% using the VITEK system when compared to disk diffusion testing (7). Furthermore, when antibiotic susceptibility testing of S. aureus clinical isolates (blood, sputum, or urine) is performed in conjunction with the oxacillin screening plate, it can take up to 3 days for confirmatory results from different clinical samples.
The use of PCR for the detection of mecA has been previously described (4, 7, 15, 19, 20, 23, 27, 37, 41, 45, 47, 48, 51); many different techniques have been utilized to generate template DNA from various sources for PCR (M. Dion, C. Menard, F. J. Picard, L. Gernier, P. H. Roy, M. Ouellette, and M. G. Bergeron, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. C-481, 1999). In the clinical laboratory, additional steps may be required for cell lysis or removal of potential inhibitors from some clinical specimens. These steps can add to the cost per test, increase sample processing time, and eliminate the formation of a general extraction procedure that can be easily and rapidly applied to identify a variety of microorganisms in a variety of clinical samples (blood, sputum, or urine). Many procedures are too time-consuming to allow easy assimilation into the clinical microbiology laboratory. Also, these PCR methods require a pure sample from a subculture, delaying result turnaround time (TAT) and necessitating the use of special media.
This paper describes a simple and rapid extraction method for DNA from gram-positive cocci as well as a simple PCR procedure for the direct identification of MRS. The entire procedure can be performed with results available within 4 h following the detection of positive blood culture bottles or urine samples. In this report, we compared our method for TAT, cost, and use of template in PCR to the QIAGEN (Santa Clarita, Calif.) DNA purification kit, which provides a reliable method to purify DNA for PCR (33). The total supply cost (including PCR amplification of the target) was as little as $2.15 per sample. A complete clinical sample processing procedure is also described. The purpose of this study was to determine whether direct identification of MRS from blood cultures, utilizing PCR, would agree with standard identification methods. A cost analysis was done to show that PCR detection optimized for use in the clinical laboratory could be cost-effective compared to standard methods.
MATERIALS AND METHODS
Bacterial strains.
To develop the rapid DNA extraction method, five S. aureus strains were used: methicillin-resistant clinical isolates 450M, N315, Col, and BMSI as well as methicillin-sensitive clinical isolate RN4220 (31). To test the limiting dilution of the new extraction method, an Escherichia coli isolate was used. To test the specificity and sensitivity of our PCR primer sets, a total of 416 clinical isolates of Staphylococcus were studied; all were obtained from Kaiser Permanente Reference Laboratories (Berkeley, Calif.). Additionally, 10 clinical isolates, including Enterococcus faecalis, Streptococcus pyogenes, Pseudomonas aeruginosa, and E. coli, obtained from the clinical laboratory at Davis Grant USAF Medical Center were also tested. Each sample was from a positive blood culture bottle, and staphylococcal species were identified by Gram stain, tube coagulase test, and use of the MicroScan system (Dade Behring, Deerfield, Ill.) and oxacillin screening plates. Staphylococcal isolates were placed into one of four groups: MRSA, methicillin-sensitive S. aureus (MSSA), oxacillin-resistant CoNS (OxRCoNS), and oxacillin-sensitive CoNS (OxSCoNS). Upon receipt of the blood culture bottle by our institution, a subculture of each sample was performed on sheep blood agar plates and incubated 37°C overnight. An inoculum (104 CFU/ml) of each sample was mixed with 5 ml of anticoagulated blood and then inoculated into a new blood culture bottle that was incubated in a BACTEC 9240 system (Becton Dickinson, Sparks, Md.) for 14 to 18 h. Blood samples that did not contain bacteria were also inoculated into blood culture bottles and incubated as well for negative controls. All samples inoculated with bacteria were positive after 14 to 18 h in the BACTEC 9240 system. The next day, 0.2-ml aliquots of the positive and negative blood culture bottles were used for DNA extraction.
Rapid extraction method.
S. aureus strains 450M, N315, Col, and BMSI (31) were used to compare the methods of DNA extractions. The S. aureus strains were taken from an overnight subculture and resuspended into 1 ml of 1× phosphate-buffered saline. One milliliter containing 109 CFU of bacterial cells per ml was centrifuged at 7,500 × g for 3 min. This was the starting point for both of the extraction procedures.
(i) QIAGEN.
The first procedure used was a modification of a QIAamp blood and tissue kit (QIAGEN). The lysostaphin incubation required for lysis of gram-positive bacteria and proteinase K enzyme incubation was reduced from 30 to 10 min. The QIAamp procedure was then followed per the manufacturer's instructions. Briefly, 109 CFU of bacterial cells per ml and white blood cells were lysed with lysis buffer, and proteinase K enzyme and ethanol were added to the resultant lysate. The total solution was added to a QIAamp spin column, which absorbs the DNA onto the QIAamp silica membrane during the brief centrifugation steps. The column was washed to remove any residual contaminants, and the bound DNA was eluted in concentrated form with water or Tris-EDTA (TE) buffer.
(ii) Bead beating with Chelex (BB+C).
Bacterial cells (109 CFU/ml) were mixed with 0.5 ml of EDTA-anticoagulated blood, 1 ml of urine, or 500 μl of 10 mM TE buffer (pH 8.4). For whole blood, the sample was mixed, centrifuged for 3 min at 10,000 rpm, and washed consecutively with 1 ml of 4% glacial acetic acid, 1 ml of 1× PBS, and 500 μl of 10 mM TE buffer (pH 8.4). After the addition of the TE buffer, 1 g of 0.1-mm-diameter glass beads (Biospec Products, Inc., Bartlesville, Okla.) and ∼0.25 g of Chelex-100 (Bio-Rad, Hercules, Calif.) were added to the sample mixture. The samples were mixed and processed in the bead beater (Biospec Products; Inc.) at three-quarters speed for 5 min and then boiled for 5 min. The samples were then centrifuged for 5 min at 10,000 × g, and the supernatants were removed to clean 1.7-ml Eppendorf tubes. All DNA samples were measured for concentration using a DNA/RNA calculator (Pharmacia Biotech, Piscataway, N.J.).
Lower limiting dilution experiment.
To determine the lower limits of detection of the target sequences, extracted DNA was diluted to 1 ng by serial dilutions and PCR was performed. To determine the lower limit of detection (in CFU per milliliter), dilutions of S. aureus 450M from 1010 to 100 CFU/ml were extracted by both QIAGEN and BB+C methods and PCR was performed.
Direct amplification of staphylococcus from extracted DNA with PCR.
The extracted staphylococcal DNA samples were used to PCR amplify different target sites in the genome (Table 1), to include the S. aureus mecA gene and staphylococcus and bacterial 16S rRNA genes. For the whole-blood sample, the wild-type brain-derived neurotrophic factor (bdnf) gene was also amplified as a positive extraction control. Life Technologies-Gibco BRL (Gaithersburg, Md.) synthesized the primers for PCR. One microliter of the extracted DNA (50-ng minimum) was added to the Ready-To-Go (RTG) PCR beads (Pharmacia Biotech). When brought to a final volume of 25 μl, the thermocycling mix contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, and ∼1.5 U of Taq DNA polymerase along with a 0.5 μM concentration of each primer set. The thermocycling conditions were as follows: 94°C for 5 min for 1 cycle and then 94°C for 1 min, 57°C for 1 min, and 72°C for 2 min for 35 cycles on a Progene (Princeton, N.J.) Thermocycler. The reaction was then incubated for an additional 10 min at 72°C and was maintained at 4°C for up to 48 h. After thermocycling, 5 μl was removed and subjected to agarose gel electrophoresis to determine the quantity, quality, purity, and appropriate size of products. The resultant amplicons were resolved by agarose gel electrophoresis (1.5% agarose) at 120 V for 30 min and were run along with molecular weight markers (Life Technologies-Gibco BRL). The gel was stained with ethidium bromide, and the amplicons were visualized using a UV light box. All PCR testing was performed by dedicated personnel in a physical location distinct from the rest of the laboratory. Contaminant primer controls were included with the substitution of deionized water for template DNA. Positive and negative controls were included with each run.
TABLE 1.
Primer sets used in this study
| DNA target | Primer paira | Size (bp) | Reference |
|---|---|---|---|
| mecA gene | 5′-CAT TTT GAG TTC TGC ACT ACC-3′ | 967 | 36 |
| 5′-GCA ATA CAA TCG CAC ATA CAT TAA TAG-3′ | |||
| 16S rRNA | 5′-GCG GAT CCT GAC TGC AGT GCC AGC AGC CGC GGT AA-3′ | 292 | 34 |
| 5′-GCG GAT CCG CGG CCG CGG ACT ACC AGG GTA TCT AAT-3′ | |||
| Staphylococcus 16S rRNA | 5′-GTT ATT AGG GAA GAA CAT ATG TG-3′ | 750 | Takahashi et al.b |
| 5′-CCA CCT TCC TCC GGT TTG TCA CC-3′ | |||
| bdnf | 5′-AUG GAG AUC UCU GGA TCC ATG ACC ATC CTT TTC CTT-3′ | 764 | 16 |
| 5′-ACG CGU ACU AGU GGA TCC CTA TCT TCC CCT TTT AAT-3′ |
For each target, the first primer is positive sense and the second primer is negative sense.
T. Takahashi, M. Kaneko, Y. Mori, M. Tsuji, N. Kikuchi, and T. Hiramune, unpublished data (GenBank accession no. D83355).
Susceptibility test.
Resistance to methicillin was determined by automated susceptibility testing using the MicroScan system for bacterial strains that were identified by PCR as mecA negative (and MRSA by MicroScan) or mecA positive (but MSSA by MicroScan). MICs of methicillin were also determined by broth microdilution methods recommended by the National Committee for Clinical Laboratory Standards (29) with minor modifications. The final inoculum was adjusted to 108 CFU/ml in Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) to increase the detection of methicillin resistance. Bacteria were incubated at 35°C for 24 h before MICs were determined. Disk diffusion tests, when used, were performed using 30-μg amoxicillin-clavulanate disks (Becton Dickinson, Cockeysville, Md.) for susceptibility and 0.4-U Taxo A disks (Hardy Diagnostics, Santa Maria, Calif.) for the identification of Micrococcus species (13). Next, a bacterial suspension, with turbidity approximately equal to a 0.5 McFarland standard, of each isolate was inoculated by being swabbed onto Mueller-Hinton agar and incubated at 35°C for 24 h. If a zone of inhibition was present, the organism was presumed to not be a Staphylococcus. β-Lactamase production, when tested, was detected using cefinase disks (Becton Dickinson).
Result TAT measurement and cost-per-test determination.
Result TAT was measured from the time the positive blood culture bottle was removed from the BACTEC 9240 system for extraction of the DNA to the resolution of the PCR amplicons by agarose gel electrophoresis and the final observance of results. Cost per test was calculated by determining the one-time use of each individual item required to perform the test, including the supplies and media.
RESULTS
Rapid extraction method.
Five S. aureus strains were used to test the BB+C DNA extraction method to generate sufficient, quality DNA for PCR. Table 2 compares the concentration and purity ranges of the DNA BB+C extraction method for the five S. aureus strains tested. The median concentration of isolated DNA ranged from 87.3 to 385 ng/μl, and the median purity based on the 260 nm/280 nm ratio ranged from 0.81 to 1.22 ng/μl. A consistent result with PCR was obtained for all five S. aureus strains with DNA obtained by the BB+C method. S. aureus 450M had the highest concentration and was used for comparison with the QIAGEN method. The median concentration of S. aureus 450M using the QIAGEN method was 78.5 ng/μl, with a purity of 1.04. DNA extracted by the QIAGEN method gave consistent results in PCR, and the resultant amplicons generated from each method were separated by agarose gel electrophoresis. These results are shown in Fig. 1.
TABLE 2.
DNA concentrations and purity determined using BB+C DNA extraction method
| Staphylococcus strain | DNA concn (ng/μl) determined by BB+C
|
DNA purity (ng/μl)
|
||
|---|---|---|---|---|
| Median | Interquartile range | Median | Interquartile range | |
| 450M | 385.0 | 358.5–472.0 | 1.22 | 1.20–1.24 |
| N315 | 87.3 | 58.3–142.0 | 0.81 | 0.72–0.95 |
| BMS1 | 350.0 | 129.8–427.5 | 1.18 | 1.05–1.23 |
| Col | 139.5 | 99.3–190.5 | 0.92 | 0.79–0.99 |
| 4220 | 128.0 | 55.3–192.8 | 0.87 | 0.70–0.93 |
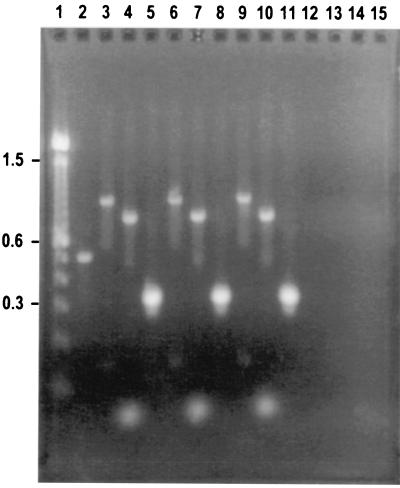
FIG. 1.
Agarose gel electrophoresis of genomic DNA extracted by different methods from S. aureus 450M, amplified with MRSA primer sets. Lane 1, 100-bp DNA ladder (100 to 15,000 bp and 2,072-bp fragment) (Life Technologies); lane 2, DNA PCR control from RTG PCR beads (Pharmacia Biotech) (500 bp); lanes 3 to 5, QIAGEN method-extracted DNA bacterial culture; lanes 6 to 8, BB+C-extracted DNA; lanes 9 to 11, previously made amplicons (positive control) generated from the mecA gene (997 bp); S. aureus 16S rRNA gene (750 bp), and bacterial 16S rRNA gene (292 bp); lanes 12 to 14, no DNA template (negative control) with each primer set. Lanes 3 and 6 contain amplicons PCR generated with the mecA gene primer set, lanes 4 and 7 contain amplicons PCR generated with the S. aureus 16S rRNA gene primer set, and lanes 5 and 8 contain amplicons PCR generated with the bacterial 16S rRNA gene primer set.
The BB+C and QIAGEN methods were tested for lower limits of detection of the target sequences. As little as 5 ng of DNA was required to amplify the target sequences using DNA generated from either method. The lower limits of detection (in CFU per milliliter) for both methods were also determined. S. aureus 450M at dilutions of 1010 100 CFU/ml was extracted by both methods, the DNA was measured, and PCR was performed. The minimum number of target sequences to be amplified by PCR was 109 CFU/ml when DNA was extracted by both methods. To test if this was strain specific, S. aureus BMS1 was tested, and it gave similar results. E. coli ATCC 25922 was also tested; however, the target sequences from both methods could be amplified at a lower limit of 103 CFU/ml. To determine the time frame for the minimum concentration required to test positive in the BACTEC 9240 system, aliquots of anticoagulated whole blood were prepared with concentrations of S. aureus 450M ranging from 102 to 108 CFU/ml. These spiked samples were then used to inoculate blood culture bottles, which were incubated in the BACTEC 9240 system. The time frame required for the blood culture bottles to test positive was 5.3 h for the 108-CFU/ml concentration, 10 h for the 106-CFU/ml concentration, 10.1 h for the 104-CFU/ml concentration, and 11.75 h for the 102-CFU/ml concentration. The bacterial density of the blood culture bottle when it became positive was between 1.25 × 109 and 4.0 × 109 CFU/ml when the starting concentration was between 102 and 104 CFU/ml.
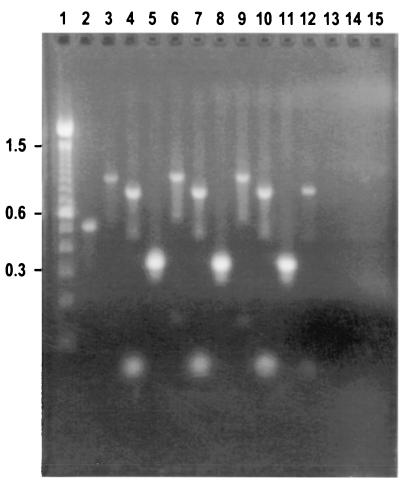
The extraction method was tested with different types of clinical samples. Figure 2 shows that the S. aureus mecA gene and Staphylococcus and bacterial 16S rRNA genes were all easily amplified from S. aureus DNA obtained by the BB+C extraction method from different sample sources: urine (lanes 3 to 5), whole blood (lanes 6 to 8), and bacterial culture (lanes 9 to 11). The bdnf gene was amplified from DNA extracted by the BB+C method from whole blood (lane 12). All primer sets were tested for contamination, and no amplicons were detected after electrophoresis (lanes 13 to 15). As negative controls, 10 clinical isolates obtained from our clinical laboratory, including E. faecalis, S. pyogenes, P. aeruginosa, and E. coli, were mixed with blood and urine and then tested. Only the bacterial 16S rRNA gene target sequence in each of these samples could be amplified by PCR (data not shown).
FIG. 2.
Agarose gel electrophoresis of S. aureus 450M genomic DNA extracted from different sample sources by the BB+C method, amplified with MRSA primer sets. Lane 1, 100-bp DNA ladder (100 to 15,000 bp, and 2,072-bp fragment) (Life Technologies); lane 2, DNA PCR control from RTG PCR beads (Pharmacia Biotech) (500 bp); lanes 3 to 5, from urine; lanes 6 to 8, from whole blood; lanes 9 to 11, from bacterial culture; lanes 13 to 15, no DNA template (negative control) with each primer set. Lanes 3, 6, and 9 contain amplicons PCR generated with the mecA gene primer set (997 bp); lanes 4, 7, and 10 contain amplicons PCR generated with the S. aureus 16S rRNA gene primer set (750 bp); lanes 5, 8, and 11 contain amplicons PCR generated with the bacterial 16S rRNA gene primer set (292 bp); and lane 12 contains amplicons PCR generated with the bdnf gene primer set (764 bp).
Amplification of MRS.
Next we tested for the presence of mecA and bacterial and staphylococcal 16S rRNA genes in each of the 416 clinical isolates. Only the expected amplified DNA products were produced from the bacterial (292-bp) and staphylococcal (750-bp) 16S rRNA primer sets in every sample. An amplified DNA product (997 bp) was seen in 106 of 106 isolates from the MRSA group and in 2 of 134 isolates from the MSSA group when the mecA primer sets were used (Table 1). The two discrepant MSSA samples showed an amplified product (997 bp) upon retesting. To further test for the presence of phenotypic resistance, two MSSA samples were tested by the broth microdilution assay. The results showed that the MIC of methicillin for the two MSSA samples was 8 μg/ml, indicating unequivocally that they did possess methicillin resistance, and that the MIC of oxacillin for these strains was 8 μg/ml, and therefore they had been falsely classified as MSSA by the oxacillin screening plate.
Detection of mecA gene in CoNS.
The presence of the mecA gene in 111 OxRCoNS isolates and 65 OxSCoNS isolates was tested. All strains tested positive for the bacterial and staphylococcal 16S rRNA genes. The mecA gene (997 bp) was detected in 97 of 111 OxRCoNS and in 4 of 65 OxSCoNS. Upon retesting the 14 discrepant OxRCoNS isolates, 10 of 14 OxRCoNS isolates again showed an amplified product. To further test for the presence of phenotypic resistance, the four OxRCoNS isolates lacking mecA were tested by the broth microdilution assay. The results showed that the MIC of methicillin for all four OxRCoNS isolates was 8 μg/ml, indicating that these strains did possess methicillin resistance, and the MIC of oxacillin for these strains was 8 μg/ml. The four OxRCoNS isolates lacking mecA were also tested for the production of β-lactamase by using cefinase disks and for susceptibility to amoxicillin-clavulanate and Taxo A by the disk diffusion test. All four OxRCoNS isolates lacking mecA were positive for the production of β-lactamase and were susceptible to amoxicillan-clavulanate, while two OxRCoNS isolates lacking mecA were sensitive to Taxo A, indicating that they were Micrococcus species.
Upon retesting of the four discrepant mecA-positive OxSCoNS isolates, two of four of the isolates again showed an amplified product (997 bp) indicating the presence of the mecA gene. To further test for phenotypic resistance, these two mecA-positive OxSCoNS isolates were tested by the broth microdilution assay. The results showed that the MIC of methicillin for both of the mecA-positive OxSCoNS isolates was 8 μg/ml, indicating that they were methicillin resistant, and the MIC of oxacillin for these strains was 8 μg/ml, and therefore they had been falsely classified as methicillin sensitive. Of the two OxSCoNS isolates lacking mecA, one was positive by the broth microdilution method (MICs of methicillin and oxacillin, 8 μg/ml), indicating that it was methicillin resistant. No further identification was performed on these two isolates.
DISCUSSION
We have developed a procedure for rapid extraction of microorganism DNA directly from select clinical samples for molecular testing in our laboratory. Our mechanical lysis procedure generated DNA from the bacterial agent directly from the clinical sample within 20 min of sample submission. Significant progress has been made in the development of commercial extraction kits that can be used for rapid nucleic acid extraction from microbial cultures for PCR. However, they require multiple steps (5 to 40) and extended times (15 to 150 min). They may require a pure culture and may be cost-prohibitive for large numbers of samples (Dion et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol.). The QIAGEN procedure for DNA extraction, against which our preparation method was compared, requires the use of these lysozyme, lysostaphin, and proteinase K enzymes specifically with gram-positive organisms (33). The most important component of an extraction method is the ability to obtain quality DNA for PCR. Even with the wide range of concentrations from the five S. aureus strains, the DNA was easily amplified by standard PCR and the target sites selected were readily amplified using a battery of primer sets that specifically identified MRSA from different sample sources (Fig. 2). The results were available in less than 4 h, confirming the identification of the microorganism as well as determining the presence of antibiotic resistance markers.
Our BB+C method does have one significant limitation. The minimum amount of organism determined to be necessary for extraction of the DNA and detection of the target sequences by PCR was 109 CFU/ml. However, when E. coli ATCC 25922 was tested, the target sequences could be amplified utilizing only 103 CFU/ml. The QIAGEN procedure states that its lower limit of detection is 103 CFU/ml; however, we could not duplicate this with our S. aureus strain. Nevertheless, our results indicate that a 104-CFU/ml concentration of bacteria requires a 10.1-h incubation to test positive in the BACTEC 9240 system. In our experience, once the blood culture bottle becomes positive (bacterial density, 1.25 × 109 to 4.0 × 109 CFU/ml), we can detect MRS with our system. However, in our clinical microbiology laboratory, the blood culture bottles are monitored continuously but the positives are not worked up until the next morning. When we tested our positive blood culture bottles the next day, all had more than 109 CFU/ml. Therefore, there will be a sufficient concentration of bacteria for extraction and detection by our method.
The cost of the rapid extraction and PCR-based method is affordable, and setup is readily applicable to the clinical laboratory. Standard identification methods, which include VITEK cards, media, inoculating loops, antibiotic disks, and reagents, can cost more than $7.00 per sample for confirmation. If it is determined that the identity of a clinical isolate with a positive blood culture bottle result needs to be confirmed using the MRS primer set (bacterial and Staphylococcus 16S rRNA gene and mecA gene), the cost could be less than that of standard identification. The cost per test for the RTG tubes is $1.50/tube plus $0.10 for the set of primers, totaling $1.60/PCR for each primer set. If a panel consisting of three primer sets were to be used, then the cost would be $1.60 × 3 (for three different primer sets for identification), or $4.80. Adding the cost of the BB+C extraction method ($0.55/extraction) makes the total cost per test $5.35 (Table 3). Technician time can also be calculated to include exact sample processing time. One sample takes approximately 30 min for DNA extraction and PCR setup. Electrophoresis setup is about 10 min. If five samples were to be tested the DNA extraction and PCR setup time would be under 40 min for all five samples. Therefore, the reported method was determined to be both time- and cost-effective compared to standard clinical procedures.
TABLE 3.
Cost comparison of rapid extraction methods
| Method | Reagent product(s) | Relative cost ($)/testa | Extraction time (min) |
|---|---|---|---|
| Modified QIAGEN | QIAamp tissue kit | 2.18 | 40 |
| BB+C | Glass beads + Chelex | 0.55 | 20 |
Relative cost/test includes one-time use of acetic acid ($0.01), phosphate-buffered saline ($0.03), TE buffer ($0.01), pipette tips ($0.04), Eppendorf tubes ($0.03), glass beads ($0.06), Chelex ($0.24), or lysostaphin ($0.36). One QIAGEN kit test was determined to cost $1.42.
The presence of the mecA gene, as detected by our PCR procedures, had a 99% agreement with clinical findings of methicillin resistance in the 416 Staphylococcus isolates tested. Interestingly, during the validation process we determined that there were two isolates that were classified as MSSA and two isolates classified as OxSCoNS by the MicroScan system in which we were able to detect the presence of the mecA gene by PCR. Upon further testing of these four isolates by broth microdilution, it was shown that all of these samples were phenotypically methicillin and oxacillin resistant. There were four additional isolates classified as OxRCoNS in which we were not able to detect the presence of the mecA gene, but all of these isolates were positive by broth microdilution for methicillin and oxacillin resistance (MIC, 8 μg/ml). Further investigation of these four OxRCoNS isolates lacking mecA revealed that they produced β-lactamase and were all susceptible to amoxicillin-clavulanate. In addition, one isolate classified as OxSCoNS also lacked mecA but was broth microdilution positive. Borderline resistant strains that do not contain the mecA gene have been hypothesized to result from modification of normal PBP genes or overproduction of staphylococcal β-lactamase (8, 26, 43). However, the role of β-lactamase overproduction in borderline resistance is less clear, and no clinical data have suggested that the level of resistance expressed by borderline resistant strains lacking mecA leads to treatment failure (8). Further testing revealed that two of the four OxRCoNS isolates lacking mecA were species of the genus Micrococcus, which is known to be more closely related to Arthrobacter, a genus of environmental coryneforms, than to Staphylococcus (7). Therefore, the PCR method did discriminate between high-level β-lactam (methicillin) resistance by the presence of the mecA gene in Staphylococcus isolates and related strains that were β-lactamase producers only.
PCR detection of mecA should be considered as the potential “gold standard” for staphylococcal methicillin resistance. Previous studies have reported discrepancies, noting that some strains lacking mecA displayed phenotypic resistance to methicillin while others containing mecA showed phenotypic susceptibility (1, 14, 22, 49). Additionally, mecA transcriptional activity does not correlate with phenotypic methicillin resistance (31). Until new sets of recommendations are established, a combination of methods should be used routinely in detecting MRSA and OxRCoNS (8, 12, 35, 48, 49).
A rapid PCR method that utilizes capillary air thermal cyclers to improve TAT has been published (7, 19, 20). Air-thermocycling–real-time PCR holds great promise as a rapid diagnostic tool, but instrumentation cost may be prohibitive, being 10 times that of regular thermocyclers. We further investigated this approach and recently presented research data utilizing this new technology with our rapid extraction method (D. M. Niemeyer, G. Veltri, and R. I. Jaffe, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 876, 1999). Our procedure took less than 40 min from DNA extraction to confirmation with our method specific for methicillin resistance and had a 100% correlation with methicillin sensitivity testing and mecA determination by standard PCR.
The sensitivity of PCR coupled with the speed of our procedure can assist the provider in making a prudent and timely selection of chemotherapeutic agents. This approach for identifying antibiotic resistance markers has great potential for augmenting standard microbiological methods. The primary value of the rapid testing approach is that it will work well where standard microbiological testing capabilities are limited but agent identification is critical (remote, deployed medical facilities), although it may find good utility in clinical laboratories in the near future as these laboratories incorporate PCR into their normal work flow. We evaluated a field PCR laboratory set up in both a deployable medical system at an army reserve training facility in Dublin, Calif., and an air force field hospital (30; D. M. Niemeyer, M. Dempsey, W. Hamilton, J. McAvy, J. Benjack, J. Ruiz, L. Lim, and K. Lohman, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1560, 1999). Additionally, to continue evaluation of real-time PCR use, an air force laboratory has been set up in Southwest Asia (D. M. Niemeyer, M. Corkern, W. J. Barnes, D. White, W. Johnson, M. Dempsey, and K. Lohman, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., 2000). In many military field hospitals, microbiological testing capabilities are limited and may not include standard culture and identification capabilities. As such, routine samples are shipped to regional or stateside reference laboratories for testing, which increases result TAT to a week or longer. PCR would provide select on-site preliminary test capabilities to assist the health care provider in patient treatment decisions under work conditions that normally may not afford standard microbiological testing.
ACKNOWLEDGMENTS
We thank Arlene Reiss, Maya Murashima, Charlene Crigger, and Judy Fusco for their help in obtaining the clinical samples from Kaiser Permenente. We also thank James W. Smith, Michael Climo, and Jennifer Brustrom for critical review of the manuscript.
This work was performed under U.S. Air Force Surgeon General-approved Clinical Investigation FDG1998021E.
REFERENCES
- 1.Araj G F, Talhouk R S, Simaan C J, Maasad M J. Discrepancies between mecA PCR and conventional tests used for detection of methicillin resistant Staphylococcus aureus. Int J Antimicrob Agents. 1997;11:47–52. doi: 10.1016/s0924-8579(98)00047-8. [DOI] [PubMed] [Google Scholar]
- 2.Archer G A, Climo M W. Antimicrobial susceptibility of coagulase-negative staphylococci. Antimicrob Agents Chemother. 1994;38:2231–2237. doi: 10.1128/aac.38.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer G A, Niemeyer D M. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. TEM. 1994;2:343–348. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 4.Barski P, Piechowicz L, Galinski J, Kur J. Rapid assay for detection of methicillin-resistant Staphylococcus aureus using multiplex PCR. Mol Cell Probes. 1996;10:471–475. doi: 10.1006/mcpr.1996.0066. [DOI] [PubMed] [Google Scholar]
- 5.Beck W D, Berger-Bachi B, Kayser F H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning mecA-specific DNA. J Bacteriol. 1986;165:373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger-Bachi B, Barberis-Maino L, Strassle A, Kayser F H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989;219:263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- 7.Carroll K C, Leonard R B, Newcomb-Gayman P L, Hillyard D R. Rapid detection of the Staphylococcus mecA gene from BACTEC blood culture bottles by the polymerase chain reaction. Clin Microbiol Infect Dis. 1996;106:600–605. doi: 10.1093/ajcp/106.5.600. [DOI] [PubMed] [Google Scholar]
- 8.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers H F. Coagulase-negative staphylococci resistant to β-lactam antibiotics in vivo produce penicillin-binding protein 2a. Antimicrob Agents Chemother. 1987;31:1919–1924. doi: 10.1128/aac.31.12.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crossley K, Loesch D, Landesman B, Mead K, Chern M, Strate R. An outbreak of infection caused by strains of Staphylococcus aureus resistant to methicillin and aminoglycosides. I. Clinical studies. J Infect Dis. 1979;139:273–279. doi: 10.1093/infdis/139.3.273. [DOI] [PubMed] [Google Scholar]
- 11.Doebbling B N. The epidemiology of methicillin-resistant Staphylococcus aureus infection and colonization. J Chemother. 1995;7(Suppl. 3):99–103. [PubMed] [Google Scholar]
- 12.Farrell D J. The reliability of Microscan conventional and rapid panels to identify Staphylococcus aureus and detect methicillin resistance: an evaluation using the tube coagulase test and mecA PCR. Pathology. 1997;29:406–410. doi: 10.1080/00313029700169405. [DOI] [PubMed] [Google Scholar]
- 13.Forbes B A, Sahm D F, Weisfield A S. Staphylococcus, Micrococcus, and similar organisms. In: Forbes B A, Sahm D F, Weissfeld A S, editors. Bailey & Scott's diagnostic microbiology. 10th ed. St. Louis, Mo: Mosby, Inc.; 1994. pp. 603–618. [Google Scholar]
- 14.Frebourg N B, Nouet D, Lemee L, Martin E, Lemeland J F. Comparison of ATB Staph, Rapid ATB Staph, Vitek, and E-test methods for detection of oxacillin heteroresistance in staphylococci possessing mecA. J Clin Microbiol. 1998;36:52–57. doi: 10.1128/jcm.36.1.52-57.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geha D J, Uhl J R, Gustaferro C A, Persing D H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GIBCO-BRL. Life Technologies-Gibco Bethesda Research lab catalogue, item 10198-018. Gaithersburg, Md: GIBCO-BRL; 1994. [Google Scholar]
- 17.Hartman B J, Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman B J, Tomasz A. A low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kearns A M, Seiders P R, Wheeler J, Freeman R, Steward M. Rapid detection of methicillin-resistant staphylococci by multiplex PCR. J Hosp Infect. 1999;43:33–37. doi: 10.1053/jhin.1999.0631. [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa Y, Ueda M, Ando N, Endo M, Ishibiki K, Kobayashi Y, Arai T, Kitajima M. Rapid diagnosis of methicillin-resistant Staphylococcus aureus bacteremia by nested polymerase chain reaction. Ann Surg. 1996;224:665–671. doi: 10.1097/00000658-199611000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klimek J J, Marsik F J, Bartlett R C, Weir B, Shea P, Quintilliani R. Clinical epidemiologic and bacteriologic observations of an outbreak of methicillin-resistant Staphylococcus aureus at a large community hospital. Am J Med. 1976;61:340–345. doi: 10.1016/0002-9343(76)90370-3. [DOI] [PubMed] [Google Scholar]
- 22.Kohner P, Uhl J, Kolbert C, Persing D, Cockerill F., III Comparison of susceptibility testing methods with mecA gene analysis for determining oxacillin (methicillin) resistance in clinical isolates of Staphylococcus aureus and coagulase-negative staphylococcus species. J Clin Microbiol. 1999;37:2952–2961. doi: 10.1128/jcm.37.9.2952-2961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolbert C P, Arruda J, Varga-Delmore P, Zheng X, Lewis M, Kolberg J, Persing D H. Branched-DNA assay for detection of the mecA gene in oxacillin-resistant and oxacillin-sensitive staphylococci. J Clin Microbiol. 1998;36:2640–2644. doi: 10.1128/jcm.36.9.2640-2644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M A. Methicillin-resistant Staphylococcus aureus: the persistent, resistant nosocomial pathogen. In: Remington J S, Schwartz M N, editors. Current clinical topics in infectious disease. Boston, Mass: Blackwell Scientific Publications; 1994. pp. 170–191. [PubMed] [Google Scholar]
- 25.Matsuhashi M, Song M D, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to β-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986;167:975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDougal L K, Thornsberry C. The role of β-lactamase in staphylococcal resistance to penicillinase-resistant penicillin and cephalosporin. J Clin Microbiol. 1986;23:832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami K, Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989;171:874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 30.Niemeyer D M, Jaffe R I, Wiggins L B. Feasibility determination for use of polymerase chain reaction (PCR) in the USAF air transportable hospital (ATH) field environment: lessons learned. Mil Med. 2000;165(11):11. [PubMed] [Google Scholar]
- 31.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Predari S C, Ligozzi M, Fontana R. Genotypic identification of methicillin-resistant coagulase-negative staphylococci by polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:2568–2573. doi: 10.1128/aac.35.12.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiagen. QIAamp blood and tissue DNA extraction handbook. Santa Clarita, Calif: Qiagen; 1996. [Google Scholar]
- 34.Relman D A, Schmidt T M, MacDermont R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;323:1573–1580. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 35.Ribeiro J, Vieira F D, King T, D'Arezzo J B, Boyce J M. Misclassification of susceptible strains of Staphylococcus aureus as methicillin-resistant S. aureus by rapid automated susceptibility testing system. J Clin Microbiol. 1999;37:1619–1620. doi: 10.1128/jcm.37.5.1619-1620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryffel C, Kayser F H, Berger-Bachi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992;36:25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salisbury S M, Sabatini L M, Spiegal C A. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction. Am J Clin Pathol. 1997;107:368–373. doi: 10.1093/ajcp/107.3.368. [DOI] [PubMed] [Google Scholar]
- 38.Seligman S J. Penicillinase-negative variants of methicillin-resistant Staphylococcus aureus. Nature (London) 1966;209:994–996. doi: 10.1038/209994a0. [DOI] [PubMed] [Google Scholar]
- 39.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;94:137–138. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki E, Hiramatsu K, Yokota T. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob Agents Chemother. 1992;36:429–434. doi: 10.1128/aac.36.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomasz A. Multiple antibiotic resistant pathogenic bacteria. N Engl J Med. 1994;330:1247–1251. doi: 10.1056/NEJM199404283301725. [DOI] [PubMed] [Google Scholar]
- 43.Tomasz A, Drugeon H B, de Lencastre H M, Jabes D, McDougal L, Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989;33:1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ubukata K, Yamashita N, Konno M. Occurrence of a β-lactam-inducible penicillin-binding protein in methicillin-resistant (heteroresistant) staphylococci. J Clin Microbiol. 1985;27:851–857. doi: 10.1128/aac.27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unal S, Hoskins J, Flokowitsch J E, Wu C Y, Preston D A, Skatrud P L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985;28:851–857. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vannuffel P, Laterre P F, Bouyer M, Gigi J, Vandercam B, Reynaert M, Gala J L. Rapid and specific molecular identification of methicillin-resistant Staphylococcus aureus in endotracheal aspirates from mechanically ventilated patients. J Clin Microbiol. 1998;36:2366–2368. doi: 10.1128/jcm.36.8.2366-2368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wautters G, Gala J L. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallet F, Roussel-Delvallez M, Courcol R J. Choice of a routine method for detecting methicillin-resistance in staphylococci. J Antimicrob Chemother. 1996;37:901–909. doi: 10.1093/jac/37.5.901. [DOI] [PubMed] [Google Scholar]
- 50.York M K, Gibbs L, Chehab F, Brooks G F. Comparison of PCR detection of mecA with standard susceptibility testing methods to determine methicillin resistance in coagulase-negative staphylococci. J Clin Microbiol. 1996;34:249–253. doi: 10.1128/jcm.34.2.249-253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zambardi G, Reverdy M E, Bland S, Bes M, Freney J, Fleurette J. Laboratory diagnosis of oxacillin resistance in Staphylococcus aureus by a multiplex-polymerase chain reaction assay. Diagn Microbiol Infect Dis. 1994;19:25–31. doi: 10.1016/0732-8893(94)90047-7. [DOI] [PubMed] [Google Scholar]